Introduction
Breast cancer (BC) is a prevalent tumor that
threatens the lives and health of patients (1). The latest global cancer statistics
indicated that 310,720 female patients were diagnosed with BC in
2024 in the United States (2).
Globally, BC remains the most common type of malignancy in female
patients, with at least 500,000 BC-associated mortalities each
year. In recent years, with changes in lifestyle and environment,
the incidence of BC is increasing year by year, and the onset age
is becoming younger (3). Although
early diagnosis and novel treatment methods such as surgery,
postoperative chemotherapy and hormone and targeted therapy have
rapidly progressed, the prevalence and mortality rates of BC remain
high with the 5-year survival rate of patients with metastatic BC
<30% (4–6). Moreover, certain patients still
encounter rapid tumor progression following treatment and exhibit a
poor prognosis (7). Therefore, it
is key to evaluate novel and reliable molecular markers and
therapeutic targets for BC treatment.
Ferroptosis is a non-apoptotic form of regulated
cell death characterized by iron dependency and lipid peroxidation
(8). When the levels of
intracellular iron ions increase, the production of lipid reactive
oxygen species (ROS) also increases. GPX4 is required to maintain
the balance between ROS production and clearance; when this is not
sustained, ferroptosis occurs (9).
System Xc is an antioxidant system widely distributed in the
phospholipid bilayer, composed of two SLC7A11 subunits and a SLC3A2
heterodimer (10). Increases in
SLC7A11 and GPX4 inhibit occurrence of ferroptosis. Moreover,
circulating iron binds to TF in the form of Fe3+ and
enters the cell via TFR1. ACSL4 is required for ROS accumulation,
so elevated levels of TFR1 and ACSL4 promote ferroptosis (11). Emerging studies have shown that
ferroptosis pathway can repress tumor growth and kill tumor cells,
which may be a new idea for antitumor treatment (9,11).
In particular, ferroptosis is closely involved in the development
of BC (12).
Solute carrier family 12 member 5 (SLC12A5) is
primarily responsible for transporting chloride ions in and out of
the cell and serves a role in regulating cell volume (13). Initially, researchers focused on
the role of SLC12A5 in the nervous system and it was reported that
SLC12A5 is upregulated in glioblastoma, where it enhances chloride
ion transport ability of glioma cells and facilitates alteration of
cell size and morphology as well as metastasis (14). With advancement of biological
research, further studies reported that SLC12A5 is upregulated in a
number of types of cancer and that SLC12A5 upregulation promotes
numerous tumor characteristics and indicates poor patient prognosis
(14–17). A previous study reported that
increased levels of SLC12A5 are associated with poor prognosis in
patients with hepatocellular carcinoma (HCC) and SLC12A5 inhibits
ferroptosis in HCC to promote tumorigenesis through upregulating
expression of the cystine transporter xCT (18). Moreover, SLC12A5 is highly
expressed in human bladder tumors and linked to poor survival in
patients with uroepithelial carcinoma of the bladder. Upregulation
of SLC12A5/SRY-box transcription factor 18 facilitates tumor
invasion and metastasis (19).
Furthermore, SLC12A5 is amplified, which is accompanied with the
concurrent amplification of an 8-gene Signature (TNF-α, IL-1β,
IL-6, MMP1, MMP9, TGF-β1, TGF-βRII, EGFR) derived based on these
macrophage-tumor interactions in BC (20).
Avian erythroblastosis virus E-26 transformation
specific (ETS) translocation variant 4 (ETV4) is a member of the
PEA3 subfamily of ETS transcription factor (21). PEA3 subfamily influences cancer
progression and metastasis by regulating cell cycle, apoptosis,
epithelial-mesenchymal transition, cell migration and invasion,
development of cancer stem cell phenotypes and chemotherapy
resistance (22). ETV4 gene is
located on chromosome 17q21 and binds to adenovirus E1A enhancer
element. ETV4 has a highly conserved 85-amino acid ETS domain that
binds to DNA, thus ETV4 can modulate expression of genes which
regulate the proliferation and metastasis of cancer cells (23). Moreover, ETV4 has been reported to
enhance glycolytic activity and stemness in BC (24). ETV4 controls HK1 expression and
glycolysis-lactate production to activate mTORC1 by relieving
Tuberous sclerosis complex 2 (TSC2) repression of Ras homolog
enriched in brain (Rheb) in non-small cell lung cancer cells by
regulating glycolysis-lactate production (25). However, to the best of our
knowledge, expression of SLC12A5 and ETV4 in BC and their role in
the development of BC remains unclear. Therefore, the present study
aimed to assess the role of SLC12A5 in BC and clarify the mechanism
underlying its effects in this disease.
Materials and methods
Bioinformatics analysis
SLC12A5 gene expression in the tissue of patients
with BC and the association of SLC12A5 with poor prognosis in these
patients were analyzed using the University of ALabama at
Birmingham CANcer data analysis Portal (UALCAN) database
(ualcan.path.uab.edu) from TCGA database (26). The binding site of ETV4 and the
SLC12A5 promoter was predicted using HumanTFDB database
(bioinfo.life.hust.edu.cn/HumanTFDB/#!/, version 3.0 (27). Moreover, expression of SLC12A5 in
BC cell lines was analyzed by Cancer Cell Line Encyclopedia (CCLE)
project (depmap.org/portal/ccle/) (28).
Cell lines
The human normal mammary epithelial cell line MCF10A
and BC cell lines MCF-7, Hs578T, T47D and BT-549 were purchased
from Cellverse Bioscience Technology Co., Ltd. MCF10A cells were
cultured in DMEM/F12 with L-glutamine and 5% horse serum (Thermo
Fisher Scientific, Inc.), 20 ng/ml epidermal growth factor, 0.5
µg/ml hydrocortisone, 100 ng/ml cholera toxin and 10 µg/ml insulin
(all Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (1:100; Thermo Fisher Scientific, Inc.) at
37°C with 5% CO2. BC cell lines were cultivated in DMEM
(Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin at 37°C with 5% CO2.
Cell transfection
ETV4-overexpressing plasmid were constructed by
inserting the ETV4 coding sequence into a pcDNA3.1 plasmid (General
Biosystems, Inc.). An empty pcDNA3.1 vector was used as the
negative control (oe-NC). The specific short hairpin (sh)RNA
sequences targeting SLC12A5 (sh-SLC12A5-1,
5′-GCAATGCAATGAAGTTGAA-3′ and sh-SLC12A5-2,
5′-GGAGAGGTTGCAAACCAAA-3′), ETV4 (sh-ETV4-1,
5′-GGTGGTGATCAAACAGGAA-3′ and sh-ETV4-2,
5′-GGAATGGAGTTCAAGCTCA-3′), negative control (sh-NC,
5′-CCGGCAACAAGATGAAGAGCACCAACTC-3′) were cloned into the
pLKO.1-puro vector (Sigma, St. Louis, USA). An empty pLKO.1-puro
vector was used as the control. These plasmids and shRNAs were
constructed by Shanghai GenePharma Co., Ltd. A total of 5 µg
plasmid or 5 µg shRNA was transfected into MCF7 cells using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at
37°C for 48 h, according to the manufacturer's instructions. After
culturing for 2 days at 37°C, cells were used for the following
experiments. Untransfected cells were referred to as the Control
group.
Cell Counting Kit-8 (CCK-8) assay
MCF-7 cells were seeded in 96-well plates at
3×103 cells/well and incubated for 24, 48 and 72 h at
37°C with 5% CO2 and saturated humidity. Subsequently,
10 µl of CCK-8 solution (Beijing Solarbio Science & Technology
Co., Ltd.) was added to each well and incubated for 1 h. The OD
value at 450 nm was then measured using a microplate reader
(Biochrom, Ltd.).
Colony formation assay
Transfected MCF-7 cells were inoculated into 6-well
plates (500 cells/well). Following a 2-week incubation in DMEM with
10% FBS at 37°C, the cells were fixed with 4% paraformaldehyde at
room temperature for 25 min and stained with 0.1% crystal violet
for 10 min at room temperature. Colonies (>50 cells) were
counted manually in five fields of view using a light microscope
(Olympus Corporation; magnification, ×10). Each group was
replicated for five times.
Wound healing assay
MCF-7 cells transfected with sh-SLC12A5 in the
presence or absence of oe-ETV4 or oe-NC were seeded into 6-well
plates at 5×105 cells/well and incubated in DMEM with
10% FBS at 37°C until 90% confluency was reached. A straight
scratch in the cell monolayer was made to create a denuded zone
using a pipette tip. The cells were then incubated for 24 h in
serum-free DMEM medium and images of the wound surface and number
of migrated cells were captured under an inverted microscope
(Olympus Corporation; magnification, ×100). Five fields were
randomly chosen and analyzed in each well. The relative migration
rate was calculated as follows: (wound width at 0 h-wound width at
24 h)/wound width at 0 h ×100.
Transwell assay
MCF-7 cells were collected and suspended at a final
concentration of 2×105 cells/ml in serum-free DMEM
(Thermo Fisher Scientific, Inc.). A total of 200 µl cell suspension
was transferred to the upper wells of Transwell chambers (Corning,
Inc.) coated with 0.1 ml Matrigel (Becton, Dickinson and Company)
at 37°C for 1 h and DMEM containing 10% FBS was placed in the lower
chamber. Following 24 h incubation at 37°C, a cotton swab was used
to remove cells in the upper chamber, while cells in the lower
chamber were fixed with 100% methanol at room temperature for 10
min and stained with 0.5% crystal violet for 10 min at room
temperature. Finally, a light microscope (Olympus Corporation;
magnification ×100) was used for cell counting. Five randomly
chosen fields were counted for each group.
Measurement of lipid peroxidation
Thiobarbituric acid reactive substances (TBARS)
assay was performed to estimate lipid peroxidation in MCF-7 cells.
For this, 7 µl 500 mM butylated hydroxyanisole and 0.25 ml 15%
(w/v) trichloroacetic acid were added to the cell lysate, which was
centrifuged at 1,000 × g for 5 min at 4°C. The supernatant was
collected and 0.5 ml 0.375% (w/v) TB was added. After 10 min
boiling at 95°C, the levels of TBARS were estimated using a
microplate reader (Thermo Fisher Scientific, Inc.) at 532 nm.
BODIPY 581/591 C11 probe (Thermo Fisher Scientific,
Inc.) was also used to detect lipid peroxidation. Transfected MCF-7
cells were incubated with 10 µM C11 BODIPY 581/591 probe for 10 min
at 37°C. The presence of green fluorescence indicated oxidized
probe; red fluorescence indicated non-oxidized probe. ImageJ
(version 1.8.0; National Institutes of Health) was used to assess
the levels of BODIPY 581/591 C11, calculated as the ratio of green
fluorescence/total fluorescence.
Detection of Fe2+
levels
MCF-7 cells were transfected with sh-SLC12A5 in the
presence or absence of oe-ETV4 or oe-NC. Fe2+ levels
were measured using an Iron Assay kit purchased from Abcam (cat.
no. ab83366) at 593 nm, according to the manufacturer's
instructions.
Extracellular acidification rate
(ECAR) analysis
XF96 Extracellular Flux Analyzer (Agilent
Technologies, Inc.) with Seahorse XFp Glycolysis Stress Test kit
(Agilent Technologies, Inc.) was used to measure ECAR in MCF-7
cells. Transfected MCF-7 cells were inoculated into wells of the
Seahorse XF plate at 1×104 cells/well and exposed to
glucose (1 µM), oligomycin (1 µM) and 2-deoxyglucose (500 mM) at
37°C for 1 h. Finally, the results were analyzed by Seahorse XF96
Wave software (version 2.6; Seahorse Bioscience; Agilent
Technologies) and ECAR was calculated as mpH/min.
Evaluation of oxygen consumption rate
(OCR)
The OCR was measured using a Seahorse XF Cell Mito
Stress Test kit (cat. no. 103010-100; Seahorse Bioscience; Agilent
Technologies, Inc). The sensor cartridge of the XFp analyzer was
calibrated for 24 h in a non-CO2 incubator at 37°C.
Cells were cultured in XFp cell culture plates at 5,000 cells/well
at 37°C for 24 h. After transfection, MCF-7 cells were incubated in
180 µl assay medium (XF Base Medium, 1 mM pyruvate, 5.5 mM glucose
and 2 mM L-glutamine, pH 7.4) for 1 h at 37°C in a
non-CO2 incubator, according to the manufacturer's
protocol. The results were analyzed by Seahorse XF96 Wave software
(version 2.6; Seahorse Bioscience; Agilent Technologies).
Measurement of lactate production and
glucose consumption
MCF-7 cells were transfected with sh-SLC12A5 in the
presence or absence of oe-ETV4. Cells were cultured for 24 h at
37°C and the cell medium was collected for lactate and glucose
measurement using Lactate Assay Kit (cat. no. MAK064;
Sigma-Aldrich; Merck KGaA) and Glucose Assay kit (cat. no. MAK476;
Sigma-Aldrich; Merck KGaA), respectively according to the
manufacturer's instructions.
Dual-luciferase reporter assay
Activity of SLC12A5 promoter was evaluated using a
dual-luciferase reporter assay. Briefly, SLC12A5 promoter fragments
including the wild-type (WT) or mutant (MUT) target sites for ETV4
were cloned into the pGL3-Control vector (Promega Corporation) to
create the reporter vectors SLC12A5-WT or SLC12A5-MUT, respectively
(Data S1). Luciferase reporter
vectors and oe-NC or oe-ETV4 were co-transfected into MCF-7 cells
for 48 h at 37°C using Lipofectamine 2000. After 48 h incubation at
37°C, Dual-Luciferase Reporter Assay (Promega Corporation) was used
for estimation of relative luciferase activities; luciferase
activity was normalized to Renilla.
Chromatin immunoprecipitation (ChIP)
assay
MCF-7 Cells were cross-linked with 1% formaldehyde
for 10 min at 37°C, then quenched with 2.5 M glycine at room
temperature for 5 min. Subsequently, cells were harvested by
centrifugation at 300 × g for 3 min at room temperature, washed
with PBS, and lysed in SDS lysis buffer (Upstate Biotechnology,
Inc.), and the chromatin from the cell lysates was sonicated with a
10-sec on and 10-sec off mode for 12 cycles on ice to shear DNA
into fragments at 20 kHz. Following sonication, the samples were
centrifuged at 13,000 × g for 10 min at 4°C. Subsequently, the
supernatant (100 µg) was pre-absorbed by 50 µl protein G beads and
was incubated with magnetic beads conjugated to 5 µg ETV4 antibody
(1/200; cat. no. 65763; Cell Signaling Technology), Next, the
mixture was washed with eluate buffer. The cross-linking was
reversed by 5 M NaCl followed by incubation at 65°C overnight. The
precipitated DNA was analyzed by PCR to amplify the ETV4 binding
site. The results were normalized to the DNA precipitated by 5 µg
IgG (1/100; cat. no. ab172730; Abcam).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
The concentration of total RNA isolated from
1×106 MCF10A and BC cell lines using TRIzol (Invitrogen;
Thermo Fisher Scientific, Inc.) was quantified using a NanoDrop
3000 spectrophotometer (Thermo Fisher Scientific, Inc.). RNA was
reverse-transcribed into cDNA using PrimeScript RT Master Mix
(Takara Bio, Inc.) at 25°C for 5 min, 42°C for 30 min, 85°C for 5
min and 4°C for 5 min. qPCR was performed using SYBR Premix Ex Taq™
II kit (Takara Bio, Inc.). The thermocycling conditions were as
follows: Initial denaturation at 95°C for 3 min, followed by 35
cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 1 min, with
final extension step at 72°C for 7 min. The following primer pairs
were used for qPCR: SLC12A5 forward (F), 5′-TCCCTCCTAGAGCCTGGTTG-3′
and reverse (R), 5′-TTGGGGTTGCCATCACCTTT-3′; ETV4 F,
5′-GAAAAACAAGTCGGTGCGCT-3′ and R, 5′-TTGCTGCTGAAGGTGTAGGG-3′ and
GAPDH F, 5′-GGGAAACTGTGGCGTGAT-3′ and R, 5′-GAGTGGGTGTCGCTGTTGA-3′.
mRNA level was quantified using the 2−ΔΔCq method
(29) and normalized to the
internal reference gene GAPDH.
Western blot assay
The isolation of total protein from sample MCF10A
and BC cell lines was conducted using RIPA buffer (Auragene
Bioscience Co.) and the proteins were quantified using the
bicinchoninic acid assay (Beyotime Institute of Biotechnology).
Following separation by 10% SDS-PAGE (Bio-Rad Laboratories, Inc.),
proteins (30 µg) were transferred to PVDF membranes
(MilliporeSigma). The membranes were blocked with 5% skimmed milk
in 0.1% tris-buffered saline with Tween-20 for 1 h at room
temperature, then incubated with primary antibodies against SLC12A5
(1:1,000; cat. no. ab259969; Abcam), SLC7A11 (1:1,000; cat. no.
ab175186; Abcam), glutathione peroxidase 4 (GPX4; 1:1,000; cat. no.
ab125066; Abcam), acyl-CoA synthetase long chain family member 4
(ACSL4; 1:1,000; cat. no. ab205197, Abcam), transferrin receptor 1
(TFR1; 1:1,000; cat. no. ab109259; Abcam), ETV4 (1:1,000; cat. no.
ab70425; Abcam) and β-actin (1:1,000, cat. no. ab8227; Abcam)
overnight at 4°C, followed by incubation with HRP-conjugated goat
anti-rabbit (1:5,000; cat. no. sc-2004; Santa Cruz Biotechnology,
Inc.) or anti-mouse secondary antibodies (1:5,000; cat. no.
sc-2005; Santa Cruz Biotechnology, Inc.) for 1 h at room
temperature. The protein bands were visualized using Amersham ECL
Prime Western blotting detection reagent (Cytiva) in accordance
with the manufacturer's instructions. Protein expression was
quantified using ImageJ software (version 1.49; National Institutes
of Health).
Statistical analysis
The data were analyzed using SPSS 23.0 software (IBM
Corp.) and presented as the mean ± standard deviation from at least
three independent experiments. For the comparison of multiple
groups, one-way ANOVA followed by Bonferroni's post hoc test was
used, while unpaired Student's t-test was applied for two groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
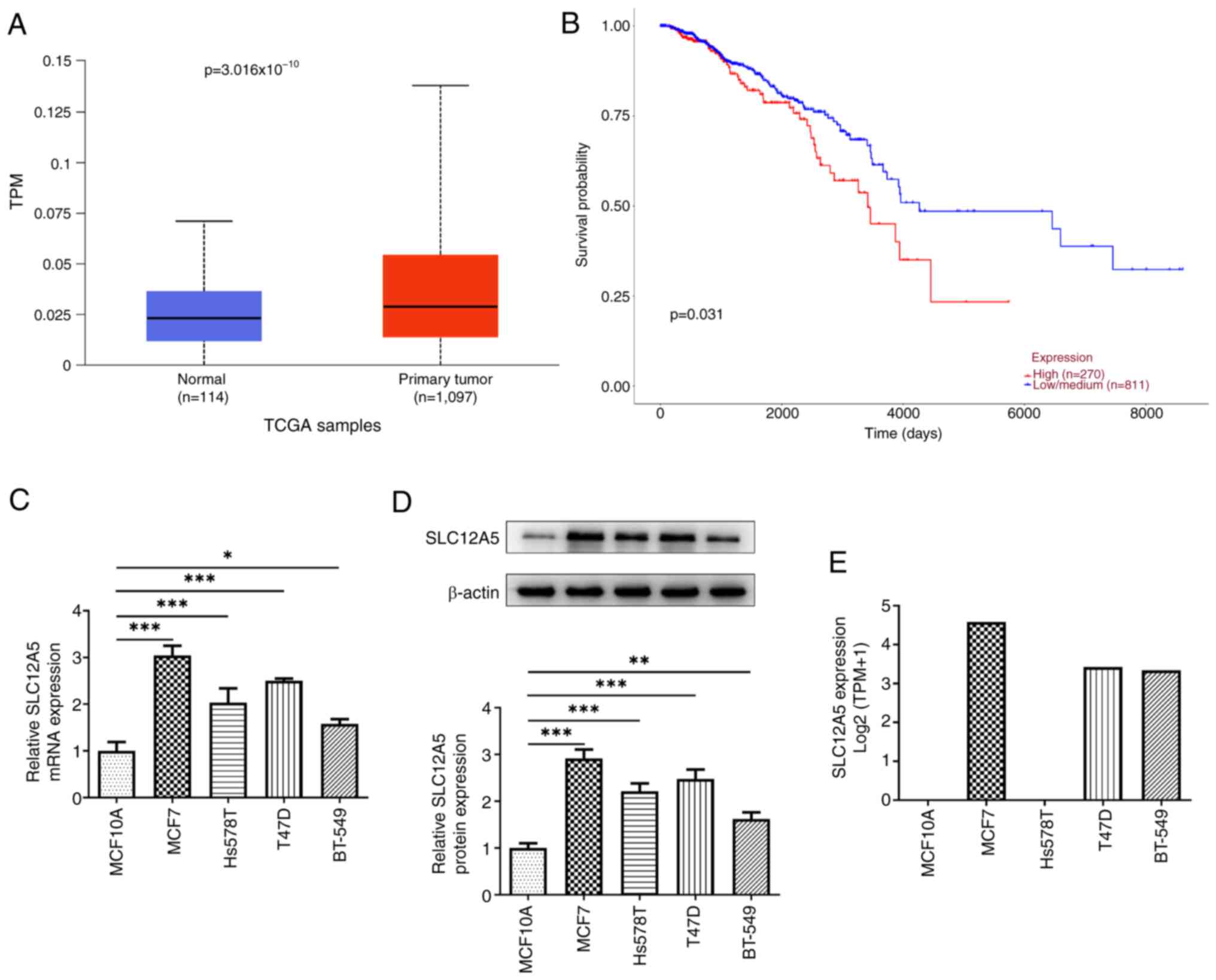
SLC12A5 is upregulated in BC tissue
and cells
To evaluate the biological role of SLC12A5 in BC,
SLC12A5 expression in BC tissue and cells was initially assessed.
The results from the UALCAN database demonstrated that SLC12A5
expression was notably increased in tissues from patients with BC
compared with normal tissues (Fig.
1A). Similarly, high expression of SLC12A5 in BC tissues was
associated with worse prognosis, compared with patients with
low/medium SLC12A5 expression according to the UALCAN database
(Fig. 1B). Moreover, RT-qPCR and
western blotting demonstrated that the mRNA and protein expression
levels of SLC12A5 were both significantly increased in BC cell
lines including MCF7, Hs578T, T47D and BT-549 cells compared with
the normal mammary epithelial cell line MCF10A (Fig. 1C and D). As MCF-7 cells had a
notably higher SLC12A5 expression than the other BC cell lines
assessed, this cell line was used for further experiments. CCLE
data also demonstrated that MCF-7 cells had the highest SLC12A5
mRNA expression of these cell lines (Fig. 1E).
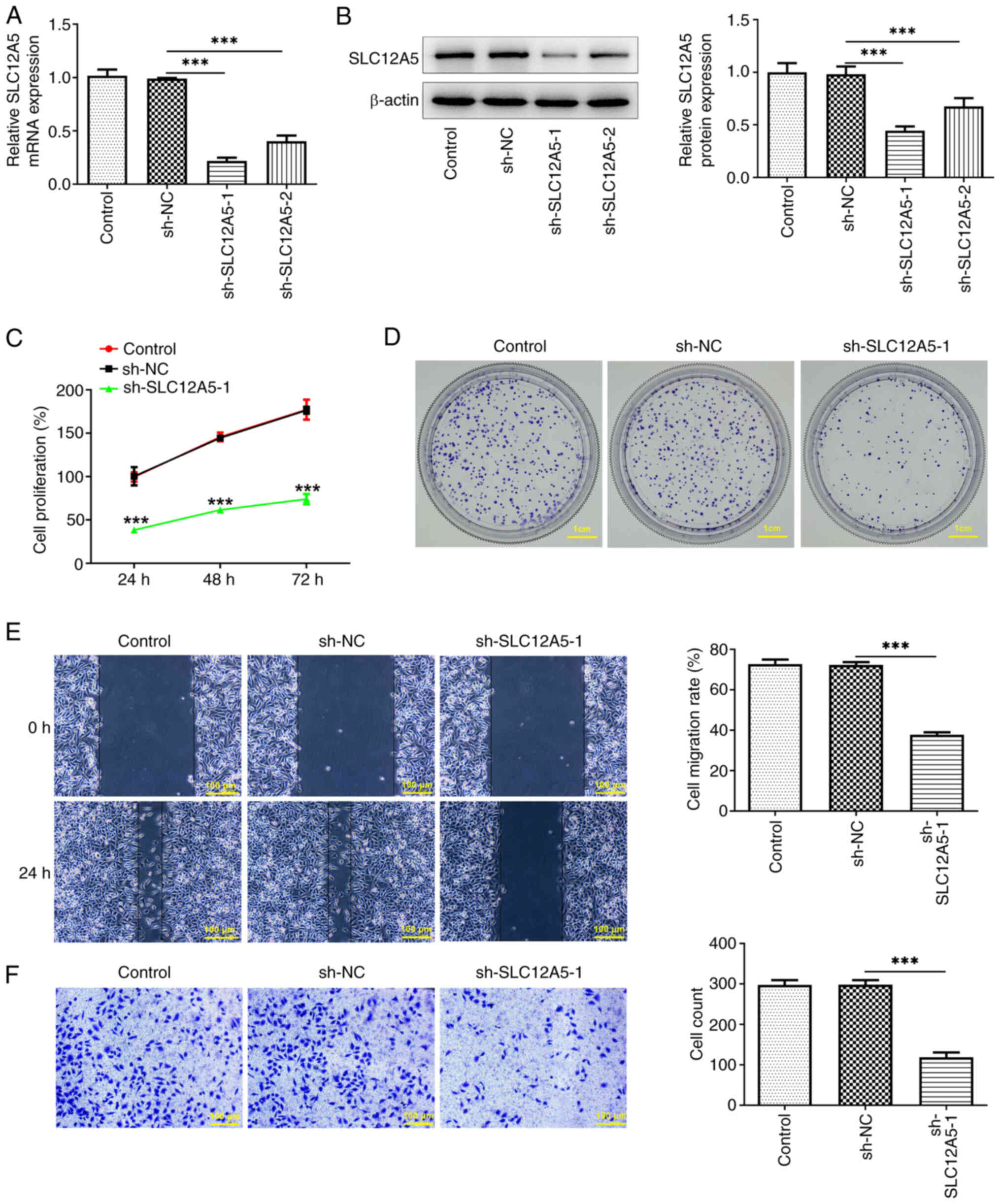
SLC12A5 knockdown attenuates
proliferation, migration and invasion of MCF-7 cells
To evaluate the biological role of SLC12A5 in BC
cells, SLC12A5 expression was knocked down by transfection with
sh-SLC12A5-1/2 (Fig. 2A and B).
sh-SLC12A5-1 had a greater knockdown effect and was selected for
subsequent assays. MCF-7 cell proliferation was significantly
reduced following SLC12A5 knockdown compared with the sh-NC group
(Fig. 2C). The colony formation
assay also demonstrated that the number of cell colonies was
notably decreased by sh-SLC12A5-1 transfection compared with sh-NC
group (Fig. 2D). Furthermore,
wound healing assay demonstrated that cell migration decreased in
cells transfected with sh-SLC12A5-1 (Fig. 2E). Also, Transwell assay
demonstrated that the number of invaded cells was significantly
reduced when SLC12A5 was down-regulated (Fig. 2F).
SLC12A5 knockdown alleviates
ferroptosis resistance in MCF-7 cells
To evaluate the effects of SLC12A5 knockdown on
ferroptosis resistance in BC cells, TBARS assay was conducted.
SLC12A5 knockdown significantly increased production of TBARS in
MCF-7 cells compared with the sh-NC group (Fig. 3A). Furthermore, the levels of
oxidized C11 were markedly increased while the levels of
non-oxidized C11 were reduced following the knockdown of SLC12A5
(Fig. 3B). Moreover, SLC12A5
knockdown led to increased levels of Fe2+ (Fig. 3C). Similarly, western blotting
demonstrated that levels of ferroptosis-related proteins SLC7A11
and GPX4 significantly decreased, whereas levels of ASCL4 and TFR1
significantly increased in SLC12A5-knockdown cells compared with
the sh-NC group (Fig. 3D).
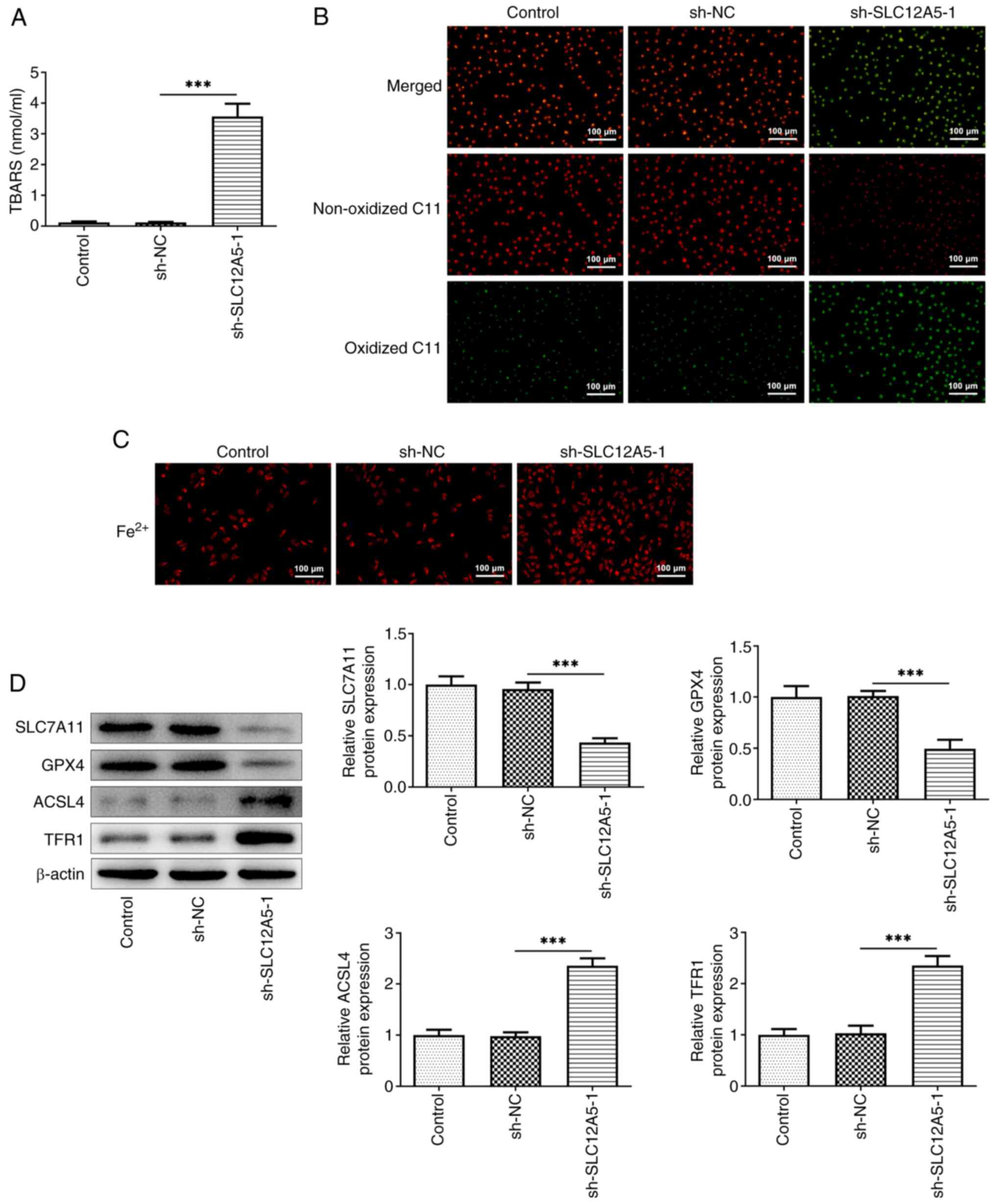 | Figure 3.SLC12A5 knockdown alleviates
ferroptosis resistance in MCF-7 cells. (A) TBARS assay and (B) C11
BODIPY 581/591 probe were used to assess lipid peroxidation. (C)
Levels of Fe2+ in MCF-7 cells with or without
transfection with sh-SLC12A5. Magnification, ×100. (D) Western
blotting was used to evaluate protein levels of SLC7A11, GPX4,
ACSL4 and TFR1. ***P<0.001. TBARS, thiobarbituric acid reactive
substances; sh, short hairpin; NC, negative control; SLC12A5,
solute carrier family 12 member 5; GPX4, glutathione peroxidase 4;
ACSL4, Acyl-CoA synthetase long-chain family 4; TFR1, transferrin
receptor 1. |
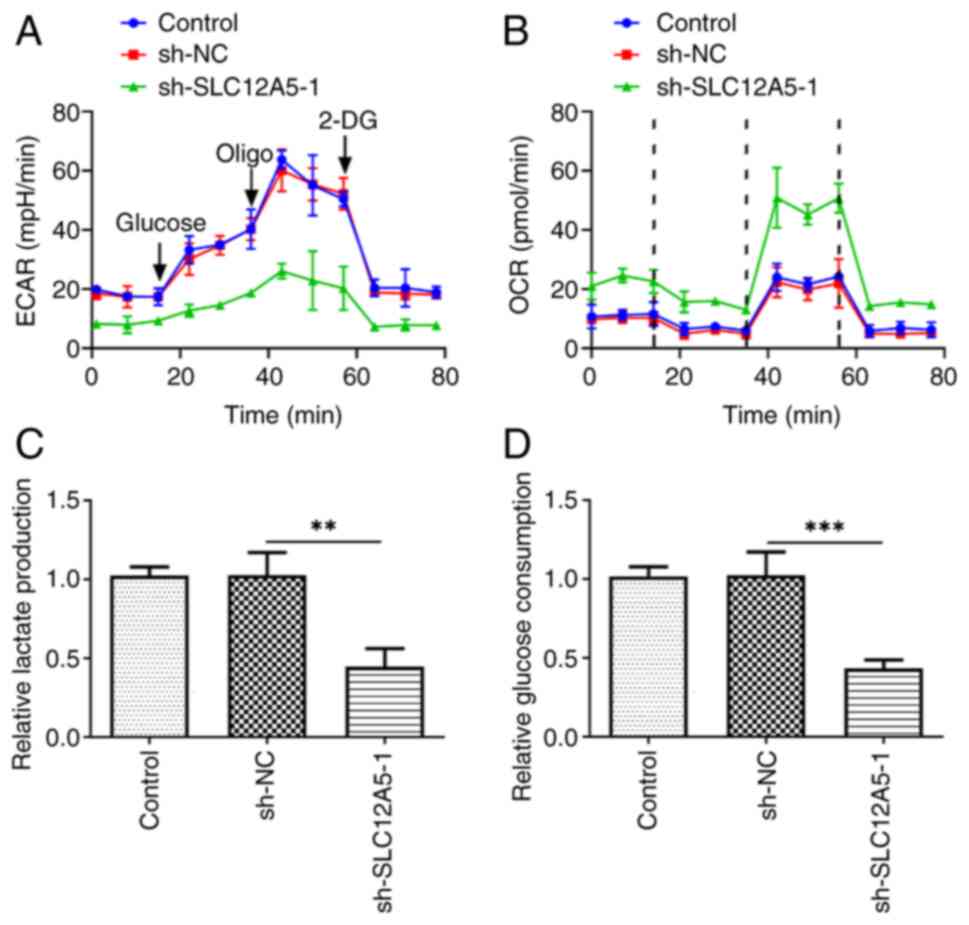
Knockdown of SLC12A5 inhibits
reprogramming of glucose metabolism in MCF-7 cells
Transfection with sh-SLC12A5 decreased ECAR compared
with the NC group (Fig. 4A).
Moreover, SLC12A5 knockdown markedly increased OCR in MCF-7 cells
compared with the sh-NC group (Fig.
4B). Moreover, knockdown of SLC12A5 significantly repressed
lactate release (Fig. 4C). Also,
down-regulation of SLC12A5 reduced glucose uptake (Fig. 4D).
ETV4 transcription factor binds to
SLC12A5 promoter and upregulates SLC12A5 expression
Both ETV4 mRNA and protein expression were
significantly increased in MCF-7 cells compared with MCF10A cells
(Fig. 5A and B). To identify the
role of ETV4 in BC cells, oe-ETV4 and ETV4-knockdown plasmids were
transfected into MCF-7 cells. The transfection efficiency was
assessed using RT-qPCR and western blotting, which demonstrated
that the ETV4 mRNA and protein expression significantly increased
in the oe-EVT4 group compared with the oe-NC group (Fig. 5C and D). sh-ETV4-1 had a greater
knockdown effect and was selected for the following assays (named
as sh-ETV4-1). There was a significant increase in SLC12A5
expression following ETV4 overexpression and a significant decrease
in SLC12A5 mRNA and protein expression following ETV4 knockdown
compared with the corresponding NC (Fig. 5E and F). To verify the interaction
between ETV4 and SLC12A5, the binding site of ETV4 on the SLC12A5
promoter sequence was predicted using HumanTFDB database (SLC12A5
WT: CTCCACTCACTCTCTTCCAGACACAATG; SLC12A5 MUT:
CTCACAGACAGAGAGGAACTCACACATG) (Fig.
5G). Moreover, luciferase reporter assay demonstrated that the
luciferase activity in SLC12A5-WT cells was significantly increased
by ETV4 overexpression compared with oe-NC group, while no
significant change in luciferase activity was observed in the
SLC12A5-MUT groups with ETV4 overexpression (Fig. 5H). ChIP assay also demonstrated a
significant enrichment of SLC12A5 in the ETV4 group compared with
the IgG group (Fig. 5I).
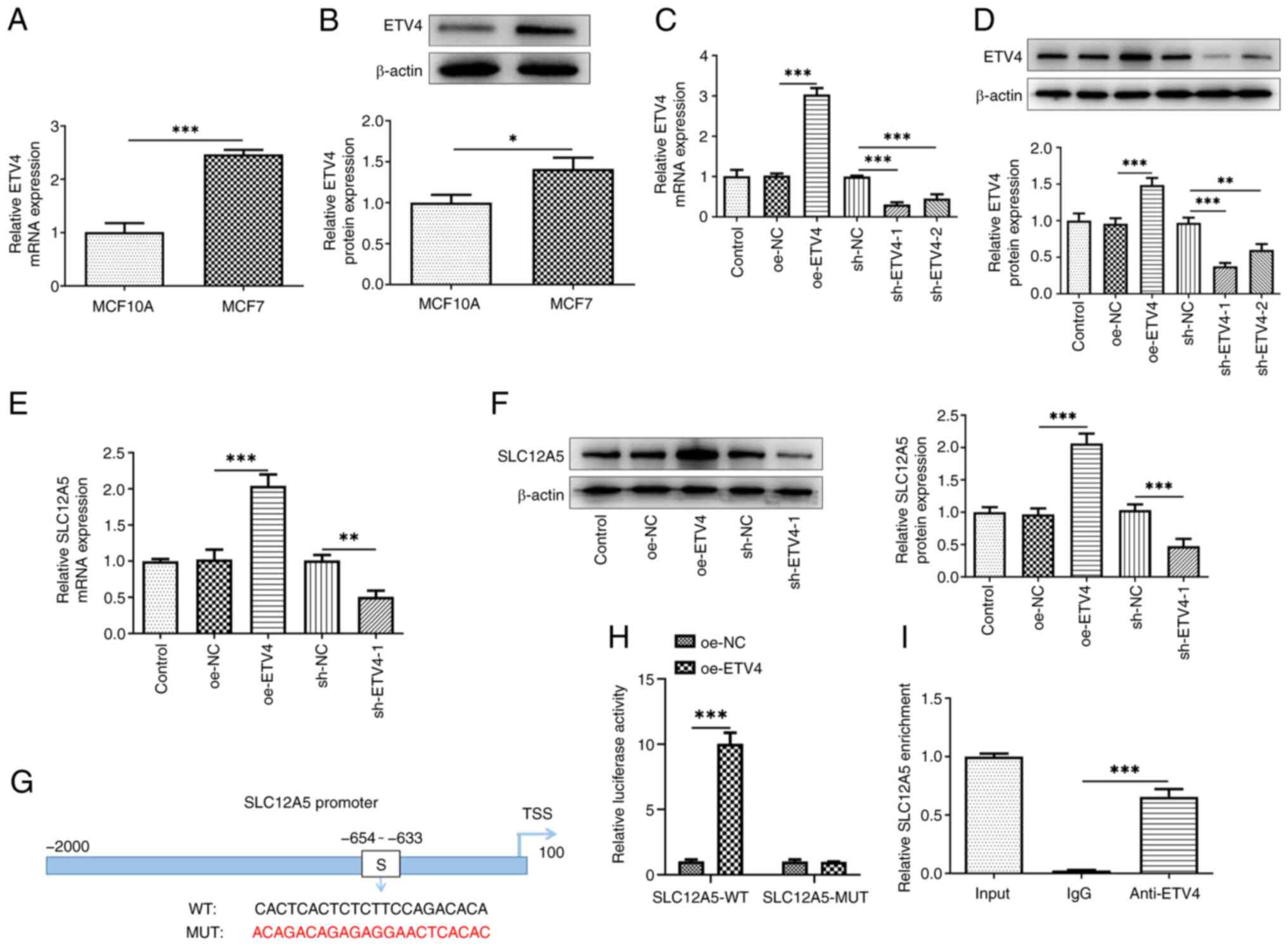 | Figure 5.Transcription factor ETV4 binds to
SLC12A5 promoter and upregulates SLC12A5 expression. (A) mRNA and
(B) protein levels of ETV4 in normal mammary epithelial cell line
MCF10A and MCF-7 cells were measured using RT-qPCR and western
blotting. (C) mRNA and (D) protein levels of ETV4 and (E) mRNA and
(F) protein levels of SLC12A5 were detected by RT-qPCR and western
blotting in MCF-7 cells after ETV4 was overexpressed or knocked
down. (G) Binding site of ETV4 and SLC12A5 promoter. (H) SLC12A5
promoter activity was evaluated by luciferase reporter assay. (I)
Chromatin immunoprecipitation assay was performed to detect the
binding of ETV4 to the WT and MUT SLC12A5 promotor. *P<0.05,
**P<0.01 and ***P<0.001. sh, short hairpin; NC, negative
control; oe, overexpression; WT, wild-type; MUT, mutant; RT-qPCR,
reverse transcription-quantitative PCR; sh, short hairpin; NC,
negative control; SLC12A5, solute carrier family 12 member 5; TSS,
transcription start site; ETV4, E-twenty-six-specific sequence
variant 4. |
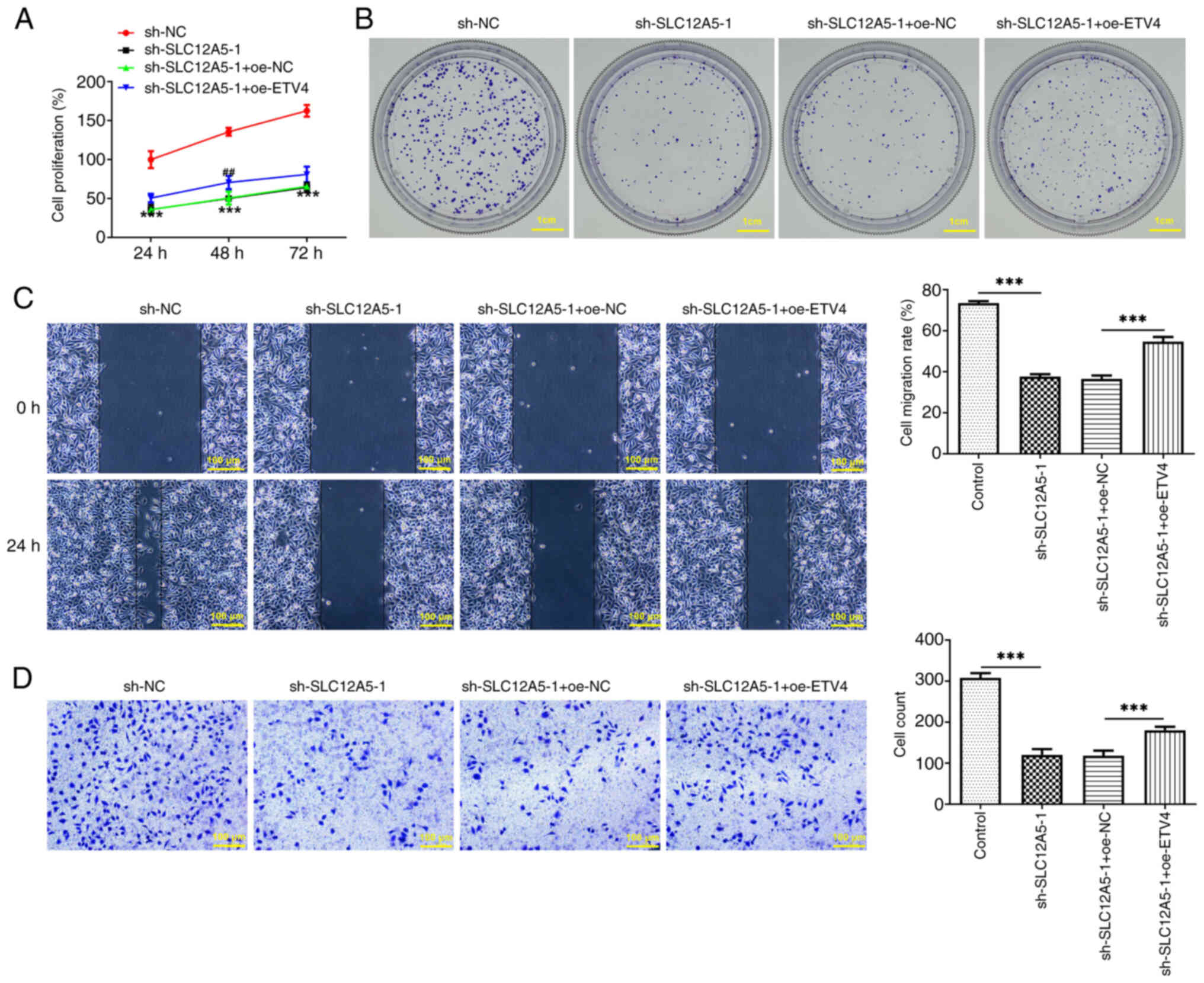
Overexpression of ETV4 partially
reverses the inhibitory effect of SLC12A5 knockdown on migration,
invasion, ferroptosis resistance and glucose metabolism of BC
cells
The role of ETV4 in SLC12A5-modulated migration and
invasion, ferroptosis resistance and glucose metabolism in MCF-7
cells was determined. CCK-8 assay demonstrated that in
SLC12A5-silencing cells, co-transfection of sh-SLC12A5-1 and
oe-ETV4 increased cell proliferation rate again (Fig. 6A). Colony formation assay
demonstrated that relative to the sh-SLC12A5-1 + oe-NC group,
concurrent down-regulation of SLC12A5 and overexpression of ETV4
enhanced the colony numbers (Fig.
6B). The reduced cell migration rate caused by SLC12A5
knockdown was enhanced again by co-transfection of sh-SLC12A5-1 and
oe-ETV4 (Fig. 6C). Cell invasion
ability was weakened in SLC12A5-depleting cells and was promoted
again by further ETV4 overexpression (Fig. 6C and D).
Furthermore, overexpression of ETV4 significantly
decreased the production of TBARS (Fig. 7A) and notably decreased the levels
of Fe2+ (Fig. 7C)
compared with SLC12A5-knockdown cells without ETV4 overexpression.
Moreover, an increase in red fluorescence and a decrease in green
fluorescence was observed in MCF-7 cells co-transfected with
sh-SLC12A5-1 and oe-ETV4 compared with sh-SLC12A5-1 alone (Fig. 7B). Consistently, ETV4
overexpression significantly increased protein levels of SLC7A11
and GPX4 and significantly decreased protein levels of ASCL4 and
TFR1 compared with SLC12A5 knockdown alone (Fig. 7D). Finally, further overexpression
of ETV4 increased the ECAR in SLC12A5-silencing cells (Fig. 8A). Compared with the sh-SLC12A5-1 +
oe-NC group, OCR value was decreased in the sh-SLC12A5-1 + oe-ETV4
group (Fig. 8B). Also, SLC12A5
knockdown inhibited lactate production, which was partially
reversed by further up-regulation of ETV4 (Fig. 8C). Similarly, overexpression of
ETV4 promoted glucose consumption that was declined in cells
transfected with sh-SLC12A5-1 (Fig.
8D).
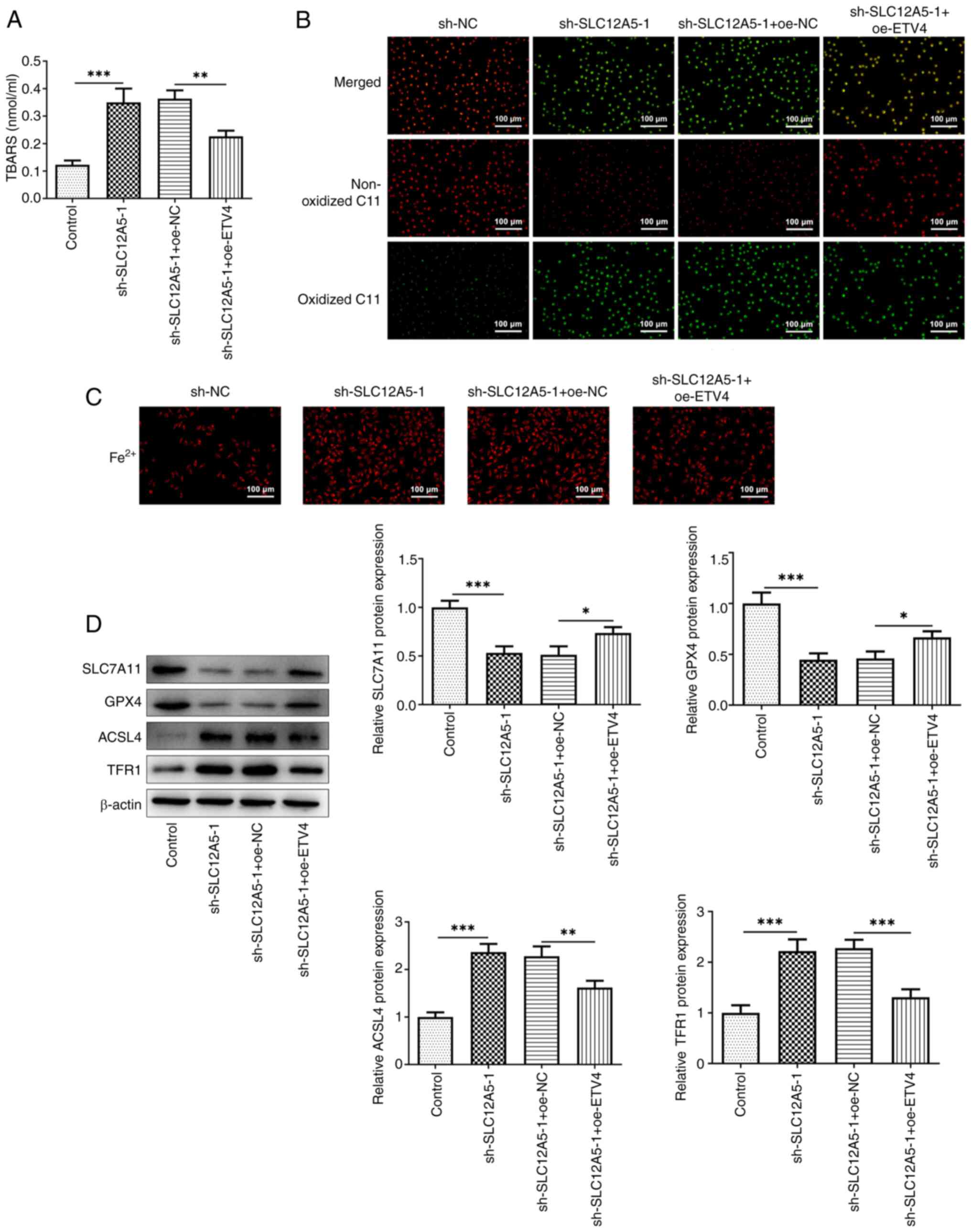 | Figure 7.Overexpression of ETV4 partially
reverses the inhibitory effect of SLC12A5 knockdown on ferroptosis
resistance of BC cells. (A) TBARS assay and (B) C11 BODIPY 581/591
probe were used to assess lipid peroxidation. (C) Levels of
Fe2+ in MCF-7 cells transfected with sh-SLC12A5 with or
without oe-ETV4. Magnification, ×100. (D) Western blotting was used
to evaluate protein levels of SLC7A11, GPX4, ACSL4 and TFR1.
*P<0.05, **P<0.01 and ***P<0.001. TBARS, thiobarbituric
acid reactive substances; sh, short hairpin; NC, negative control;
oe, overexpression; ETV4, E-26-specific sequence variant 4;
SLC12A5, solute carrier family 12 member 5; GPX4, glutathione
peroxidase 4; ACSL4, Acyl-CoA synthetase long-chain family 4; TFR1,
transferrin receptor 1. |
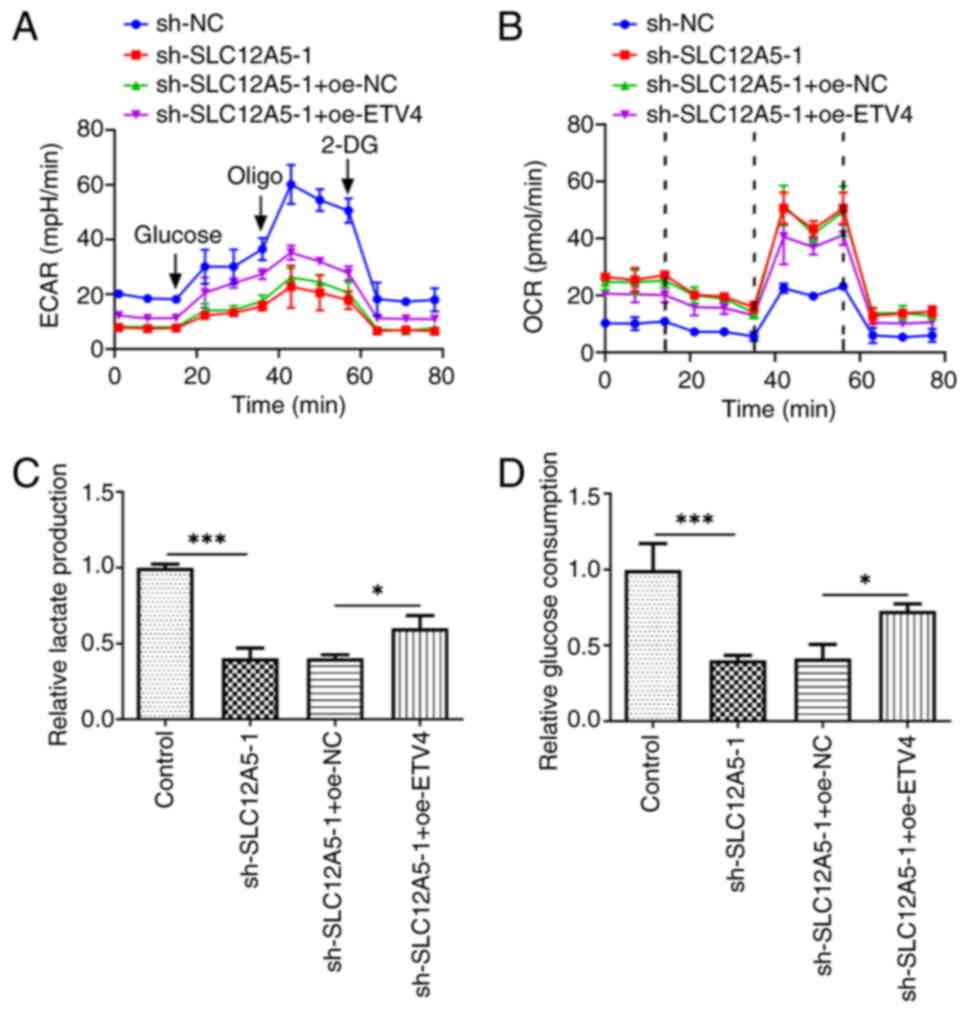 | Figure 8.Overexpression of ETV4 partially
reverses the inhibitory effect of SLC12A5 silencing on glucose
metabolism of BC cells. (A) ECAR, (B) OCR, (C) lactic acid
production and (D) glucose consumption in MCF-7 cells transfected
with sh-SLC12A5 with or without oe-ETV4 were assessed. *P<0.05
and ***P<0.001. ECAR, extracellular acidification rate; OCR,
oxygen consumption rate; sh, short hairpin; NC, negative control;
ETV4, E-twenty-six-specific sequence variant 4; SLC12A5, Solute
carrier family 12 member 5; 2-DG, 2-deoxyglucose; oligo,
oligonucleotide. |
Discussion
BC is the most commonly diagnosed malignancy in
female patients worldwide and the second highest contributor to
cancer-related mortality in female patients (30). The incidence and mortality rates of
BC continue to rise worldwide, posing a threat to the physical and
mental health and lives of patients (31). Hence, early diagnosis and
monitoring of BC is key. The demand for identification of effective
biomarkers for BC, particularly novel molecular therapeutic
targets, has been highlighted (32). In the present study, it was
demonstrated that SLC12A5 was upregulated in BC tissue and cells
and ETV4 was overexpressed in BC cells and high SLC12A5 expression
was positively associated with poor prognosis in patients with BC.
SLC12A5 knockdown ameliorated MCF-7 cell proliferation, migration,
ferroptosis resistance and glucose metabolism reprogramming.
Moreover, ETV4 overexpression promoted SLC12A5 transcription and
ETV4 overexpression reversed the anticancer effects of SLC12A5
knockdown in MCF-7 cells.
SLC12A5 can promote intracellular transport of
K+ and Cl− ions (33). Previous studies have shown that
SLC12A5 expression is elevated in numerous tumor types and high
expression is typically associated with a poor prognosis (34,35).
Yang et al (36) reported
that FEZF1 antisense RNA 1 (FEZF1-AS1) is highly expressed in
cervical cancer (CC) cells and that increased levels of FEZF1-AS1
increases the proliferation, migration and invasion capabilities of
CC cells. FEZF1-AS1 also decreases apoptosis via the
microRNA-367-3p/SLC12A5 signaling axis. Another study reported that
high expression of SLC12A5 protein indicates an aggressive and/or
invasive phenotype in ovarian cancer (37). In the present study, SLC12A5 was
increased in BC tissues and was associated with poor prognosis
according to the analysis of data from the UALCAN database.
Increased SLC12A5 expression in BC cell lines was verified by in
vitro experiments.
HumanTFDB website was used to evaluate the upstream
and downstream mechanisms of SLC12A5. Numerous transcription
factors are reported to bind to the SLC12A5 promoter, among which
only ETV4 is reported to be associated with ferroptosis resistance
and glucose metabolic reprogramming (24,38).
Therefore, the role of ETV4 was further assessed. Yuan et al
(39) reported that high ETV4
expression increases the risk of distant metastasis in patients
with triple-negative BC, leading to poor prognosis. Furthermore,
ETV4 facilitates the proliferation, migration, invasion and
anchorage-independent growth of mammary tumors in mice (40). In the present study, it was
demonstrated that ETV4 expression was elevated in BC compared with
the control cells. To evaluate the interaction between ETV4 and
SLC12A5, the binding site of ETV4 in the SLC12A5 promoter was
predicted using the HumanTFDB database and confirmed experimentally
by luciferase reporter and ChIP assays.
Wang et al (38) reported that ETV4 knockdown
facilitates ferroptosis to inhibit the progression of papillary
thyroid carcinoma through the downregulation of SLC7A11. Verma
et al (41) used RNA
sequencing to evaluate the role of genes in the metabolic
reprogramming and drug resistance of prostate cancer; upregulation
of SLC12A5 was associated with higher lactate/citrate uptake and
lower glucose uptake in drug-resistant cells. In human BC tissues,
ETV4 expression is associated with the glycolytic signaling pathway
and ETV4 deficiency markedly suppresses the expression of
glycolytic enzymes such as hexokinase 2 and lactate dehydrogenase A
and decreases glucose uptake and lactate release in BC cells
(24). Similarly, the present
study demonstrated that SLC12A5 knockdown in MCF-7 cells promoted
ferroptosis and decreased glucose metabolism reprogramming, which
were reversed following overexpression of ETV4.
However, the present experiments were performed in
cell lines only, with no verification from patient samples.
Moreover, the association between the expression of SLC12A5 and
ETV4 in BC and healthy controls should be assessed. Future studies
should assess the SLC12A5 expression by immunofluorescence
staining, immunohistochemistry staining and evaluate the
association between SLC12A5 and ETV4 in clinical samples. Tumor
microenvironment, including immune cell infiltration, serves a role
in cancer progression and outcomes (42). It is possible that the role of
SLC12A5 may exert an effect on immune filtration. Thus, the
association between SLC12A5 expression and immune cell infiltration
in BC should be assessed.
In conclusion, the present study reported a
mechanism of SLC12A5 regulated by ETV4 in BC cells, which may serve
a role in ferroptosis and glucose metabolism. The present study may
therefore contribute to the understanding of BC pathogenesis and
offer prospective therapeutic targets for patients with BC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
HW and FW designed the study and wrote and revised
the manuscript. HW and YD analyzed the data and searched the
literature. All authors performed experiments. HW and FW confirm
the authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zannetti A: Breast cancer: From
pathophysiology to novel therapeutic Approaches 2.0. Int J Mol Sci.
24:25422023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Giaquinto AN and Jemal A:
Cancer statistics, 2024. CA Cancer J Clin. 74:12–49. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Houghton SC and Hankinson SE: Cancer
progress and priorities: Breast cancer. Cancer Epidemiol Biomarkers
Prev. 30:822–844. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
De Rose F, Meduri B, De Santis MC, Ferro
A, Marino L, Colciago RR, Gregucci F, Vanoni V, Apolone G, Di
Cosimo S, et al: Rethinking breast cancer follow-up based on
individual risk and recurrence management. Cancer Treat Rev.
109:1024342022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Parisi S, Gambardella C, Conzo G, Ruggiero
R, Tolone S, Lucido FS, Iovino F, Fisone F, Brusciano L,
Parmeggiani D and Docimo L: Advanced localization technique for
non-palpable breast cancer: Radiofrequency alone VS combined
technique with ultrasound. J Clin Med. 12:50762023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Parisi S, Ruggiero R, Gualtieri G, Volpe
ML, Rinaldi S, Nesta G, Bogdanovich L, Lucido FS, Tolone S,
Parmeggiani D, et al: Combined LOCalizer™ and intraoperative
ultrasound localization: First experience in localization of
non-palpable breast cancer. In Vivo. 35:1669–1676. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kawiak A: Molecular research and treatment
of breast cancer. Int J Mol Sci. 23:96172022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jiang X, Stockwell BR and Conrad M:
Ferroptosis: Mechanisms, biology and role in disease. Nat Rev Mol
Cell Biol. 22:266–282, 2021.9. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen X, Kang R, Kroemer G and Tang D:
Broadening horizons: The role of ferroptosis in cancer. Nat Rev
Clin Oncol. 18:280–296. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu J, Kang R and Tang D: Signaling
pathways and defense mechanisms of ferroptosis. Febs J.
289:7038–7050. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun S, Shen J, Jiang J, Wang F and Min J:
Targeting ferroptosis opens new avenues for the development of
novel therapeutics. Signal Transduct Target Ther. 8:3722023.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li Z, Chen L, Chen C, Zhou Y, Hu D, Yang
J, Chen Y, Zhuo W, Mao M, Zhang X, et al: Targeting ferroptosis in
breast cancer. Biomark Res. 8:582020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fukuda A and Watanabe M: Pathogenic
potential of human SLC12A5 variants causing KCC2 dysfunction. Brain
Res. 1710:1–7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Damanskienė E, Balnytė I, Valančiūtė A,
Alonso MM and Stakišaitis D: Different effects of valproic acid on
SLC12A2, SLC12A5 and SLC5A8 gene expression in pediatric
glioblastoma cells as an approach to personalised therapy.
Biomedicines. 10:9682022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang Y, Liao HL and Chen LY: A pan-cancer
analysis of SLC12A5 reveals its correlations with tumor immunity.
Dis Markers. 2021:30626062021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yuan S, He SH, Li LY, Xi S, Weng H, Zhang
JH, Wang DQ, Guo MM, Zhang H, Wang S, et al: A potassium-chloride
co-transporter promotes tumor progression and castration resistance
of prostate cancer through m(6)A reader YTHDC1. Cell Death Dis.
14:72023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Peng L, Cao Z, Wang Q, Fang L, Yan S, Xia
D, Wang J and Bi L: Screening of possible biomarkers and
therapeutic targets in kidney renal clear cell carcinoma: Evidence
from bioinformatic analysis. Front Oncol. 12:9634832022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tong Q, Qin W, Li ZH, Liu C, Wang ZC, Chu
Y and Xu XD: SLC12A5 promotes hepatocellular carcinoma growth and
ferroptosis resistance by inducing ER stress and cystine transport
changes. Cancer Med. 12:8526–8541. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang L, Zhang Q, Wu P, Xiang W, Xie D,
Wang N, Deng M, Cao K, Zeng H, Xu Z, et al: SLC12A5 interacts and
enhances SOX18 activity to promote bladder urothelial carcinoma
progression via upregulating MMP7. Cancer Sci. 111:2349–2360. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Singh R, Dagar P, Pal S, Basu B and
Shankar BS: Significant alterations of the novel 15 gene signature
identified from macrophage-tumor interactions in breast cancer.
Biochim Biophys Acta Gen Subj. 1862:669–683. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Neri P, Barwick BG, Jung D, Patton JC,
Maity R, Tagoug I, Stein CK, Tilmont R, Leblay N, Ahn S, et al:
ETV4-dependent transcriptional plasticity maintains MYC expression
and results in IMiD resistance in multiple myeloma. Blood Cancer
Discov. 5:56–73. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qi D, Lu M, Xu P, Yao X, Chen Y, Gan L, Li
Y, Cui Y, Tong X, Liu S, et al: Transcription factor ETV4 promotes
the development of hepatocellular carcinoma by driving hepatic
TNF-α signaling. Cancer Commun (Lond). 43:1354–1372. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cosi I, Moccia A, Pescucci C, Munagala U,
Di Giorgio S, Sineo I, Conticello SG, Notaro R and De Angioletti M:
Identification and characterization of novel ETV4 splice variants
in prostate cancer. Sci Rep. 13:52672023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu T, Zheng J, Zhuo W, Pan P, Li M, Zhang
W, Zhou H, Gao Y, Li X and Liu Z: ETV4 promotes breast cancer cell
stemness by activating glycolysis and CXCR4-mediated sonic Hedgehog
signaling. Cell Death Discov. 7:1262021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu B, Zhang J, Meng X, Xie SM, Liu F,
Chen H, Yao D, Li M, Guo M, Shen H, et al: HDAC6-G3BP2 promotes
lysosomal-TSC2 and suppresses mTORC1 under ETV4 targeting-induced
low-lactate stress in non-small cell lung cancer. Oncogene.
42:1181–1195. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chandrashekar DS, Bashel B, Balasubramanya
SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658, 2017.27.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hu H, Miao YR, Jia LH, Yu QY, Zhang Q and
Guo AY: AnimalTFDB 3.0: A comprehensive resource for annotation and
prediction of animal transcription factors. Nucleic Acids Res.
47:D33–D38, 2019.28. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nusinow DP, Szpyt J, Ghandi M, Rose CM,
McDonald ER III, Kalocsay M, Jané-Valbuena J, Gelfand E, Schweppe
DK, Jedrychowski M, et al: Quantitative proteomics of the cancer
cell line encyclopedia. Cell. 180:387–402e16. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Trayes KP and Cokenakes SEH: Breast cancer
treatment. Am Fam Physician. 104:171–178. 2021.PubMed/NCBI
|
|
31
|
Zhang YN, Xia KR, Li CY, Wei BL and Zhang
B: Review of breast cancer pathologigcal image processing. Biomed
Res Int. 2021:19947642021.PubMed/NCBI
|
|
32
|
Criscitiello C and Corti C: Breast cancer
genetics: Diagnostics and treatment. Genes (Basel). 13:15932022.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kontou G, Josephine Ng SF, Cardarelli RA,
Howden JH, Choi C, Ren Q, Rodriguez Santos MA, Bope CE, Dengler JS,
Kelley MR, et al: KCC2 is required for the survival of mature
neurons but not for their development. J Biol Chem. 296:1003642021.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tang Y, Qing C, Wang J and Zeng Z: DNA
methylation-based diagnostic and prognostic biomarkers for
glioblastoma. Cell Transplant. 29:9636897209332412020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gao JL, Peng K, Shen MW, Hou YH, Qian XB,
Meng XW, Ji FH, Wang LN and Yang JP: Suppression of WNK1-SPAK/OSR1
attenuates bone cancer pain by regulating NKCC1 and KCC2. J Pain.
20:1416–1428. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang X, Qu Y and Zhang J: Up-regulated
LncRNA FEZF1-AS1 promotes the progression of cervical carcinoma
cells via MiR-367-3p/SLC12A5 signal axis. Arch Med Res. 53:9–19.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang GP, He WP, Tan JF, Yang ZX, Fan RR,
Ma NF, Wang FW, Chen L, Li Y, Li Y, et al: Overexpression of
SLC12A5 is associated with tumor progression and poor survival in
ovarian carcinoma. Int J Gynecol Cancer. 29:1280–1284. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang L, Zhang Y, Yang J, Liu L, Yao B,
Tian Z and He J: The knockdown of ETV4 inhibits the papillary
thyroid cancer development by promoting ferroptosis upon SLC7A11
downregulation. DNA Cell Biol. 40:1211–1221. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yuan ZY, Dai T, Wang SS, Peng RJ, Li XH,
Qin T, Song LB and Wang X: Overexpression of ETV4 protein in
triple-negative breast cancer is associated with a higher risk of
distant metastasis. Onco Targets Ther. 7:1733–1742. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dumortier M, Ladam F, Damour I, Vacher S,
Bièche I, Marchand N, de Launoit Y, Tulasne D and Chotteau-Lelièvre
A: ETV4 transcription factor and MMP13 metalloprotease are
interplaying actors of breast tumorigenesis. Breast Cancer Res.
20:732018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Verma S, Shankar E, Chan ER and Gupta S:
Metabolic reprogramming and predominance of solute carrier genes
during acquired enzalutamide resistance in prostate cancer. Cells.
9:25352020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
de Visser KE and Joyce JA: The evolving
tumor microenvironment: From cancer initiation to metastatic
outgrowth. Cancer Cell. 41:374–403. 2023. View Article : Google Scholar : PubMed/NCBI
|