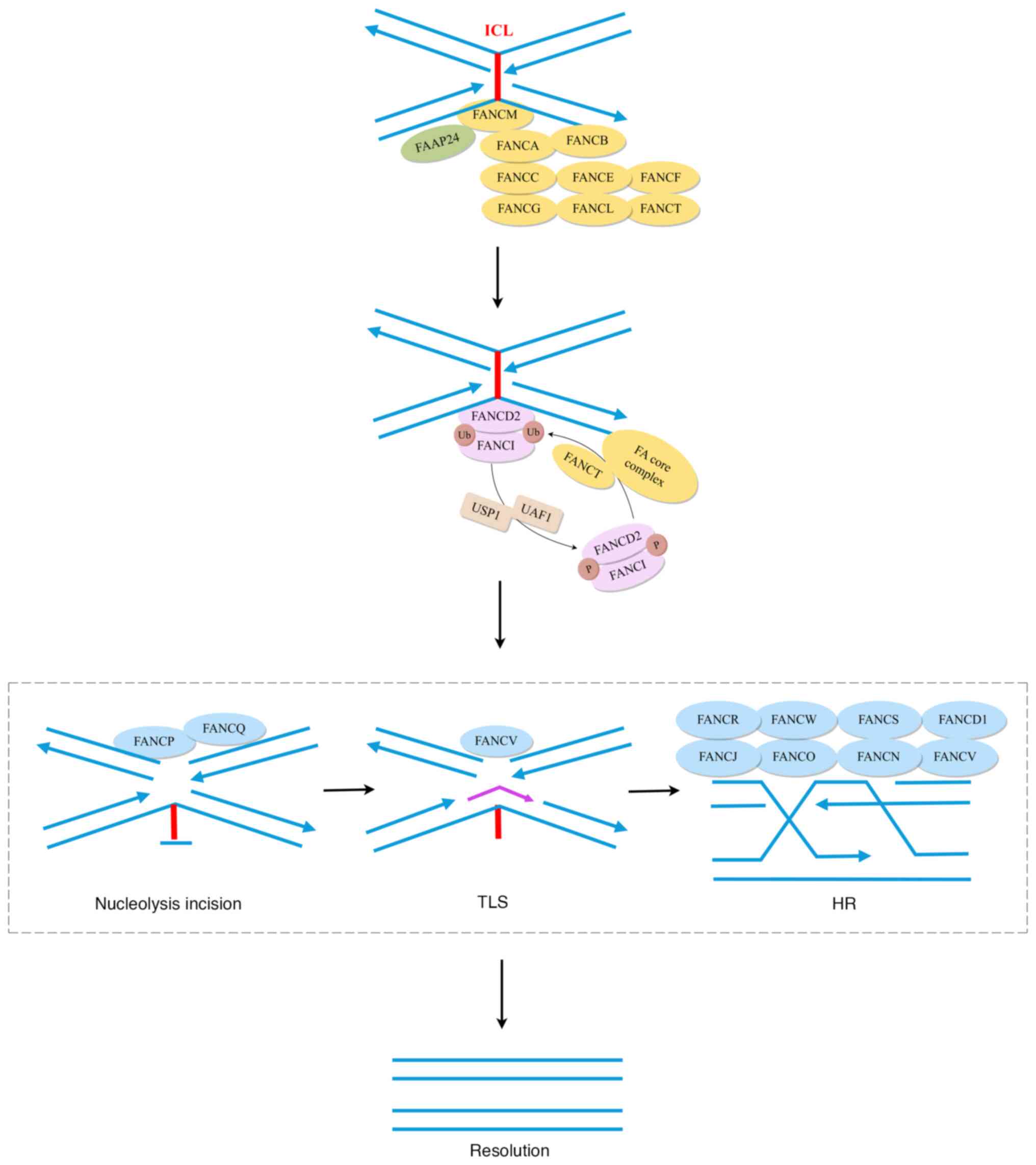
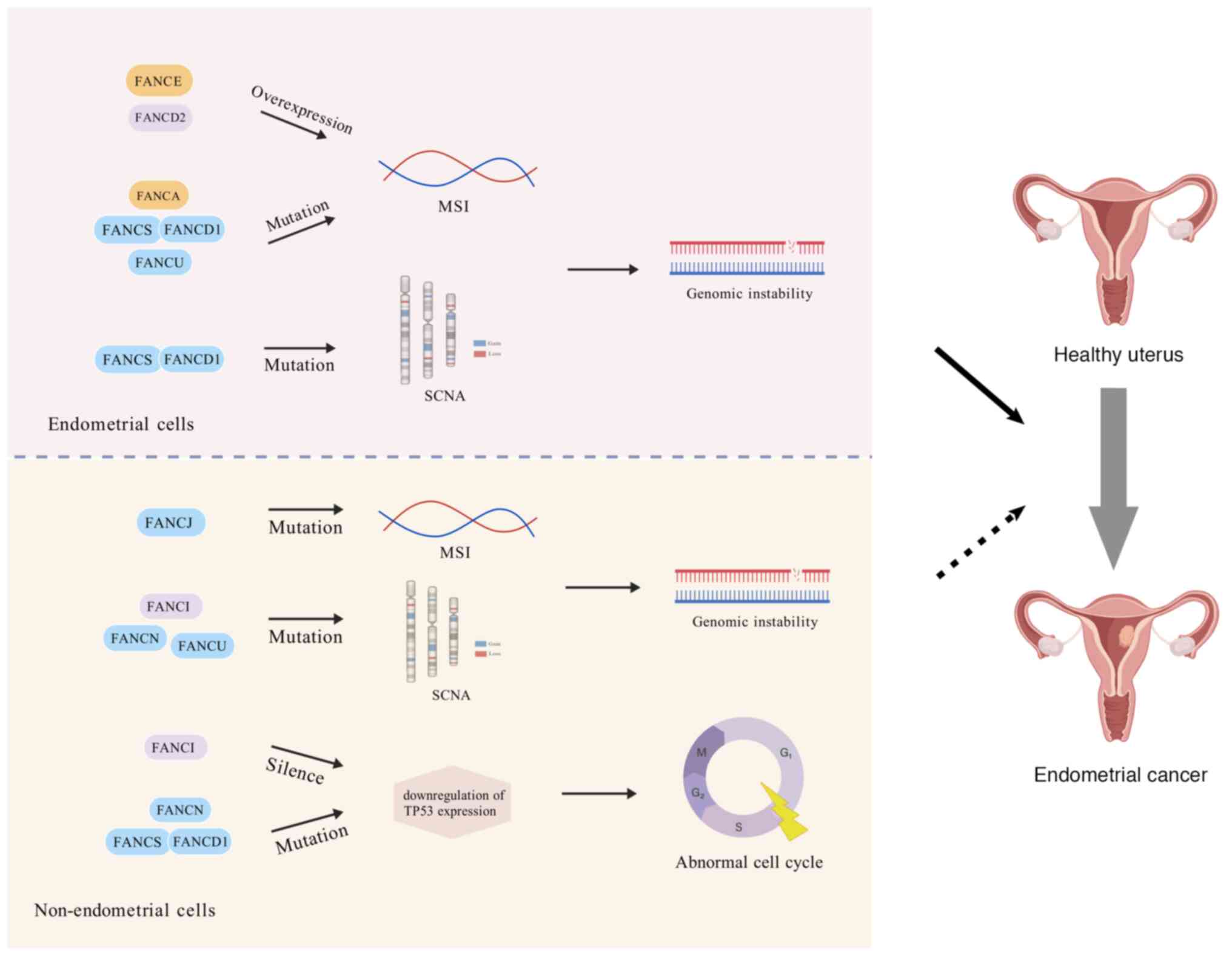
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
2
|
Feng J, Lin R, Li H, Wang J and He H:
Global and regional trends in the incidence and mortality burden of
endometrial cancer, 1990–2019: Updated results from the global
burden of disease study, 2019. Chin Med J (Engl). 137:294–302.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Abdol Manap N, Ng BK, Phon SE, Abdul Karim
AK, Lim PS and Fadhil M: Endometrial cancer in pre-menopausal women
and younger: Risk factors and outcome. Int J Environ Res Public
Health. 19:90592022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gu B, Shang X, Yan M, Li X, Wang W, Wang Q
and Zhang C: Variations in incidence and mortality rates of
endometrial cancer at the global, regional, and national levels,
1990–2019. Gynecol Oncol. 161:573–580. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bokhman JV: Two pathogenetic types of
endometrial carcinoma. Gynecol Oncol. 15:10–17. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bae HS, Kim H, Kwon SY, Kim KR, Song JY
and Kim I: Should endometrial clear cell carcinoma be classified as
type II endometrial carcinoma? Int J Gynecol Pathol. 34:74–84.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Suarez AA, Felix AS and Cohn DE: Bokhman
Redux: Endometrial cancer ‘types’ in the 21st century. Gynecol
Oncol. 144:243–249. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Amant F, Moerman P, Neven P, Timmerman D,
Van Limbergen E and Vergote I: Endometrial cancer. Lancet.
366:491–505. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mhawech-Fauceglia P, Wang D, Kim G,
Sharifian M, Chen X, Liu Q, Lin YG, Liu S and Pejovic T: Expression
of DNA repair proteins in endometrial cancer predicts disease
outcome. Gynecol Oncol. 132:593–598. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cancer Genome Atlas Research Network, .
Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H,
Robertson AG, Pashtan I, Shen R, et al: Integrated genomic
characterization of endometrial carcinoma. Nature. 497:67–73. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Matthews HK, Bertoli C and de Bruin RAM:
Cell cycle control in cancer. Nat Rev Mol Cell Biol. 23:74–88.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Milletti G, Strocchio L, Pagliara D,
Girardi K, Carta R, Mastronuzzi A, Locatelli F and Nazio F:
Canonical and noncanonical roles of fanconi anemia proteins:
Implications in cancer predisposition. Cancers (Basel).
12:26842020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang X, Joseph S, Wu D, Bowser JL and
Vaziri C: The DNA Damage Response (DDR) landscape of endometrial
cancer defines discrete disease subtypes and reveals therapeutic
opportunities. NAR Cancer. 6:zcae0152024. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nalepa G and Clapp DW: Fanconi anemia and
the cell cycle: New perspectives on aneuploidy. F1000Prime Rep.
6:232014. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
García-de-Teresa B, Rodríguez A and Frias
S: Chromosome instability in Fanconi anemia: From breaks to
phenotypic consequences. Genes (Basel). 11:15282020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang W: Emergence of a DNA-damage response
network consisting of Fanconi anaemia and BRCA proteins. Nat Rev
Genet. 8:735–748. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang Y, Leung JWC, Lowery M, Matsushita
N, Wang Y, Shen X, Huong D, Takata M, Chen J and Li L: Modularized
functions of the Fanconi anemia core complex. Cell Rep.
7:1849–1857. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ciccia A, Ling C, Coulthard R, Yan Z, Xue
Y, Meetei AR, Laghmani el H, Joenje H, McDonald N, de Winter JP, et
al: Identification of FAAP24, a Fanconi anemia core complex protein
that interacts with FANCM. Mol Cell. 25:331–343. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang CC, Li Z, Lopez-Martinez D,
Nicholson WV, Vénien-Bryan C and Cohn MA: The FANCD2-FANCI complex
is recruited to DNA interstrand crosslinks before
monoubiquitination of FANCD2. Nat Commun. 7:121242016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim H and D'Andrea AD: Regulation of DNA
cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev.
26:1393–1408. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Salomão VMR, de Almeida AM, Matuo R and
Sousa FG: In silico studies of molecular alterations in fanconi
anemia genes from cancer cell lines and samples. Obs Econ Latinoam.
21:13067–13087. 2023.
|
|
22
|
Pace P, Johnson M, Tan WM, Mosedale G, Sng
C, Hoatlin M, de Winter J, Joenje H, Gergely F and Patel KJ: FANCE:
The link between Fanconi anaemia complex assembly and activity.
EMBO J. 21:3414–3423. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bouffard F, Plourde K, Bélanger S,
Ouellette G, Labrie Y and Durocher F: Analysis of a FANCE splice
isoform in regard to DNA repair. J Mol Biol. 427:3056–3073. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
de Angelis de Carvalho N, Niitsuma BN,
Kozak VN, Costa FD, de Macedo MP, Kupper BEC, Silva MLG, Formiga
MN, Volc SM, Aguiar Junior S, et al: Clinical and molecular
assessment of patients with Lynch syndrome and sarcomas
underpinning the association with MSH2 germline pathogenic
variants. Cancers (Basel). 12:18482020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng C, Ren Z, Chen H, Yuan X, Suye S,
Yin H and Fu C: Reduced FANCE confers genomic instability and
malignant behavior by regulating cell cycle progression in
endometrial cancer. J Cancer. 14:2670–2685. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou Z, Yin H, Suye S, He J and Fu C:
Pan-cancer analysis of the prognostic and immunological role of
Fanconi anemia complementation group E. Front Genet.
13:10249892023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin B, Li H, Zhang T, Ye X, Yang H and
Shen Y: Comprehensive analysis of macrophage-related multigene
signature in the tumor microenvironment of head and neck squamous
cancer. Aging (Albany NY). 13:5718–5747. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Niedzwiedz W, Mosedale G, Johnson M, Ong
CY, Pace P and Patel KJ: The Fanconi anaemia gene FANCC promotes
homologous recombination and error-prone DNA repair. Mol Cell.
15:607–620. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Heald B, Mokhtary S, Nielsen SM, Rojahn S,
Yang S, Michalski ST and Esplin ED: Unexpected actionable genetic
variants revealed by multigene panel testing of patients with
uterine cancer. Gynecol Oncol. 166:344–350. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Luebben SW, Kawabata T, Johnson CS,
O'sullivan MG and Shima N: A concomitant loss of dormant origins
and FANCC exacerbates genome instability by impairing DNA
replication fork progression. Nucleic Acids Res. 42:5605–5615.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Heinrich MC, Silvey KV, Stone S, Zigler
AJ, Griffith DJ, Montalto M, Chai L, Zhi Y and Hoatlin ME:
Posttranscriptional cell cycle-dependent regulation of human FANCC
expression. Blood. 95:3970–3977. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gallmeier E, Calhoun ES, Rago C, Brody JR,
Cunningham SC, Hucl T, Gorospe M, Kohli M, Lengauer C and Kern SE:
Targeted disruption of FANCC and FANCG in human cancer provides a
preclinical model for specific therapeutic options.
Gastroenterology. 130:2145–2154. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li X, Plett PA, Yang Y, Hong P, Freie B,
Srour EF, Orschell CM, Clapp DW and Haneline LS: Fanconi anemia
type C-deficient hematopoietic stem/progenitor cells exhibit
aberrant cell cycle control. Blood. 102:2081–2084. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Singh DK, Gamboa RS, Singh AK, Walkemeier
B, Van Leene J, De Jaeger G, Siddiqi I, Guerois R, Crismani W and
Mercier R: The FANCC-FANCE-FANCF complex is evolutionarily
conserved and regulates meiotic recombination. Nucleic Acids Res.
51:2516–2528. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ahsan MD, Webster EM, Qazi M, Weiss J,
Levi S, Cantillo E, Chapman-Davis E, Holcomb K, Sharaf R and Frey
MK: #1090 The mutational landscape of uterine sarcoma: Is there
rationale for targeted therapies? Int J Gynecol Cancer. 33 (Suppl
3):A202.2–A202. 2023.
|
|
36
|
Nasioudis D, Latif NA, Ko EM, Cory L, Kim
SH, Martin L, Simpkins F and Giuntoli R II: Next generation
sequencing reveals a high prevalence of pathogenic mutations in
homologous recombination DNA damage repair genes among patients
with uterine sarcoma. Gynecol Oncol. 177:14–19. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Williams SA, Wilson JB, Clark AP,
Mitson-Salazar A, Tomashevski A, Ananth S, Glazer PM, Semmes OJ,
Bale AE, Jones NJ and Kupfer GM: Functional and physical
interaction between the mismatch repair and FA-BRCA pathways. Hum
Mol Genet. 20:4395–4410. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Drusbosky L, Haynes G, Grant B, Sobernis P
and Lheureux S: Abstract A012: Genomic landscape of somatic
alterations identified in endometrial cancer using liquid biopsy.
Clin Cancer Res. 30 (5 Suppl):A0122024. View Article : Google Scholar
|
|
39
|
Kral J, Jelinkova S, Zemankova P, Vocka M,
Borecka M, Cerna L, Cerna M, Dostalek L, Duskova P, Foretova L, et
al: Germline multigene panel testing of patients with endometrial
cancer. Oncol Lett. 25:2162023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu Y, Gusev A and Kraft P: Germline
cancer gene expression quantitative trait loci influence local and
global tumor mutations. medRxiv. 2022.2008.2023.22279002. 2022.
|
|
41
|
Wepy C, Nucci MR and Parra-Herran C:
Atypical endometriosis: Comprehensive characterization of
clinicopathologic, immunohistochemical, and molecular features. Int
J Gynecol Pathol. 43:70–77. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lopez-Martinez D, Kupculak M, Yang D,
Yoshikawa Y, Liang CC, Wu R, Gygi SP and Cohn MA: Phosphorylation
of FANCD2 inhibits the FANCD2/FANCI complex and suppresses the
Fanconi anemia pathway in the absence of DNA damage. Cell Rep.
27:2990–3005.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zheng C, Ren Z, Chen H, Yuan X, Suye S,
Yin H, Zhou Z and Fu C: FANCD2 promotes the malignant behavior of
endometrial cancer cells and its prognostic value. Exp Cell Res.
421:1133882022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhao Z, Wang R, Wang R, Song J, Ma F, Pan
H, Gao C, Wang D, Chen X and Fan X: Pancancer analysis of the
prognostic and immunological role of FANCD2: A potential target for
carcinogenesis and survival. BMC Med Genomics. 17:692024.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lin HH, Zeng WH, Yang HK, Huang LS, Pan R
and Lei NX: Fanconi anemia complementation group D2 promotes
sensitivity of endometrial cancer cells to chemotherapeutic agents
by inhibiting the ferroptosis pathway. BMC Womens Health.
24:412024. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ishiai M, Kitao H, Smogorzewska A, Tomida
J, Kinomura A, Uchida E, Saberi A, Kinoshita E, Kinoshita-Kikuta E,
Koike T, et al: FANCI phosphorylation functions as a molecular
switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol.
15:1138–1146. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kaljunen H, Taavitsainen S, Kaarijärvi R,
Takala E, Paakinaho V, Nykter M, Bova GS and Ketola K: Fanconi
anemia pathway regulation by FANCI in prostate cancer. Front Oncol.
13:12608262023. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhao Y, Li Q, Li J, Cui Y and Lu Z:
Expression and clinical significance of FANCI gene in pan-cancer: A
comprehensive analysis based on multi-omics data. Front Genet.
16:15428882025. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Fierheller CT, Alenezi WM, Serruya C,
Revil T, Amuzu S, Bedard K, Subramanian DN, Fewings E, Bruce JP,
Prokopec S, et al: Molecular genetic characteristics of FANCI, a
proposed new ovarian cancer predisposing gene. Genes (Basel).
14:2772023. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wen H, Xu Q, Sheng X, Li H, Wang X and Wu
X: Prevalence and landscape of pathogenic or likely pathogenic
germline variants and their association with somatic phenotype in
unselected Chinese patients with gynecologic cancers. JAMA Netw
Open. 6:e23264372023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dong L, Wang T, Li N, Yao H, Ying J, Wu L
and Yuan G: Prevalence and prognostic relevance of homologous
recombination repair gene mutations in uterine serous carcinoma.
Cells. 11:35632022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bi J, Areecheewakul S, Li Y, Yang S, Zhang
Y, Ebeid K, Li L, Thiel KW, Zhang J, Dai D, et al: MTDH/AEG-1
downregulation using pristimerin-loaded nanoparticles inhibits
Fanconi anemia proteins and increases sensitivity to platinum-based
chemotherapy. Gynecol Oncol. 155:349–358. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Becker AE, Hernandez YG, Frucht H and
Lucas AL: Pancreatic ductal adenocarcinoma: Risk factors,
screening, and early detection. World J Gastroenterol.
20:11182–11198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Buisson R and Masson JY: PALB2
self-interaction controls homologous recombination. Nucleic Acids
Res. 40:10312–10323. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Foo TK and Xia B: BRCA1-dependent and
independent recruitment of PALB2-BRCA2-RAD51 in the DNA damage
response and cancer. Cancer Res. 82:3191–3197. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Huang KL, Mashl RJ, Wu Y, Ritter DI, Wang
J, Oh C, Paczkowska M, Reynolds S, Wyczalkowski MA, Oak N, et al:
Pathogenic germline variants in 10,389 adult cancers. Cell.
173:355–370.e14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Teo ZL, Park DJ, Provenzano E, Chatfield
CA, Odefrey FA, Nguyen-Dumont T kConFab, Dowty JG, Hopper JL,
Winship I, et al: Prevalence of PALB2 mutations in Australasian
multiple-case breast cancer families. Breast Cancer Res.
15:R172013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Susswein LR, Marshall ML, Nusbaum R, Vogel
Postula KJ, Weissman SM, Yackowski L, Vaccari EM, Bissonnette J,
Booker JK, Cremona ML, et al: Pathogenic and likely pathogenic
variant prevalence among the first 10,000 patients referred for
next-generation cancer panel testing. Genet Med. 18:823–832. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fulk K, Milam MR, Li S, Yussuf A, Black
MH, Chao EC, LaDuca H and Stany MP: Women with breast and uterine
cancer are more likely to harbor germline mutations than women with
breast or uterine cancer alone: A case for expanded gene testing.
Gynecol Oncol. 152:612–617. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Heeke AL, Pishvaian MJ, Lynce F, Xiu J,
Brody JR, Chen WJ, Baker TM, Marshall JL and Isaacs C: Prevalence
of homologous recombination-related gene mutations across multiple
cancer types. JCO Precis Oncol. 2018.PO.17.00286. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chan GHJ, Ong PY, Low JJH, Kong HL, Ow
SGW, Tan DSP, Lim YW, Lim SE and Lee SC: Clinical genetic testing
outcome with multi-gene panel in Asian patients with multiple
primary cancers. Oncotarget. 9:306492018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Johnatty SE, Pesaran T, Dolinsky J, Yussuf
A, LaDuca H, James PA, O'Mara TA and Spurdle AB: Case-case analysis
addressing ascertainment bias for multigene panel testing
implicates BRCA1 and PALB2 in endometrial cancer. Hum Mutat.
42:1265–1278. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Tian W, Bi R, Ren Y, He H, Shi S, Shan B,
Yang W, Wang Q and Wang H: Screening for hereditary cancers in
patients with endometrial cancer reveals a high frequency of
germline mutations in cancer predisposition genes. Int J Cancer.
145:1290–1298. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kondrashova O, Shamsani J, O'Mara TA,
Newell F, McCart Reed AE, Lakhani SR, Kirk J, Pearson JV, Waddell N
and Spurdle AB: Tumor signature analysis implicates hereditary
cancer genes in endometrial cancer development. Cancers (Basel).
13:17622021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Cilento MA, Poplawski NK, Paramasivam S,
Thomas DM and Kichenadasse G: Germline PALB2 variants and PARP
inhibitors in endometrial cancer. J Natl Compr Canc Netw.
19:1212–1217. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Foo TK, Tischkowitz M, Simhadri S, Boshari
T, Zayed N, Burke KA, Berman SH, Blecua P, Riaz N, Huo Y, et al:
Compromised BRCA1-PALB2 interaction is associated with breast
cancer risk. Oncogene. 36:4161–4170. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bowman-Colin C, Xia B, Bunting S, Klijn C,
Drost R, Bouwman P, Fineman L, Chen X, Culhane AC, Cai H, et al:
Palb2 synergizes with Trp53 to suppress mammary tumor formation in
a model of inherited breast cancer. Proc Natl Acad Sci USA.
110:8632–8637. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Somyajit K, Subramanya S and Nagaraju G:
Distinct roles of FANCO/RAD51C protein in DNA damage signaling and
repair: Implications for Fanconi anemia and breast cancer
susceptibility. J Biol Chem. 287:3366–3380. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kolinjivadi AM, Chong ST, Choudhary R,
Sankar H, Chew EL, Yeo C, Chan SH and Ngeow J: Functional analysis
of germline RAD51C missense variants highlight the role of RAD51C
in replication fork protection. Hum Mol Genet. 32:1401–1409. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Chun J, Buechelmaier ES and Powell SN:
Rad51 paralog complexes BCDX2 and CX3 act at different stages in
the BRCA1-BRCA2-dependent homologous recombination pathway. Mol
Cell Biol. 33:387–395. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ring KL, Bruegl AS, Allen BA, Elkin EP,
Singh N, Hartman AR, Daniels MS and Broaddus RR: Germline
multi-gene hereditary cancer panel testing in an unselected
endometrial cancer cohort. Mod Pathol. 29:1381–1389. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Hirshfield KM, Rebbeck TR and Levine AJ:
Germline mutations and polymorphisms in the origins of cancers in
women. J Oncol. 2010:2976712010. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Nath S and Nagaraju G: FANCJ helicase
promotes DNA end resection by facilitating CtIP recruitment to DNA
double-strand breaks. PLoS Genet. 16:e10087012020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Alayev A, Salamon RS, Manna S, Schwartz
NS, Berman AY and Holz MK: Estrogen induces RAD51C expression and
localization to sites of DNA damage. Cell Cycle. 15:3230–3239.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Muhseena NK, Mathukkada S, Das SP and Laha
S: The repair gene BACH1-a potential oncogene. Oncol Rev.
15:5192021. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Long B, Lilyquist J, Weaver A, Hu C,
Gnanaolivu R, Lee KY, Hart SN, Polley EC, Bakkum-Gamez JN, Couch FJ
and Dowdy SC: Cancer susceptibility gene mutations in type I and II
endometrial cancer. Gynecol Oncol. 152:20–25. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
de Jonge MM, Auguste A, van Wijk LM,
Schouten PC, Meijers M, Ter Haar NT, Smit VTHBM, Nout RA, Glaire
MA, Church DN, et al: Frequent homologous recombination deficiency
in high-grade endometrial carcinomas. Clin Cancer Res.
25:1087–1097. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Nakamura K, Aimono E, Tanishima S, Imai M,
Nagatsuma AK, Hayashi H, Yoshimura Y, Nakayama K, Kyo S and
Nishihara H: Olaparib monotherapy for BRIP1-mutated high-grade
serous endometrial cancer. JCO Precis Oncol. 4:PO.19.00368.
2020.
|
|
79
|
Matsuzaki K, Borel V, Adelman CA,
Schindler D and Boulton SJ: FANCJ suppresses microsatellite
instability and lymphomagenesis independent of the Fanconi anemia
pathway. Genes Dev. 29:2532–2546. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Liu Y and Lu LY: BRCA1 and homologous
recombination: Implications from mouse embryonic development. Cell
Biosci. 10:492020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Kolinjivadi AM, Sannino V, de Antoni A,
Técher H, Baldi G and Costanzo V: Moonlighting at replication
forks-a new life for homologous recombination proteins BRCA1, BRCA2
and RAD51. FEBS Lett. 591:1083–1100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Deng CX: BRCA1: Cell cycle checkpoint,
genetic instability, DNA damage response and cancer evolution.
Nucleic Acids Res. 34:1416–1426. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Lee H: Cycling with BRCA2 from DNA repair
to mitosis. Exp Cell Res. 329:78–84. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
de Jonge MM, de Kroon CD, Jenner DJ,
Oosting J, de Hullu JA, Mourits MJE, Gómez Garcia EB, Ausems MGEM,
Margriet Collée J, van Engelen K, et al: Endometrial cancer risk in
women with germline BRCA1 or BRCA2 mutations: Multicenter cohort
study. J Natl Cancer Inst. 113:1203–1211. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Jamieson A, Sobral de Barros J, Cochrane
DR, Douglas JM, Shankar S, Lynch BJ, Leung S, Martin S, Senz J, Lum
A, et al: Targeted and shallow whole-genome sequencing identifies
therapeutic opportunities in p53abn endometrial cancers. Clin
Cancer Res. 30:2461–2474. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Smith ES, Da Cruz Paula A, Cadoo KA,
Abu-Rustum NR, Pei X, Brown DN, Ferrando L, Sebastiao APM, Riaz N,
Robson ME, et al: Endometrial cancers in BRCA1 or BRCA2 germline
mutation carriers: Assessment of homologous recombination DNA
repair defects. JCO Precis Oncol. 3:PO.19.00103. 2019.
|
|
87
|
Ito M, Fujita Y and Shinohara A: Positive
and negative regulators of RAD51/DMC1 in homologous recombination
and DNA replication. DNA Repair (Amst). 134:1036132024. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Auguste A, Genestie C, De Bruyn M, Adam J,
Le Formal A, Drusch F, Pautier P, Crosbie EJ, MacKay H, Kitchener
HC, et al: Refinement of high-risk endometrial cancer
classification using DNA damage response biomarkers: A TransPORTEC
initiative. Mod Pathol. 31:1851–1861. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Smolarz B and Romanowicz H: Association
between single nucleotide polymorphism of DNA repair genes and
endometrial cancer: A case-control study. Int J Clin Exp Pathol.
11:1732–1738. 2018.PubMed/NCBI
|
|
90
|
Romanowicz-Makowska H, Smolarz B, Połać I
and Sporny S: Single nucleotide polymorphisms of RAD51 G135C, XRCC2
Arg188His and XRCC3 Thr241Met homologous recombination repair genes
and the risk of sporadic endometrial cancer in Polish women. J
Obstet Gynaecol Res. 38:918–924. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zeng X, Zhang Y, Yang L, Xu H, Zhang T, An
R and Zhu K: Association between RAD51 135 G/C polymorphism and
risk of 3 common gynecological cancers: A meta-analysis. Medicine
(Baltimore). 97:e112512018. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Michalska MM, Samulak D, Romanowicz H and
Smolarz B: Association of polymorphisms in the 5′ untranslated
region of RAD51 gene with risk of endometrial cancer in the Polish
population. Arch Gynecol Obstet. 290:985–991. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Krupa R, Sobczuk A, Popławski T, Wozniak K
and Blasiak J: DNA damage and repair in endometrial cancer in
correlation with the hOGG1 and RAD51 genes polymorphism. Mol Biol
Rep. 38:1163–1170. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Park JY, Virts EL, Jankowska A, Wiek C,
Othman M, Chakraborty SC, Vance GH, Alkuraya FS, Hanenberg H and
Andreassen PR: Complementation of hypersensitivity to DNA
interstrand crosslinking agents demonstrates that XRCC2 is a
Fanconi anaemia gene. J Med Genet. 53:672–680. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Johnson RD, Liu N and Jasin M: Mammalian
XRCC2 promotes the repair of DNA double-strand breaks by homologous
recombination. Nature. 401:397–399. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Griffin CS, Simpson PJ, Wilson CR and
Thacker J: Mammalian recombination-repair genes XRCC2 and XRCC3
promote correct chromosome segregation. Nat Cell Biol. 2:757–761.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Webster ALH, Sanders MA, Patel K, Dietrich
R, Noonan RJ, Lach FP, White RR, Goldfarb A, Hadi K, Edwards ME, et
al: Fanconi anemia pathway deficiency drives copy number variation
in squamous cell carcinomas. bioRXiv. 2021.2008. 2014.456365.
2021.PubMed/NCBI
|
|
98
|
Nero C, Pasciuto T, Cappuccio S, Corrado
G, Pelligra S, Zannoni GF, Santoro A, Piermattei A, Minucci A,
Lorusso D, et al: Further refining 2020 ESGO/ESTRO/ESP molecular
risk classes in patients with early-stage endometrial cancer: A
propensity score-matched analysis. Cancer. 128:2898–2907. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Taylor NP, Gibb RK, Powell MA, Mutch DG,
Huettner PC and Goodfellow PJ: Defective DNA mismatch repair and
XRCC2 mutation in uterine carcinosarcomas. Gynecol Oncol.
100:107–110. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Michalska MM, Samulak D, Bieńkiewicz J,
Romanowicz H and Smolarz B: Association between-41657C/T single
nucleotide polymorphism of DNA repair gene XRCC2 and endometrial
cancer risk in Polish women. Pol J Pathol. 66:67–71. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Romanowicz H, Brys M, Forma E and Smolarz
B: Lack of association between the 4234G/C X-ray repair
cross-complementing 2 (XRCC2) gene polymorphism and the risk of
endometrial cancer among Polish population. J Gynecol Res Obstet.
2:47–50. 2016. View Article : Google Scholar
|
|
102
|
Inano S, Sato K, Katsuki Y, Kobayashi W,
Tanaka H, Nakajima K, Nakada S, Miyoshi H, Knies K, Takaori-Kondo
A, et al: RFWD3-mediated ubiquitination promotes timely removal of
both RPA and RAD51 from DNA damage sites to facilitate homologous
recombination. Mol Cell. 66:622–634.e8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Fu X, Yucer N, Liu S, Li M, Yi P, Mu JJ,
Yang T, Chu J, Jung SY, O'Malley BW, et al: RFWD3-Mdm2 ubiquitin
ligase complex positively regulates p53 stability in response to
DNA damage. Proc Natl Acad Sci USA. 107:4579–4584. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Wang Y: Role of WD domain-containing
proteins in cell cycle progression (unpublished thesis). University
of Illinois at Urbana-Champaign; 2017
|