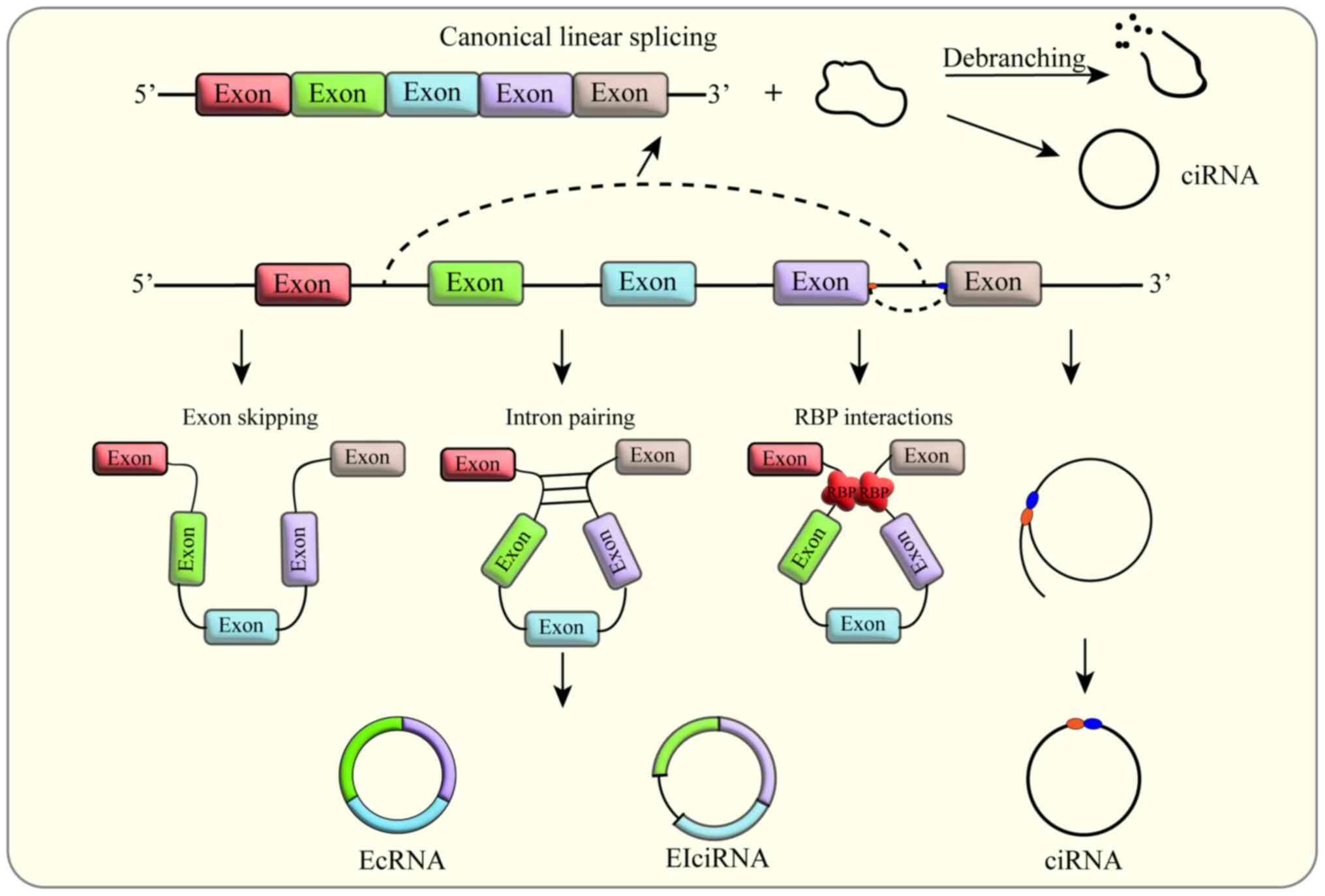
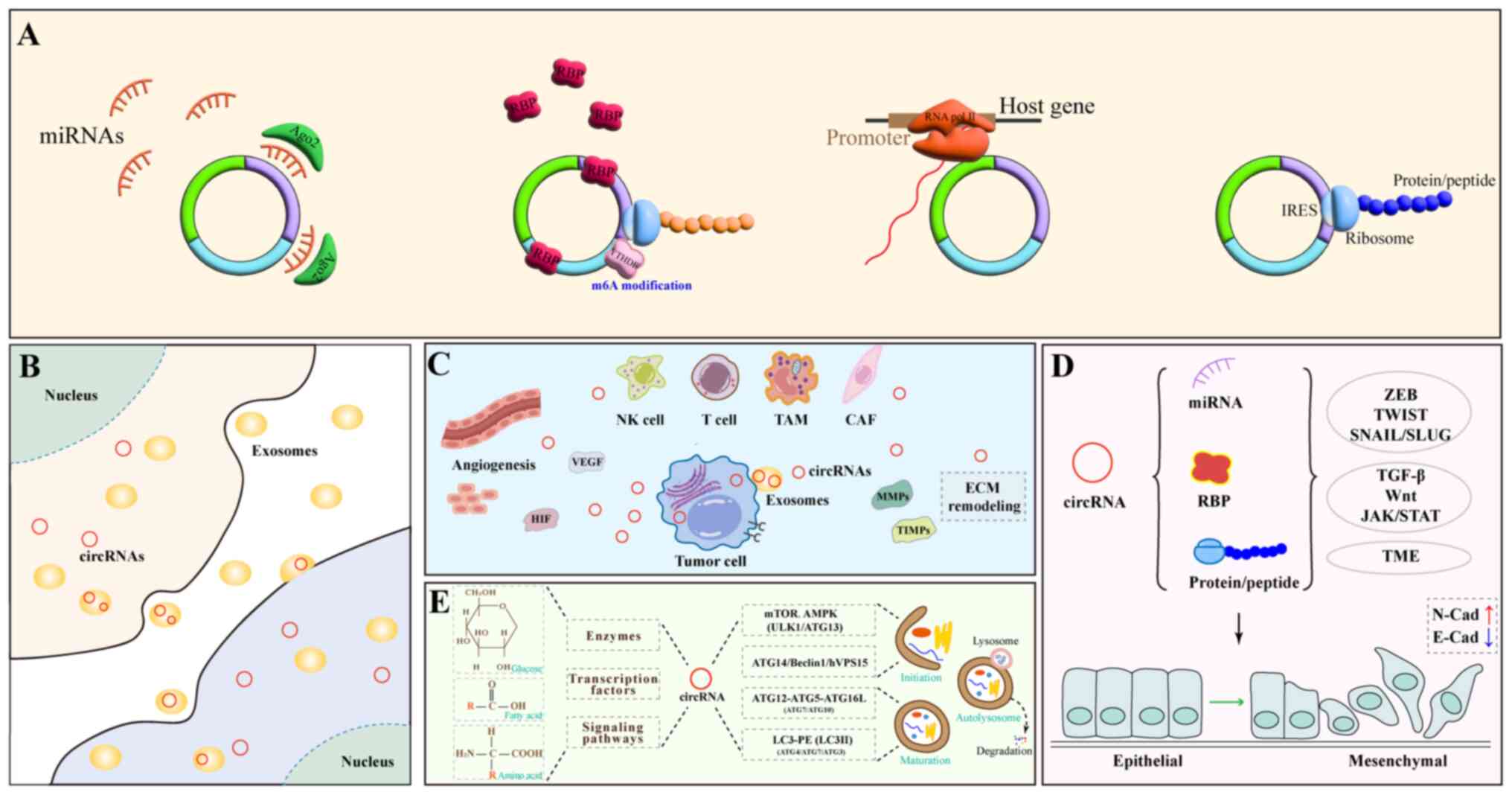
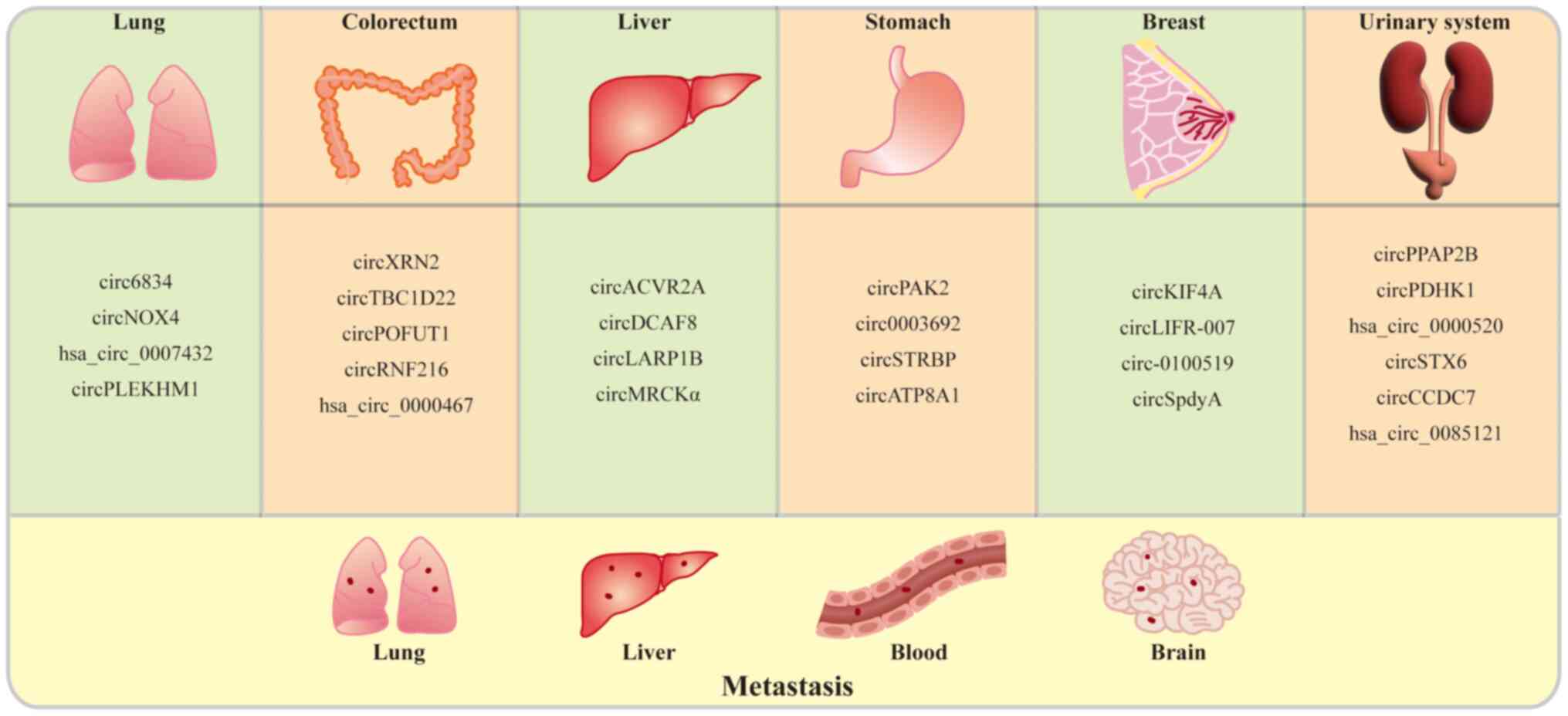
|
1
|
Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ and
Xu RH: Circular RNA: Metabolism, functions and interactions with
proteins. Mol Cancer. 19:1722020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly Base-paired rod-like
structures. Proc Natl Acad Sci. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cocquerelle C, Mascrez B, Hétuin D and
Bailleul B: Mis-splicing yields circular RNA molecules. FASEB J.
7:155–160. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Beermann J, Piccoli MT, Viereck J and Thum
T: Non-coding RNAs in development and disease: Background,
mechanisms, and therapeutic approaches. Physiol Rev. 96:1297–1325.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang R, Yang B, Liu W, Tan C, Chen H and
Wang X: Emerging role of non-coding RNAs in neuroinflammation
mediated by microglia and astrocytes. J Neuroinflammation.
20:1732023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang ZY, Liu XX and Deng YF: Negative
feedback of SNRK to circ-SNRK regulates cardiac function
post-myocardial infarction. Cell Death Differ. 29:709–721. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou Z, Sun B, Huang S and Zhao L: Roles
of circular RNAs in immune regulation and autoimmune diseases. Cell
Death Dis. 10:5032019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu YL, Lin ZJ, Li CC, Lin X, Shan SK, Guo
B, Zheng MH, Wang Y, Li F and Yuan LQ: Adipose exosomal noncoding
RNAs: Roles and mechanisms in metabolic diseases. Obes Rev.
25:e137402024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kristensen LS, Jakobsen T, Hager H and
Kjems J: The emerging roles of circRNAs in cancer and oncology. Nat
Rev Clin Oncol. 19:188–206. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Conn VM, Chinnaiyan AM and Conn SJ:
Circular RNA in cancer. Nat Rev Cancer. 24:597–613. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Valastyan S and Weinberg RA: Tumor
metastasis: Molecular insights and evolving paradigms. Cell.
147:275–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu QL, Zhang Z, Wei X and Zhou ZG:
Noncoding RNAs in tumor metastasis: Molecular and clinical
perspectives. Cell Mol Life Sci. 78:6823–6850. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fidler IJ: The pathogenesis of cancer
metastasis: The ‘seed and soil’ hypothesis revisited. Nat Rev
Cancer. 3:453–458. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gerstberger S, Jiang Q and Ganesh K:
Metastasis. Cell. 186:1564–1579. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chambers AF, Groom AC and MacDonald IC:
Dissemination and growth of cancer cells in metastatic sites. Nat
Rev Cancer. 2:563–572. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo
Z, Zhang J, Chen LL and Yang L: Diverse alternative back-splicing
and alternative splicing landscape of circular RNAs. Genome Res.
26:1277–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Capel B, Swain A, Nicolis S, Hacker A,
Walter M, Koopman P, Goodfellow P and Lovell-Badge R: Circular
transcripts of the testis-determining gene Sry in adult mouse
testis. Cell. 73:1019–1030. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen H, Liu B, Xu J, Zhang B, Wang Y, Shi
L and Cai X: Circular RNAs: Characteristics, biogenesis, mechanisms
and functions in liver cancer. J Hematol Oncol. 14:1342021.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, et
al: circRNA biogenesis competes with Pre-mRNA splicing. Mol Cell.
56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang L, Wilusz JE and Chen LL: Biogenesis
and regulatory roles of circular RNAs. Annu Rev Cell Dev Biol.
38:263–289. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu CX and Chen LL: Circular RNAs:
Characterization, cellular roles, and applications. Cell.
185:2016–2034. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu H, Tang J, Wang H, Guo X, Tu C and Li
Z: The crosstalk between alternative splicing and circular RNA in
cancer: Pathogenic insights and therapeutic implications. Cell Mol
Biol Lett. 29:1422024. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li J, Sun D, Pu W, Wang J and Peng Y:
Circular RNAs in cancer: Biogenesis, function, and clinical
significance. Trends Cancer. 6:319–336. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu CY and Kuo HC: The emerging roles and
functions of circular RNAs and their generation. J Biomed Sci.
26:292019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tran AM, Chalbatani GM, Berland L, Cruz De
los Santos M, Raj P, Jalali SA, Gharagouzloo E, Ivan C, Dragomir MP
and Calin GA: A new world of biomarkers and therapeutics for female
reproductive system and breast cancers: Circular RNAs. Front Cell
Dev Biol. 8:502020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
He X, Xu T, Hu W, Tan Y, Wang D, Wang Y,
Zhao C, Yi Y, Xiong M, Lv W, et al: Circular RNAs: Their role in
the pathogenesis and orchestration of breast cancer. Front Cell Dev
Biol. 9:6477362021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang A, Zheng H, Wu Z, Chen M and Huang
Y: Circular RNA-protein interactions: Functions, mechanisms, and
identification. Theranostics. 10:3503–3517. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guo Y, Huang Q, Heng Y, Zhou Y, Chen H, Xu
C, Wu C, Tao L and Zhou L: Circular RNAs in cancer. MedComm.
6:e700792025. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nemeth K, Bayraktar R, Ferracin M and
Calin GA: Non-coding RNAs in disease: From mechanisms to
therapeutics. Nat Rev Genet. 25:211–232. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shang R, Lee S, Senavirathne G and Lai EC:
microRNAs in action: Biogenesis, function and regulation. Nat Rev
Genet. 24:816–833. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen J, Yang J, Fei X, Wang X and Wang K:
CircRNA ciRS-7: A novel oncogene in multiple cancers. Int J Biol
Sci. 17:379–389. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Su C, Han Y, Zhang H, Li Y, Yi L, Wang X,
Zhou S, Yu D, Song X, Xiao N, et al: CiRS-7 targeting miR-7
modulates the progression of non-small cell lung cancer in a manner
dependent on NF-κB signalling. J Cell Mol Med. 22:3097–3107. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang J, Hu H and Zhao Y and Zhao Y:
CDR1as is overexpressed in laryngeal squamous cell carcinoma to
promote the tumour's progression via miR-7 signals. Cell Prolif.
51:e125212018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li F, Yang Q, He AT and Yang BB: Circular
RNAs in cancer: Limitations in functional studies and diagnostic
potential. Semin Cancer Biol. 75:49–61. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Matia-González AM, Laing EE and Gerber AP:
Conserved mRNA-binding proteomes in eukaryotic organisms. Nat
Struct Mol Biol. 22:1027–1033. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang J, Liu Q and Shyr Y: Dysregulated
transcription across diverse cancer types reveals the importance of
RNA-binding protein in carcinogenesis. BMC Genomics. 16
(Suppl):S52015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yao ZT, Yang YM, Sun MM, He Y, Liao L,
Chen KS and Li B: New insights into the interplay between long
non-coding RNAs and RNA-binding proteins in cancer. Cancer Commun
(Lond). 42:117–140. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang S, Sun Z, Lei Z and Zhang HT:
RNA-binding proteins and cancer metastasis. Semin Cancer Biol.
86:748–768. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wei W, Liu K, Huang X, Tian S, Wang H,
Zhang C, Ye J, Dong Y, An Z, Ma X, et al: EIF4A3-mediated
biogenesis of circSTX6 promotes bladder cancer metastasis and
cisplatin resistance. J Exp Clin Cancer Res. 43:22024. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li R, Wang J, Xie Z, Tian X, Hou J, Wang
D, Qian H, Shen H and Xu W: CircUSP1 as a novel marker promotes
gastric cancer progression via stabilizing HuR to upregulate USP1
and Vimentin. Oncogene. 43:1033–1049. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gu C, Shi X, Dai C, Shen F, Rocco G, Chen
J, Huang Z and Chen C, He C, Huang T and Chen C: RNA m6A
modification in cancers: Molecular mechanisms and potential
clinical applications. Innovation (Camb). 1:1000662020.PubMed/NCBI
|
|
46
|
Zhu ZM, Huo FC, Zhang J, Shan HJ and Pei
DS: Crosstalk between m6A modification and alternative splicing
during cancer progression. Clin Transl Med. 13:e14602023.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jiang X, Liu B, Nie Z, Duan L, Xiong Q,
Jin Z, Yang C and Chen Y: The role of m6A modification in the
biological functions and diseases. Signal Transduct Target Ther.
6:742021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xie F, Huang C, Liu F, Zhang H, Xiao X,
Sun J, Zhang X and Jiang G: CircPTPRA blocks the recognition of RNA
N6-methyladenosine through interacting with IGF2BP1 to suppress
bladder cancer progression. Mol Cancer. 20:682021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li Z, Yang HY, Dai XY, Zhang X, Huang YZ,
Shi L, Wei JF and Ding Q: CircMETTL3, upregulated in a
m6A-dependent manner, promotes breast cancer progression. Int J
Biol Sci. 17:1178–1190. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ruan H, Gu W, Xia W, Gong Y, Zhou X, Chen
W and Xiong J: METTL3 is suppressed by circular RNA
circMETTL3/miR-34c-3p signaling and limits the tumor growth and
metastasis in triple negative breast cancer. Front Oncol.
11:7781322021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tang X, Guo M, Zhang Y, Lv J, Gu C and
Yang Y: Examining the evidence for mutual modulation between m6A
modification and circular RNAs: Current knowledge and future
prospects. J Exp Clin Cancer Res. 43:2162024. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang KS, Choo QL, Weiner AJ, Ou JH,
Najarian RC, Thayer RM, Mullenbach GT, Denniston KJ, Gerin JL and
Houghton M: Structure, sequence and expression of the hepatitis
delta (delta) viral genome. Nature. 323:508–514. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Margvelani G, Maquera KAA, Welden JR,
Rodgers DW and Stamm S: Translation of circular RNAs. Nucleic Acids
Res. 53:gkae11672025. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hwang HJ and Kim YK: Molecular mechanisms
of circular RNA translation. Exp Mol Med. 56:1272–1280. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li Y, Wang Z, Su P, Liang Y, Li Z, Zhang
H, Song X, Han D, Wang X, Liu Y, et al: circ-EIF6 encodes
EIF6-224aa to promote TNBC progression via stabilizing MYH9 and
activating the Wnt/beta-catenin pathway. Mol Ther. 30:415–430.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li Y, Wang Z, Yang J, Sun Y, He Y, Wang Y,
Chen X, Liang Y, Zhang N, Wang X, et al: CircTRIM1 encodes
TRIM1-269aa to promote chemoresistance and metastasis of TNBC via
enhancing CaM-dependent MARCKS translocation and PI3K/AKT/mTOR
activation. Mol Cancer. 23:1022024. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wei J, Li M, Xue C, Chen S, Zheng L, Deng
H, Tang F, Li G, Xiong W, Zeng Z and Zhou M: Understanding the
roles and regulation patterns of circRNA on its host gene in
tumorigenesis and tumor progression. J Exp Clin Cancer Res.
42:862023. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li X, Wang J, Zhang C, Lin C, Zhang J,
Zhang W, Zhang W, Lu Y, Zheng L and Li X: Circular RNA circITGA7
inhibits colorectal cancer growth and metastasis by modulating the
Ras pathway and upregulating transcription of its host gene ITGA7.
J Pathol. 246:166–179. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhou J, Zhang S, Chen Z, He Z, Xu Y and Li
Z: CircRNA-ENO1 promoted glycolysis and tumor progression in lung
adenocarcinoma through upregulating its host gene ENO1. Cell Death
Dis. 10:8852019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhao W, Wang S, Qin T and Wang W: Circular
RNA (circ-0075804) promotes the proliferation of retinoblastoma via
combining heterogeneous nuclear ribonucleoprotein K (HNRNPK) to
improve the stability of E2F transcription factor 3 E2F3. J Cell
Biochem. 121:3516–3525. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhang C, Zhou X, Geng X, Zhang Y, Wang J,
Wang Y, Jing J, Zhou X and Pan W: Circular RNA hsa_circ_0006401
promotes proliferation and metastasis in colorectal carcinoma. Cell
Death Dis. 12:4432021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Feng Y, Yang Y, Zhao X, Fan Y, Zhou L,
Rong J and Yu Y: Circular RNA circ0005276 promotes the
proliferation and migration of prostate cancer cells by interacting
with FUS to transcriptionally activate XIAP. Cell Death Dis.
10:7922019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Hu X, Wu D, He X, Zhao H, He Z, Lin J,
Wang K, Wang W, Pan Z, Lin H and Wang M: circGSK3β promotes
metastasis in esophageal squamous cell carcinoma by augmenting
β-catenin signaling. Mol Cancer. 18:1602019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang X, Li H, Lu Y and Cheng L: Regulatory
effects of circular RNAs on host genes in human cancer. Front
Oncol. 10:5861632021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhang F, Jiang J, Qian H, Yan Y and Xu W:
Exosomal circRNA: Emerging insights into cancer progression and
clinical application potential. J Hematol Oncol. 16:672023.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhou H, He X, He Y, Ou C and Cao P:
Exosomal circRNAs: Emerging players in tumor metastasis. Front Cell
Dev Biol. 9:7862242021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F,
Ma P, Jiang H, Wu X, Shu Y and Xu T: Exosomal circSHKBP1 promotes
gastric cancer progression via regulating the miR-582-3p/HUR/VEGF
axis and suppressing HSP90 degradation. Mol Cancer. 19:1122020.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Dai J, Su Y, Zhong S, Cong L, Liu B, Yang
J, Tao Y, He Z, Chen C and Jiang Y: Exosomes: Key players in cancer
and potential therapeutic strategy. Signal Transduct Target Ther.
5:1452020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhu C, Su Y, Liu L, Wang S, Liu Y and Wu
J: Circular RNA hsa_circ_0004277 Stimulates malignant phenotype of
hepatocellular carcinoma and Epithelial-mesenchymal transition of
peripheral cells. Front Cell Dev Biol. 8:5855652021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Xu Z, Chen Y, Ma L, Chen Y, Liu J, Guo Y,
Yu T, Zhang L, Zhu L and Shu Y: Role of exosomal non-coding RNAs
from tumor cells and tumor-associated macrophages in the tumor
microenvironment. Mol Ther. 30:3133–3154. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Li J, Zhang G, Liu CG, Xiang X, Le MTN,
Sethi G, Wang L, Goh BC and Ma Z: The potential role of exosomal
circRNAs in the tumor microenvironment: Insights into cancer
diagnosis and therapy. Theranostics. 12:87–104. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liu J, Wu S, Zheng X, Zheng P, Fu Y, Wu C,
Lu B, Ju J and Jiang J: Immune suppressed tumor microenvironment by
exosomes derived from gastric cancer cells via modulating immune
functions. Sci Rep. 10:147492020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Hosseini R, Sarvnaz H, Arabpour M, Ramshe
SM, Asef-Kabiri L, Yousefi H, Akbari ME and Eskandari N: Cancer
exosomes and natural killer cells dysfunction: Biological roles,
clinical significance and implications for immunotherapy. Mol
Cancer. 21:152022. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yang C, Wu S, Mou Z, Zhou Q, Dai X, Ou Y,
Chen X, Chen Y, Xu C, Hu Y, et al: Exosome-derived circTRPS1
promotes malignant phenotype and CD8+ T cell exhaustion in bladder
cancer microenvironments. Mol Ther. 30:1054–1070. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Chen T, Liu Y, Li C, Xu C, Ding C, Chen J
and Zhao J: Tumor-derived exosomal circFARSA mediates M2 macrophage
polarization via the PTEN/PI3K/AKT pathway to promote non-small
cell lung cancer metastasis. Cancer Treat Res Commun.
28:1004122021.PubMed/NCBI
|
|
76
|
Vahabi A, Rezaie J, Hassanpour M, Panahi Y
and Nemati M, Rasmi Y and Nemati M: Tumor Cells-derived exosomal
CircRNAs: Novel cancer drivers, molecular mechanisms, and clinical
opportunities. Biochem Pharmacol. 200:1150382022. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yang Y and Cao Y: The impact of VEGF on
cancer metastasis and systemic disease. Semin Cancer Biol.
86:251–261. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ding P, Wu H, Wu J, Li T, He J, Ju Y, Liu
Y, Li F, Deng H, Gu R, et al: N6-methyladenosine modified circPAK2
promotes lymph node metastasis via targeting IGF2BPs/VEGFA
signaling in gastric cancer. Oncogene. 43:2548–2563. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhong Z, Huang M, Lv M, He Y, Duan C,
Zhang L and Chen J: Circular RNA MYLK as a competing endogenous RNA
promotes bladder cancer progression through modulating VEGFA/VEGFR2
signaling pathway. Cancer Lett. 403:305–317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Liu Y, Yang Y, Wang Z, Fu X, Chu XM, Li Y,
Wang Q, He X, Li M, Wang K, et al: Insights into the regulatory
role of circRNA in angiogenesis and clinical implications.
Atherosclerosis. 298:14–26. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhu J, Luo Y, Zhao Y, Kong Y, Zheng H, Li
Y, Gao B, Ai L, Huang H, Huang J, et al: circEHBP1 promotes
lymphangiogenesis and lymphatic metastasis of bladder cancer via
miR-130a-3p/TGFβR1/VEGF-D signaling. Mol Ther. 29:1838–1852. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Ma Z, Shuai Y, Gao X, Wen X and Ji J:
Circular RNAs in the tumour microenvironment. Mol Cancer. 19:82020.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Duan S, Wang S, Huang T, Wang J and Yuan
X: circRNAs: Insight into their role in Tumor-associated
macrophages. Front Oncol. 11:7807442021. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Lu C, Shi W, Hu W, Zhao Y, Zhao X, Dong F,
Xin Y, Peng T and Liu C: Endoplasmic reticulum stress promotes
breast cancer cells to release exosomes circ_0001142 and induces M2
polarization of macrophages to regulate tumor progression.
Pharmacol Res. 177:1060982022. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Chen W, Tang D, Lin J, Huang X, Lin S,
Shen G and Dai Y: Exosomal circSHKBP1 participates in non-small
cell lung cancer progression through PKM2-mediated glycolysis. Mol
Ther Oncolytics. 24:470–485. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Hu Z, Zhou S, Li J, Zhou Z, Wang P, Xin H,
Mao L, Luo C, Yu S, Huang XW, et al: Circular RNA sequencing
identifies CircASAP1 as a key regulator in hepatocellular carcinoma
metastasis. Hepatology. 72:906–922. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Sarnaik AA, Hwu P, Mulé JJ and
Pilon-Thomas S: Tumor-infiltrating lymphocytes: A new hope. Cancer
Cell. 42:1315–1318. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Weng Q, Chen M, Li M, Zheng YF, Shao G,
Fan W, Xu XM and Ji J: Global microarray profiling identified
hsa_circ_0064428 as a potential immune-associated prognosis
biomarker for hepatocellular carcinoma. J Med Genet. 56:32–38.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Wei CY, Zhu MX, Lu NH, Liu JQ, Yang YW,
Zhang Y, Shi YD, Feng ZH, Li JX, Qi FZ and Gu JY: Circular RNA
circ_0020710 drives tumor progression and immune evasion by
regulating the miR-370-3p/CXCL12 axis in melanoma. Mol Cancer.
19:842020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Doroshow DB, Bhalla S, Beasley MB, Sholl
LM, Kerr KM, Gnjatic S, Wistuba II, Rimm DL, Tsao MS and Hirsch FR:
PD-L1 as a biomarker of response to immune-checkpoint inhibitors.
Nat Rev Clin Oncol. 18:345–362. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Lin X, Kang K, Chen P, Zeng Z, Li G, Xiong
W, Yi M and Xiang B: Regulatory mechanisms of PD-1/PD-L1 in
cancers. Mol Cancer. 23:1082024. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Jiang W, Pan S, Chen X, Wang Z and Zhu X:
The role of lncRNAs and circRNAs in the PD-1/PD-L1 pathway in
cancer immunotherapy. Mol Cancer. 20:1162021. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Hong W, Xue M, Jiang J, Zhang Y and Gao X:
Circular RNA circ-CPA4/let-7 miRNA/PD-L1 axis regulates cell
growth, stemness, drug resistance and immune evasion in non-small
cell lung cancer (NSCLC). J Exp Clin Cancer Res. 39:1492020.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Li Y, Wang Z, Gao P, Cao D, Dong R, Zhu M,
Fei Y, Zuo X and Cai J: CircRHBDD1 promotes immune escape via
IGF2BP2/PD-L1 signaling and acts as a nanotherapeutic target in
gastric cancer. J Transl Med. 22:7042024. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Chan IS and Ewald AJ: The changing role of
natural killer cells in cancer metastasis. J Clin Invest.
132:e1437622022. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Chen Y, McAndrews KM and Kalluri R:
Clinical and therapeutic relevance of cancer-associated
fibroblasts. Nat Rev Clin Oncol. 18:792–804. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zheng S, Hu C, Lin H, Li G, Xia R, Zhang
X, Su D, Li Z, Zhou Q and Chen R: circCUL2 induces an inflammatory
CAF phenotype in pancreatic ductal adenocarcinoma via the
activation of the MyD88-dependent NF-κB signaling pathway. J Exp
Clin Cancer Res. 41:712022. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Yang F, Ning Z, Ma L, Liu W, Shao C, Shu Y
and Shen H: Exosomal miRNAs and miRNA dysregulation in
cancer-associated fibroblasts. Mol Cancer. 16:1482017. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Gou Z, Li J, Liu J and Yang N: The hidden
messengers: Cancer associated fibroblasts-derived exosomal miRNAs
as key regulators of cancer malignancy. Front Cell Dev Biol.
12:13783022024. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Liu G, Sun J, Yang ZF, Zhou C, Zhou PY,
Guan RY, Sun BY, Wang ZT, Zhou J, Fan J, et al: Cancer-associated
fibroblast-derived CXCL11 modulates hepatocellular carcinoma cell
migration and tumor metastasis through the
circUBAP2/miR-4756/IFIT1/3 axis. Cell Death Dis. 12:2602021.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Yang K, Zhang J and Bao C: Exosomal
circEIF3K from cancer-associated fibroblast promotes colorectal
cancer (CRC) progression via miR-214/PD-L1 axis. BMC Cancer.
21:9332021. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Sleeboom JJF, van Tienderen GS,
Schenke-Layland K, van der Laan LJW, Khalil AA and Verstegen MMA:
The extracellular matrix as hallmark of cancer and metastasis: From
biomechanics to therapeutic targets. Sci Transl Med.
16:eadg38402024. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Chen Z, Zheng Z, Xie Y, Zhong Q, Shangguan
W, Zhang Y, Zhu D and Xie W: Circular RNA circPPP6R3 upregulates
CD44 to promote the progression of clear cell renal cell carcinoma
via sponging miR-1238-3p. Cell Death Dis. 13:222021. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Hong Y, Qin H, Li Y, Zhang Y, Zhuang X,
Liu L, Lu K, Li L, Deng X, Liu F, et al: FNDC3B circular RNA
promotes the migration and invasion of gastric cancer cells via the
regulation of E-cadherin and CD44 expression. J Cell Physiol.
234:19895–19910. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Zhang Q, Wang W, Zhou Q, Chen C, Yuan W,
Liu J, Li X and Sun Z: Roles of circRNAs in the tumour
microenvironment. Mol Cancer. 19:142020. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
de Almeida LGN, Thode H, Eslambolchi Y,
Chopra S, Young D, Gill S, Devel L and Dufour A: Matrix
metalloproteinases: From molecular mechanisms to physiology,
pathophysiology, and pharmacology. Pharmacol Rev. 74:712–768. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Li Q, Liu Y, Chen S, Zong Z, Du Y, Sheng X
and Zhao Y: circ-CSPP1 promotes proliferation, invasion and
migration of ovarian cancer cells by acting as a miR-1236-3p
sponge. Biomed Pharmacother. 114:1088322019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Luo YH, Zhu XZ, Huang KW, Zhang Q, Fan YX,
Yan PW and Wen J: Emerging roles of circular RNA hsa_circ_0000064
in the proliferation and metastasis of lung cancer. Biomed
Pharmacother. 96:892–898. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Bakir B, Chiarella AM, Pitarresi JR and
Rustgi AK: EMT, MET, plasticity, and tumor metastasis. Trends Cell
Biol. 30:764–776. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Liu T, Zhao M, Peng L, Chen J, Xing P, Gao
P, Chen L, Qiao X, Wang Z, Di J, et al: WFDC3 inhibits tumor
metastasis by promoting the ERβ-mediated transcriptional repression
of TGFBR1 in colorectal cancer. Cell Death Dis. 14:4252023.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Zhang L, Song X, Chen X, Wang Q, Zheng X,
Wu C and Jiang J: Circular RNA CircCACTIN promotes gastric cancer
progression by sponging MiR-331-3p and regulating TGFBR1
Expression. Int J Biol Sci. 15:1091–1103. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhang X, Wu N and Wang J: Hsa-circ_0058106
induces EMT and metastasis in laryngeal cancer via sponging miR-153
and inducing Twist1 nuclear translocation. Cell Oncol.
44:1373–1386. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Rao D, Yu C, Sheng J, Lv E and Huang W:
The emerging roles of circFOXO3 in cancer. Front Cell Dev Biol.
9:6594172021. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Shen Z, Zhou L, Zhang C and Xu J:
Reduction of circular RNA Foxo3 promotes prostate cancer
progression and chemoresistance to docetaxel. Cancer Lett.
468:88–101. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Neumann DP, Goodall GJ and Gregory PA:
Regulation of splicing and circularisation of RNA in epithelial
mesenchymal plasticity. Semin Cell Dev Biol. 75:50–60. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Zhou Y, Zhan Y, Jiang W, Liu H and Wei S:
Long noncoding RNAs and circular RNAs in the metabolic
reprogramming of lung cancer: Functions, mechanisms, and clinical
potential. Oxid Med Cell Longev. 2022:48023382022. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T
and Shu Y: CircRNAs in cancer metabolism: A review. J Hematol
Oncol. 12:902019. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Cai ZR, Hu Y, Liao K, Li H, Chen DL and Ju
HQ: Circular RNAs: Emerging regulators of glucose metabolism in
cancer. Cancer Lett. 552:2159782023. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Zhang X, Wang S, Wang H, Cao J, Huang X,
Chen Z, Xu P, Sun G, Xu J, Lv J and Xu Z: Circular RNA circNRIP1
acts as a microRNA-149-5p sponge to promote gastric cancer
progression via the AKT1/mTOR pathway. Mol Cancer. 18:202019.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Hang D, Zhou J, Qin N, Zhou W, Ma H, Jin
G, Hu Z, Dai J and Shen H: A novel plasma circular RNA circFARSA is
a potential biomarker for non-small cell lung cancer. Cancer Med.
7:2783–2791. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Liu X, Liu Y, Liu Z, Lin C, Meng F, Xu L,
Zhang X, Zhang C, Zhang P, Gong S, et al: CircMYH9 drives
colorectal cancer growth by regulating serine metabolism and redox
homeostasis in a p53-dependent manner. Mol Cancer. 20:1142021.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Luo H, Peng J and Yuan Y: CircRNA OXCT1
promotes the malignant progression and glutamine metabolism of
non-small cell lung cancer by absorbing miR-516b-5p and
upregulating SLC1A5. Cell Cycle. 22:1182–1195. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Liu S, Yao S, Yang H, Liu S and Wang Y:
Autophagy: Regulator of cell death. Cell Death Dis. 14:6482023.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Zhou X, Lin J, Wang F, Chen X, Zhang Y, Hu
Z and Jin X: Circular RNA-regulated autophagy is involved in cancer
progression. Front Cell Dev Biol. 10:9619832022. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Wang Y, Mo Y, Peng M, Zhang S, Gong Z, Yan
Q, Tang Y, He Y, Liao Q, Li X, et al: The influence of circular
RNAs on autophagy and disease progression. Autophagy. 18:240–253.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Song H, Zhao Z, Ma L, Zhao W, Hu Y and
Song Y: Novel exosomal circEGFR facilitates triple negative breast
cancer autophagy via promoting TFEB nuclear trafficking and
modulating miR-224-5p/ATG13/ULK1 feedback loop. Oncogene.
43:821–836. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Jiang Y, Zhang Y, Chu F, Xu L and Wu H:
Circ_0032821 acts as an oncogene in cell proliferation, metastasis
and autophagy in human gastric cancer cells in vitro and in vivo
through activating MEK1/ERK1/2 signaling pathway. Cancer Cell Int.
20:742020. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
He Z, Cai K, Zeng Z, Lei S, Cao W and Li
X: Autophagy-associated circRNA circATG7 facilitates autophagy and
promotes pancreatic cancer progression. Cell Death Dis. 13:2332022.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Li XX, Xiao L, Chung HK, Ma XX, Liu X,
Song JL, Jin CZ, Rao JN, Gorospe M and Wang JY: Interaction between
HuR and circPABPN1 modulates autophagy in the intestinal epithelium
by altering ATG16L1 translation. Mol Cell Biol. 40:e00492–19. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Palanisamy K, Tsai T, Yu T, Sun K, Yu S,
Lin F, Wang I and Li C: RNA-binding protein, human antigen R
regulates hypoxia-induced autophagy by targeting ATG7/ATG16L1
expressions and autophagosome formation. J Cell Physiol.
234:7448–7458. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Zhang C, Ding R, Sun Y, Huo ST, He A, Wen
C, Chen H, Du WW, Lai W and Wang H: Circular RNA in tumor
metastasis. Mol Ther Nucleic Acids. 23:1243–1257. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Wang M, Ding X, Fang X, Xu J, Chen Y, Qian
Y, Zhang J, Yu D, Zhang X, Ma X, et al: Circ6834 suppresses
non-small cell lung cancer progression by destabilizing ANHAK and
regulating miR-873-5p/TXNIP axis. Mol Cancer. 23:1282024.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Li J, Wang X, Shi L, Liu B, Sheng Z, Chang
S, Cai X and Shan G: A mammalian conserved circular RNA CircLARP1B
regulates hepatocellular carcinoma metastasis and lipid metabolism.
Adv Sci (Weinh). 11:e23059022024. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Liu YY, Zhang YY, Ran LY, Huang B, Ren JW,
Ma Q, Pan XJ, Yang FF, Liang C, Wang XL, et al: A novel protein
FNDC3B-267aa encoded by circ0003692 inhibits gastric cancer
metastasis via promoting proteasomal degradation of c-Myc. J Transl
Med. 22:5072024. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Zhao Y, Jia Y, Wang J, Chen X, Han J, Zhen
S, Yin S, Lv W, Yu F, Wang J, et al: circNOX4 activates an
inflammatory fibroblast niche to promote tumor growth and
metastasis in NSCLC via FAP/IL-6 axis. Mol Cancer. 23:472024.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Mao S, Wu D, Cheng X and Wu J:
Circ_0007432 promotes non-small cell lung cancer progression and
macrophage M2 polarization through SRSF1/KLF12 axis. iScience.
27:1098612024. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Wang D, Wang S, Jin M, Zuo Y, Wang J, Niu
Y, Zhou Q, Chen J, Tang X, Tang W, et al: Hypoxic exosomal
circPLEKHM1-mediated crosstalk between tumor cells and macrophages
drives lung cancer metastasis. Adv Sci (Weinh). 11:e23098572024.
View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Feng PF, Zhu LX, Sheng N, Li XS, Liu PG
and Chen XF: CircXRN2 accelerates colorectal cancer progression
through regulating miR-149-5p/MACC1 axis and EMT. Sci Rep.
14:24482024. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Sun J, Wu H, Luo J, Qiu Y, Li Y, Xu Y, Liu
L, Liu X and Zhang Q: CircTBC1D22A inhibits the progression of
colorectal cancer through autophagy regulated via miR-1825/ATG14
axis. iScience. 27:1091682024. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Long F, Tian B, Li L, Ma M, Chen Z, Tan G,
Yin N, Zhong C, Yu B, Guo Y, et al: CircPOFUT1 fosters colorectal
cancer metastasis and chemoresistance via decoying
miR-653-5p/E2F7/WDR66 axis and stabilizing BMI1. iScience.
27:1087292024. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Du W, Quan X, Wang C, Song Q, Mou J and
Pei D: Regulation of tumor metastasis and CD8+ T cells infiltration
by circRNF216/miR-576-5p/ZC3H12C axis in colorectal cancer. Cell
Mol Biol Lett. 29:192024. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Jiang X, Peng M, Liu Q, Peng Q, Oyang L,
Li S, Xu X, Shen M, Wang J, Li H, et al: Circular RNA
hsa_circ_0000467 promotes colorectal cancer progression by
promoting eIF4A3-mediated c-Myc translation. Mol Cancer.
23:1512024. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Fei D, Wang F, Wang Y, Chen J, Chen S, Fan
L, Yang L, Ren Q, Duangmano S, Du F, et al: Circular RNA ACVR2A
promotes the progression of hepatocellular carcinoma through
mir-511-5p targeting PI3K-Akt signaling pathway. Mol Cancer.
23:1592024. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Gong J, Han G, Chen Z, Zhang Y, Xu B, Xu
C, Gao W and Wu J: CircDCAF8 promotes the progression of
hepatocellular carcinoma through miR-217/NAP1L1 Axis, and induces
angiogenesis and regorafenib resistance via exosome-mediated
transfer. J Transl Med. 22:5172024. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Yu S, Su S, Wang P, Li J, Chen C, Xin H,
Gong Y, Wang H, Ye X, Mao L, et al: Tumor-associated
macrophage-induced circMRCKα encodes a peptide to promote
glycolysis and progression in hepatocellular carcinoma. Cancer
Lett. 591:2168722024. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Wang Y, Zou R, Li D, Gao X and Lu X:
Exosomal circSTRBP from cancer cells facilitates gastric cancer
progression via regulating miR-1294/miR-593-3p/E2F2 axis. J Cell
Mol Med. 28:e182172024. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Deng C, Huo M, Chu H, Zhuang X, Deng G, Li
W, Wei H, Zeng L, He Y, Liu H, et al: Exosome circATP8A1 induces
macrophage M2 polarization by regulating the miR-1-3p/STAT6 axis to
promote gastric cancer progression. Mol Cancer. 23:492024.
View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Wu S, Lu J, Zhu H, Wu F, Mo Y, Xie L, Song
C, Liu L, Xie X, Li Y, et al: A novel axis of
circKIF4A-miR-637-STAT3 promotes brain metastasis in
triple-negative breast cancer. Cancer Lett. 581:2165082024.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Zhang Y, Tan Y, Yuan J, Tang H, Zhang H,
Tang Y, Xie Y, Wu L, Xie J, Xiao X, et al: circLIFR-007 reduces
liver metastasis via promoting hnRNPA1 nuclear export and YAP
phosphorylation in breast cancer. Cancer Lett. 592:2169072024.
View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Zhuang M, Zhang X, Ji J, Zhang H, Shen L,
Zhu Y and Liu X: Exosomal circ-0100519 promotes breast cancer
progression via inducing M2 macrophage polarisation by USP7/NRF2
axis. Clin Transl Med. 14:e17632024. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Gao X, Sun Z, Liu X, Luo J, Liang X, Wang
H, Zhou J, Yang C, Wang T and Li J: 127aa encoded by circSpdyA
promotes FA synthesis and NK cell repression in breast cancers.
Cell Death Differ. 32:416–433. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Zheng Z, Zeng X, Zhu Y, Leng M, Zhang Z,
Wang Q, Liu X, Zeng S, Xiao Y, Hu C, et al: CircPPAP2B controls
metastasis of clear cell renal cell carcinoma via HNRNPC-dependent
alternative splicing and targeting the miR-182-5p/CYP1B1 axis. Mol
Cancer. 23:42024. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Huang B, Ren J, Ma Q, Yang F, Pan X, Zhang
Y, Liu Y, Wang C, Zhang D, Wei L, et al: A novel peptide
PDHK1-241aa encoded by circPDHK1 promotes ccRCC progression via
interacting with PPP1CA to inhibit AKT dephosphorylation and
activate the AKT-mTOR signaling pathway. Mol Cancer. 23:342024.
View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Zhang C, Hu J, Liu Z, Deng H, Xiao J, Yi
Z, He Y, Xiao Z, Huang J, Liang H, et al: Hsa_circ_0000520
suppresses vasculogenic mimicry formation and metastasis in bladder
cancer through Lin28a/PTEN/PI3K signaling. Cell Mol Biol Lett.
29:1182024. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Wang Q, Cheng B, Singh S, Tao Y, Xie Z,
Qin F, Shi X, Xu J, Hu C, Tan W, et al: A protein-encoding CCDC7
circular RNA inhibits the progression of prostate cancer by
up-regulating FLRT3. NPJ Precis Oncol. 8:112024. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Li J, Qiu H, Dong Q, Yu H, Piao C, Li Z,
Sun Y and Cui X: Androgen-targeted hsa_circ_0085121 encodes a novel
protein and improves the development of prostate cancer through
facilitating the activity of PI3K/Akt/mTOR pathway and enhancing
AR-V7 alternative splicing. Cell Death Dis. 15:8482024. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Lei K, Liang R, Liang J, Lu N, Huang J, Xu
K, Tan B, Wang K, Liang Y, Wang W, et al: CircPDE5A-encoded novel
regulator of the PI3K/AKT pathway inhibits esophageal squamous cell
carcinoma progression by promoting USP14-mediated de-ubiquitination
of PIK3IP1. J Exp Clin Cancer Res. 43:1242024. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Shen J, Ma Z, Yang J, Qu T, Xia Y, Xu Y,
Zhou M and Liu W: CircPHGDH downregulation decreases papillary
thyroid cancer progression through miR-122-5p/PKM2 axis. BMC
Cancer. 24:5112024. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Li J, Cao H, Yang J and Wang B:
IGF2BP2-m6A-circMMP9 axis recruits ETS1 to promote TRIM59
transcription in laryngeal squamous cell carcinoma. Sci Rep.
14:30142024. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Zhang D, Luo Y, Lin Y, Fang Z, Zheng H, An
M, Xie Q, Wu Z, Yu C, Yang J, et al: Endosomal trafficking bypassed
by the RAB5B-CD109 interplay promotes axonogenesis in KRAS-mutant
pancreatic cancer. Adv Sci. 11:24050922024. View Article : Google Scholar
|
|
161
|
Henry NL and Hayes DF: Cancer biomarkers.
Mol Oncol. 6:140–146. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu
H, Xu X, Liang Q, Christiani DC, Wang M, et al: Circular RNAs in
body fluids as cancer biomarkers: the new frontier of liquid
biopsies. Mol Cancer. 20:132021. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Yin WB, Yan MG, Fang X, Guo JJ, Xiong W
and Zhang RP: Circulating circular RNA hsa_circ_0001785 acts as a
diagnostic biomarker for breast cancer detection. Clin Chim Acta.
487:363–368. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Yang H, Li X, Meng Q, Sun H, Wu S, Hu W,
Liu G, Li X, Yang Y and Chen R: CircPTK2 (hsa_circ_0005273) as a
novel therapeutic target for metastatic colorectal cancer. Mol
Cancer. 19:132020. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Xu H, Wang C, Song H, Xu Y and Ji G:
RNA-Seq profiling of circular RNAs in human colorectal cancer liver
metastasis and the potential biomarkers. Mol Cancer. 18:82019.
View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Liu J, Ren L, Li S, Li W, Zheng X, Yang Y,
Fu W, Yi J, Wang J and Du G: The biology, function, and
applications of exosomes in cancer. Acta Pharm Sin B. 11:2783–2797.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Chen X, Chen RX, Wei WS, Li YH, Feng ZH,
Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ, et al: PRMT5 Circular RNA
promotes metastasis of urothelial carcinoma of the bladder through
sponging miR-30c to induce Epithelial-mesenchymal transition. Clin
Cancer Res. 24:6319–6330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Li J, Li Z, Jiang P, Peng M, Zhang X, Chen
K, Liu H, Bi H, Liu X and Li X: Circular RNA IARS (circ-IARS)
secreted by pancreatic cancer cells and located within exosomes
regulates endothelial monolayer permeability to promote tumor
metastasis. J Exp Clin Cancer Res. 37:1772018. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Lin H, Conn VM and Conn SJ: Past, present,
and future strategies for detecting and quantifying circular RNA
variants. FEBS J. Feb 11–2025.doi: 10.1111/febs.70012 (Epub ahead
of print). View Article : Google Scholar
|
|
170
|
Arnberg AC, Van Ommen G-JB, Grivell LA,
Van Bruggen EFJ and Borst P: Some yeast mitochondrial RNAs are
circular. Cell. 19:313–319. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
van der Veen R, Arnberg AC, van der Horst
G, Bonen L, Tabak HF and Grivell LA: Excised group II introns in
yeast mitochondria are lariats and can be formed by self-splicing
in vitro. Cell. 44:225–234. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Szabo L and Salzman J: Detecting circular
RNAs: Bioinformatic and experimental challenges. Nat Rev Genet.
17:679–692. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Mi Z, Zhongqiang C, Caiyun J, Yanan L,
Jianhua W and Liang L: Circular RNA detection methods: A
minireview. Talanta. 238:1230662022. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Burnett WV: Northern blotting of RNA
denatured in glyoxal without buffer recirculation. Biotechniques.
22:668–671. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Schneider T, Schreiner S, Preußer C,
Bindereif A and Rossbach O: Northern Blot Analysis of Circular
RNAs. Circular RNAs. vol. 1724. Dieterich C and Papantonis A:
Springer; New York, New York, NY: pp. 119–133. 2018, View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Panda A and Gorospe M: Detection and
analysis of circular RNAs by RT-PCR. Bio Protoc. 8:e27752018.
View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Vromman M, Yigit N, Verniers K, Lefever S,
Vandesompele J and Volders P: Validation of circular RNAs Using
RT-qPCR after effective removal of linear RNAs by Ribonuclease R.
Curr Protoc. 1:e1812021. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Dahl M, Daugaard I, Andersen MS, Hansen
TB, Grønbæk K, Kjems J and Kristensen LS: Enzyme-free digital
counting of endogenous circular RNA molecules in B-cell
malignancies. Lab Invest. 98:1657–1669. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Li H, Bai R, Zhao Z, Tao L, Ma M, Ji Z,
Jian M, Ding Z, Dai X, Bao F and Liu A: Application of droplet
digital PCR to detect the pathogens of infectious diseases. Biosci
Rep. 38:BSR201811702018. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W,
Yu R, Xiao B and Guo J: Plasma circular RNA profiling of patients
with gastric cancer and their droplet digital RT-PCR detection. J
Mol Med. 96:85–96. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Chen DF, Zhang LJ, Tan K and Jing Q:
Application of droplet digital PCR in quantitative detection of the
cell-free circulating circRNAs. Biotechnol Biotechnol Equip.
32:116–123. 2018. View Article : Google Scholar
|
|
185
|
Maheshwari Y, Selvaraj V, Hajeri S and
Yokomi R: Application of droplet digital PCR for quantitative
detection of Spiroplasma citri in comparison with real time PCR.
PLoS One. 12:e01847512017. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Hansen TB, Venø MT, Damgaard CK and Kjems
J: Comparison of circular RNA prediction tools. Nucleic Acids Res.
44:e58. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Liang M, Yao W, Shi B, Zhu X, Cai R, Yu Z,
Guo W, Wang H, Dong Z, Lin M, et al: Circular RNA hsa_circ_0110389
promotes gastric cancer progression through upregulating SORT1 via
sponging miR-127-5p and miR-136-5p. Cell Death Dis. 12:6392021.
View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Zirkel A and Papantonis A: Detecting
circular RNAs by RNA Fluorescence in situ Hybridization. Circular
RNAs. vol. 1724. Dieterich C and Papantonis A: Springer; New York,
New York, NY: pp. 69–75. 2018, View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Zhang P, Guo N, Gao K, Su F, Wang F and Li
Z: Direct recognition and sensitive detection of circular RNA with
ligation-based PCR. Org Biomol Chem. 18:3269–3273. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Liu Y, Zhang X, Liu M, Xu F, Zhang Q,
Zhang Y, Weng X, Liu S, Du Y and Zhou X: Direct detection of
circRNA in real samples using reverse transcription-rolling circle
amplification. Anal Chim Acta. 1101:169–175. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Jiao J, Xiang Y, Duan C, Liu Y, Li C and
Li G: Lighting Up CircRNA using a linear DNA NAnostructure. Anal
Chem. 92:12394–12399. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Dong J, Zeng Z, Sun R, Zhang X, Cheng Z,
Chen C and Zhu Q: Specific and sensitive detection of CircRNA based
on netlike hybridization chain reaction. Biosens Bioelectron.
192:1135082021. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Jiao J, Duan C, Zheng J, Li D, Li C, Wang
Z, Gao T and Xiang Y: Development of a two-in-one integrated assay
for the analysis of circRNA-microRNA interactions. Biosens
Bioelectron. 178:1130322021. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Zhang P, Gao K, Liang Y, Su F, Wang F and
Li Z: Ultrasensitive detection of circular RNA by accurate
recognition of the specific junction site using stem-loop primer
induced double exponential amplification. Talanta. 217:1210212020.
View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Zhou Z, Han B, Wang Y, Lin N, Zhou Z,
Zhang Y, Bai Y, Shen L, Shen Y, Zhang Y and Yao H: Fast and
sensitive multivalent spatial pattern-recognition for circular RNA
detection. Nat Commun. 15:109002024. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Wu P, Nie Z, Huang Z and Zhang X:
CircPCBL: Identification of plant CircRNAs with a CNN-BiGRU-GLT
model. Plants (Basel). 12:16522023.PubMed/NCBI
|
|
197
|
Song P, Zhang P, Qin K, Su F, Gao K, Liu X
and Li Z: CRISPR/Cas13a induced exponential amplification for
highly sensitive and specific detection of circular RNA. Talanta.
246:1235212022. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Wang X, Wu Y, Wen X, Zhu J, Bai X, Qi Y
and Yang H: Surface plasmons and SERS application of Au nanodisk
array and Au thin film composite structure. Opt Quantum Electron.
52:2382020. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Zhai WL, Li DW, Qu LL, Fossey JS and Long
YT: Multiple depositions of Ag nanoparticles on chemically modified
agarose films for surface-enhanced Raman spectroscopy. Nanoscale.
4:137–142. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Li N, Shen F, Cai Z, Pan W, Yin Y, Deng X,
Zhang X, Machuki JO, Yu Y, Yang D, et al: Target-induced
Core-satellite nanostructure assembly strategy for Dual-Signal-On
fluorescence imaging and raman quantification of intracellular
MicroRNA guided photothermal therapy. Small. 16:e20055112020.
View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Zhu R, Feng H, Li Q, Su L, Fu Q, Li J,
Song J and Yang H: Asymmetric Core-Shell gold nanoparticles and
controllable assemblies for SERS ratiometric detection of MicroRNA.
Angew Chem Int Ed. 60:12560–12568. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Chen R, Shi H, Meng X, Su Y, Wang H and He
Y: Dual-amplification Strategy-based SERS chip for sensitive and
reproducible detection of DNA methyltransferase activity in human
serum. Anal Chem. 91:3597–3603. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Guerrini L, Garcia-Rico E, O'Loghlen A,
Giannini V and Alvarez-Puebla RA: Surface-enhanced raman scattering
(SERS) Spectroscopy for sensing and characterization of exosomes in
cancer diagnosis. Cancers (Basel). 13:21792021. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Muskan M, Abeysinghe P, Cecchin R,
Branscome H, Morris KV and Kashanchi F: Therapeutic potential of
RNA-enriched extracellular vesicles: The next generation in RNA
delivery via biogenic nanoparticles. Mol Ther J Am Soc Gene Ther.
32:2939–2949. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Xu S, Xu Y, Solek NC, Chen J, Gong F,
Varley AJ, Golubovic A, Pan A, Dong S, Zheng G and Li B:
Tumor-tailored ionizable lipid nanoparticles Facilitate IL-12
circular RNA delivery for enhanced lung cancer immunotherapy. Adv
Mater. 36:e24003072024. View Article : Google Scholar : PubMed/NCBI
|