Introduction
Lung cancer remains a leading cause of global
mortality (1), with tobacco use,
occupational exposure and genetic factors being key contributors to
its high incidence (2). Lung
adenocarcinoma (LUAD) and lung squamous cell carcinoma are the two
main subtypes of non-small-cell lung cancer (NSCLC) (3). Notably, with the advent of precision
medicine, treatment strategies for LUAD have undergone
transformative changes, incorporating targeted therapy and
immunotherapy (4). However,
genetic mutations, acquired resistance and epigenetic modifications
inevitably lead to drug resistance in patients (5). Therefore, identifying novel
therapeutic markers to more accurately characterize LUAD is crucial
for overcoming treatment resistance.
Cancer cells, which exhibit uncontrolled
proliferation, require elevated cholesterol levels for membrane
synthesis and other vital functions (6), making cholesterol metabolism
reprogramming a common feature in cancer. Dysregulation of
proto-oncogenes and oncogenes results in notable increases in
cholesterol uptake and the production of its derivatives within
tumor cells (7,8). Additionally, the tumor
microenvironment (TME), characterized by factors such as acidity
and inflammation, further promotes cholesterol biosynthesis and
uptake (9,10). 22-Hydroxycholesterol, a metabolite
of cholesterol abundant in the TME (11), recruits neutrophils by binding to
CXCR2, thereby exerting immunosuppressive effects (12). Tumor-associated macrophages also
undergo functional and phenotypic alterations due to shifts in
cholesterol metabolism, contributing to tumor progression (13). In summary, cholesterol and its
metabolites drive tumor progression by enhancing tumor malignancy
and remodeling the TME. Therefore, statin-based inhibition of
active cholesterol metabolism has emerged as a promising antitumor
strategy (14).
The aim of the present study was to identify
potential biomarkers associated with cholesterol metabolism in the
diagnosis of LUAD. The study aimed to screen differentially
expressed genes related to cholesterol metabolism between LUAD and
normal lung tissues, analyze their gene functions, and prognostic
and clinical relevance, and perform cell experiments to validate
the functions of the selected genes. The current study established
a robust diagnostic and prognostic model for patients with
LUAD.
Materials and methods
Data collection and processing
RNA-sequencing data from patients with LUAD were
obtained from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/), Subsequently, R
project (v4.0.4; http://www.r-project.org/) was used for rigorous
quality control and screening to remove duplicate samples, ensuring
dataset uniqueness and reliability. Samples with a survival time of
<30 days were excluded to ensure complete and usable survival
data. A training set consisting of 485 LUAD samples and 59
paracancerous tissues was selected from TCGA, and a validation set
(GSE72094) (15) was obtained from
the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/gds), comprising 398
samples with complete clinical data. cholesterol metabolism-related
genes (CMRGs) were downloaded from MSigDB (https://www.gsea-msigdb.org/gsea/msigdb) (16), totaling 158 genes.
Consensus cluster analysis
Univariate Cox regression analysis was performed to
assess the relationship between CMRGs and prognosis. TCGA cohort
was clustered using the consensus clustering algorithm implemented
through the ‘ConsensusClusterPlus’ R package (v3.0; http://bioconductor.org/packages/3.0/bioc/html/ConsensusClusterPlus.html)
(17). Clustering was conducted
with the PAM algorithm based on 1-Pearson correlation distance,
with 80% of the samples repeated 1,000 times. The optimal number of
clusters was determined through analysis of the empirical
cumulative distribution function (CDF) plot. A Sankey diagram was
generated using the ‘ggplot2’ R package (v 3.3.0; http://cran.r-project.org/web/packages/ggplot2/index.html).
Gene set enrichment analysis
(GSEA)
GSEA was performed using ‘c5.go.v7.5.1.symbols’
(Gene Ontology terms) and ‘c2.cp.kegg.v7.5.1.symbols’ (Kyoto
Encyclopedia of Genes and Genomes) from the molecular signature
database (https://www.gsea-msigdb.org/gsea/index.jsp) to explore
biological signatures and pathways across different clusters
(18).
TME analysis
Using the single sample GSEA algorithm, the activity
levels of 28 immune cell types were evaluated for each sample (in
the GSE72094 dataset) (19). The
ESTIMATE algorithm, applied to pre-screened immune-related gene
expression data, was used to predict cell infiltration and tumor
purity in LUAD tissues (TCGA) (20).
Construction of cholesterol metabolism
gene signature
Differentially expressed genes between clusters were
identified using ‘limma’ R package (v3.42.2; http://bioconductor.org/packages/3.10/bioc/html/limma.html)
analysis of variance (21), and
genes with a P-value of <0.05 and fold change of ≥2 were further
analyzed using univariate or LASSO Cox regression. The risk index
for each patient was calculated by multiplying the expression
levels of target genes by their corresponding coefficients. Based
on the median risk index, patients with LUAD were categorized into
high-risk and low-risk groups for subsequent analyses. Kaplan-Meier
curves and log-rank tests were used to assess survival differences,
with the same analysis applied to validate the prognostic model in
the GSE72094 dataset.
Nomogram development and
validation
Univariate and multivariate Cox regression analyses
were conducted using R project to assess the independence of the
risk index from traditional clinical features. A nomogram was
created to visually represent the significant risk index
(P<0.05) alongside other risk factors. Calibration curves and
time-dependent receiver operating characteristic curves were
generated to evaluate the prognostic predictive power of the
nomogram at various time points.
Analysis of tumor mutation burden
(TMB)
Somatic mutation data for LUAD cases were sourced
from TCGA database, with TMB calculated by quantifying somatic
non-synonymous mutations within defined genomic regions. The
‘maftools_2.24.0’ R package (https://bioconductor.org/packages/release/bioc/vignettes/maftools/inst/doc/maftools.html)
was used to analyze the correlation between risk scores and TMB
using the Pearson correlation coefficient.
Prediction of immunotherapy and
chemotherapy response
The response of solid tumors to treatment with
immune checkpoint inhibitors (ICIs) was predicted using the Tumor
Immune Dysfunction and Exclusion (TIDE) (http://tide.dfci.harvard.edu) algorithm, which
evaluates immune cell infiltration and dysfunction levels (22). The TIDE score for each patient with
LUAD (from TCGA dataset) was calculated based on their expression
profile, with higher TIDE scores indicating an increased potential
for ICI resistance. Additionally, the ‘pRRophetic_0.5’ R package
(https://github.com/paulgeeleher/pRRophetic2) was
employed to estimate the half maximal inhibitory concentration
values for various chemotherapy agents in patients with LUAD (from
TCGA dataset).
Cell culture
Two human LUAD cell lines, A549 and NCI-H1975, were
purchased from Wuhan Pricella Biotechnology Co., Ltd. A549 cells
were cultured in F12K medium (Gibco; Thermo Fisher Scientific,
Inc.), while NCI-H1975 cells were cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.). All of the media were
supplemented with 10% fetal bovine serum (NEWZERUM Ltd.) and 1%
penicillin/streptomycin in a 5% CO2 incubator at 37°C.
Small interfering (si)RNAs were synthesized by Guangzhou RiboBio
Co., Ltd. with the following sequences and targets (5′-3′):
Non-targeting siRNA-negative control, antisense strand:
ACGUGACACGUUCGGAGAA, sense strand: CUCCGAACGUGUCUCGUAA;
GJB3-siRNA1, antisense strand: AGACCUUGUACCCACAGCGGG, sense strand:
CGCUGUGGGUACAAGGUCUGC (target: CCCGCUGUGGGUACAAGGUCUGC);
GJB3-siRNA2, antisense strand: AGGACUCGCCCACGCUAGUUU, sense strand:
ACUAGCGUGGGCGAGUCCUGA, (target: AAACUAGCGUGGGCGAGUCCUGA);
GJB3-siRNA3, antisense strand: UGGCGCGGGACCUUGACCGUG, sense strand:
CGGUCAAGGUCCCGCGCCAAG (target: CACGGUCAAGGUCCCGCGCCAAG).
Transfection was performed using Lipofectamine® RNAiMAX
(Invitrogen; Thermo Fisher Scientific, Inc.), and A549 and
NCI-H1975 cells were used for transfection. Briefly, when the cell
confluence was 30–50%, Opti-MEM (Gibco; Thermo Fisher Scientific,
Inc.), 10 nm siRNA and Lipofectamine RNAiMAX reagent were
successively added to a sterile microcentrifuge tube, gently mixed
and incubated for 20 min at room temperature (25°C). After 20 min,
the transfection complexes were successively added to the cell
culture dishes, gently mixed and placed in the cell incubator for 6
h at 37°C. Subsequent experiments were performed 48 h after
transfection.
Western blotting
After transfection, the cells were lysed with RIPA
buffer (Merck KGaA) containing protease inhibitors (Merck KGaA).
Protein concentrations were determined using the BCA kit (Nanjing
KeyGen Biotech Co., Ltd.), and proteins (30 µg/lane) were separated
by SDS-PAGE on 10% gels. Following transfer to PVDF membranes
(Merck KGaA), protein blocking was carried out with 5% skim milk at
room temperature for 1 h. The membranes were then incubated with
primary antibodies overnight at 4°C, followed by secondary antibody
incubation at room temperature for 1 h. Blots were detected using
Ultra-sensitive ECL chemiluminescent substrate (cat. no. BL523B;
Biosharp Life Sciences) and images were captured with a Tanon 5200
system (Tanon Science and Technology Co., Ltd.). The following
antibodies were used for western blotting: GJB3 polyclonal antibody
(cat. no. 12880-1-AP; Proteintech Group, Inc.), E-cadherin
polyclonal antibody (cat. no. 20874-1-AP; Proteintech Group, Inc.),
N-cadherin polyclonal antibody (cat. no. 22018-1-AP; Proteintech
Group, Inc.), vimentin polyclonal antibody (cat. no. 10366-1-AP;
Proteintech Group, Inc.), Snail1 polyclonal antibody (cat. no.
13099-1-AP; Proteintech Group, Inc.), β-actin recombinant antibody
(cat. no. 81115-1-RR; Proteintech Group, Inc.) and Goat Anti-Rabbit
IgG H&L antibody (cat. no. BF03008; Biodragon).
Cell viability and proliferation
assays
Cell viability was evaluated using the Cell Counting
Kit (CCK)-8 (Dojindo Laboratories, Inc.). Cells were seeded in
96-well plates at 1,000 cells/well and cultured in a 5%
CO2 incubator at 37°C. After 6 h, the cells adhered to
the surface of the plates, and measurements at this time were
considered 0-h data. Subsequently, CCK-8 values were measured every
24 h, and the data at 0, 24, 48, 72, 96 and 120 h time points were
analyzed and plotted. Cells were seeded in 96-well plates and
cultured in a 5% CO2 incubator at 37°C. Subsequently,
100 µl complete medium containing 10% CCK-8 reagent was added to
each well, and incubation was continued for 1 h. Absorbance at
OD450 was measured using the MD SpectraMax Plus 384
(Molecular Devices, Inc.).
On day 5 of culture, cell proliferation was assessed
using the Click-iT EdU-488 Cell Proliferation Detection Kit (Wuhan
Servicebio Technology Co., Ltd.), with 1 µg/ml Hoechst 33342 and 10
µM EdU used for detection. EdU staining experiments were performed
according to the manufacturer's protocol. Images were captured
using a Nikon ECLIPSE Ti-2 fluorescence microscope (Nikon
Corporation), with excitation/emission wavelengths of 346/460 nm
for Hoechst and 491/516 nm for EdU.
Colony formation assay
For colony formation assays, cells were seeded in
6-well plates at 1,000 cells/well and cultured in a 5%
CO2 incubator at 37°C, with the medium changed every 3
days. After 2 weeks, the cells were fixed in 4% paraformaldehyde at
room temperature for 60 min and stained with 0.1% crystal violet
(Beyotime Institute of Biotechnology) at room temperature for 5
min. The colonies were rinsed with tap water, dried and images were
captured using a digital camera. The data were statistically
analyzed according to the area of crystal violet staining.
Migration and invasion assays
Cell migration and invasion were assessed using
24-well Transwell Tissue Culture Plate Inserts (pore size, 8.0 µm)
(Guangzhou Jet Bio-Filtration Co., Ltd.). For the migration assay,
2,000 cells/well were seeded in the upper chamber of an uncoated
Transwell insert, whereas for the invasion assay, the upper chamber
was coated with VitroGel™ 3D hydrogel (cat. no. VHM01;
Shanghai XP Biomed Ltd.) at room temperature for 10–15 min. The
cells were seeded in the upper chamber in serum-free medium,
whereas the lower chamber contained complete medium; the cells were
cultured in a 5% CO2 incubator at 37°C. After 24 h of
incubation, the cells were fixed with 4% paraformaldehyde at room
temperature for 60 min and stained with 0.1% crystal violet at room
temperature for 5 min. After rinsing with tap water and drying,
images were captured using Nikon ECLIPSE Ti-2 light microscope
channel (Nikon Corporation).
Statistical analysis
Data analysis and image processing were performed
using R software (version 4.1.3) (https://www.r-project.org/) and GraphPad Prism 9
(Dotmatics). Differences between continuous variables were analyzed
using the Wilcoxon rank-sum test, while categorical variables were
assessed using the χ2 test. Survival analysis was
conducted using the log-rank test. Spearman's rank correlation was
applied for correlation analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
Features of cholesterol metabolism in
LUAD identified by consensus clustering
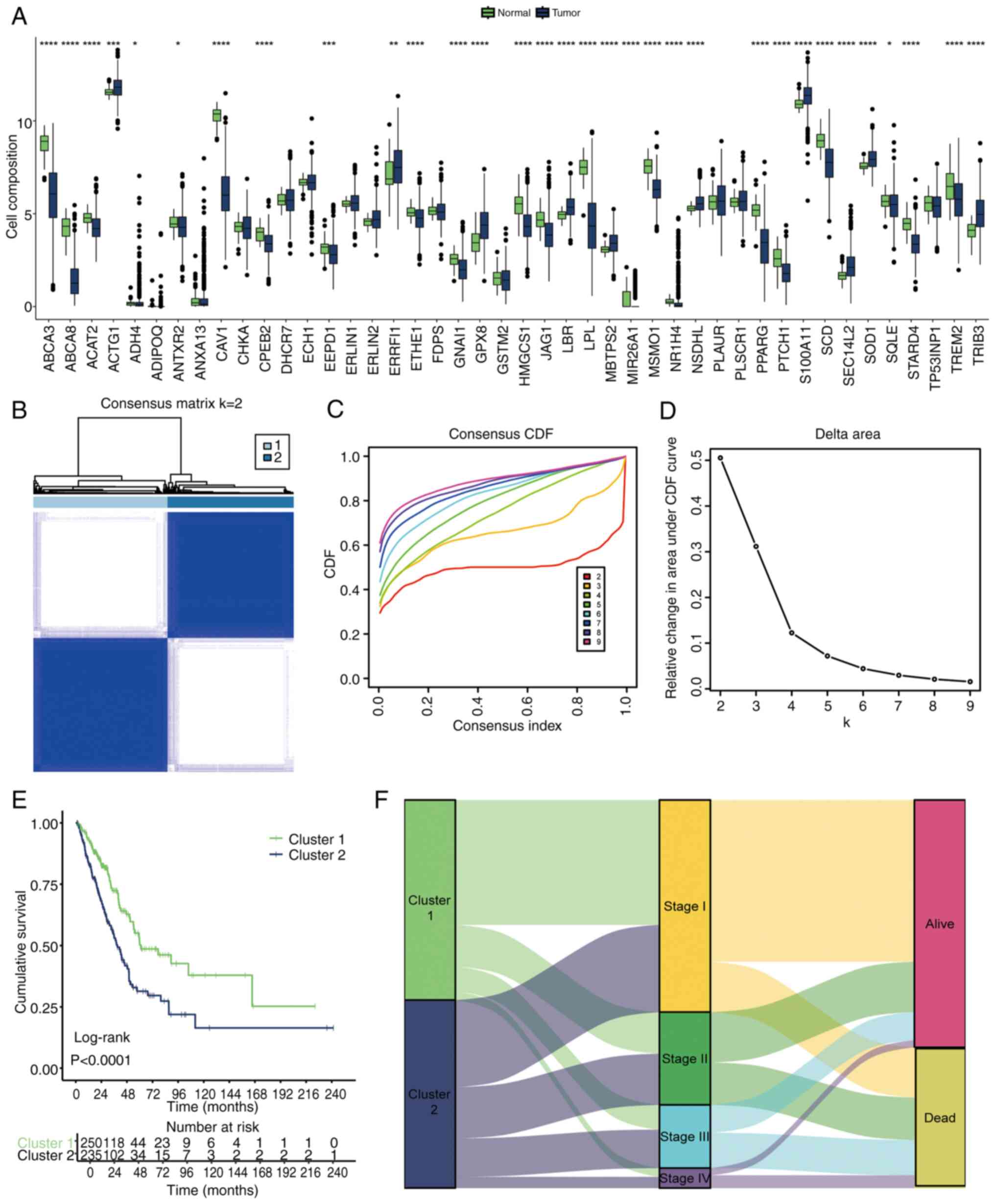
Univariate Cox regression analysis was performed on
158 CMRGs, revealing that 44 of these genes were significantly
associated with prognosis in LUAD (Table SI). Most of these 44 genes
exhibited marked differential expression between LUAD and normal
tissues (Fig. 1A). These 44 genes
were further used to classify LUAD into two distinct cholesterol
metabolism clusters based on their expression patterns (Fig. 1B). This clustering was validated by
observing relative changes in the CDF curves (Fig. 1C) and the area under the curve
(AUC) of CDF curves (Fig. 1D),
with the optimal division (k=2) indicating the ideal number of
clusters. When k=2, the delta area of CDF began to markedly
decrease, thus choosing k=2 gave the best clustering effect
(Fig. 1C and D). Survival analysis
showed a significant difference in prognosis between the two
clusters (Fig. 1E), with the
Sankey diagram highlighting that cluster 1 contained more patients
with lower Tumor-Node-Metastasis stages (23) and a better prognosis (Fig. 1F).
Potential functional pathways
analysis
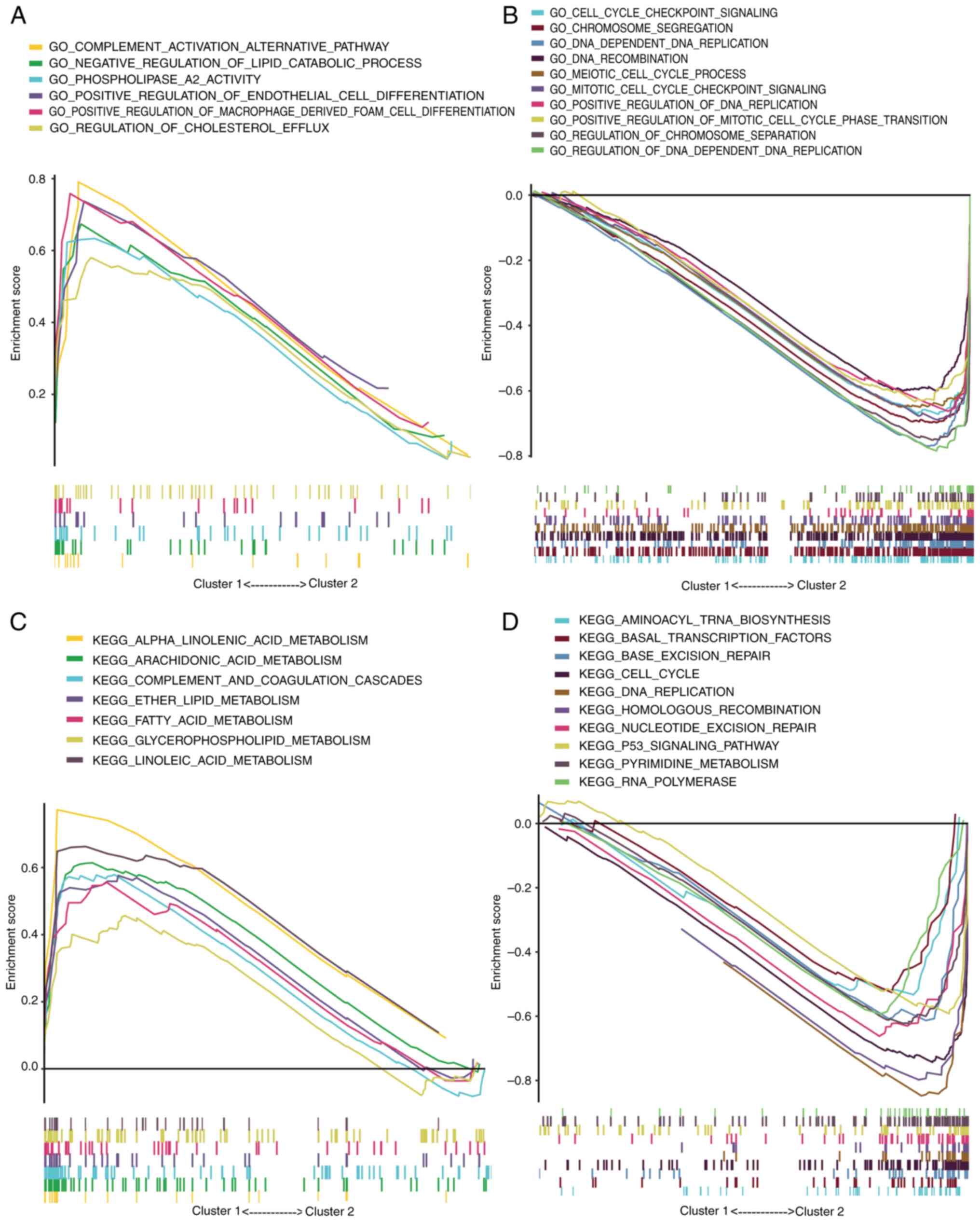
GSEA of Gene Ontology terms revealed that cluster 1
was primarily involved in ‘POSITIVE REGULATION OF ENDOTHELIAL CELL
DIFFERENTIATION’, ‘REGULATION OF CHOLESTEROL EFFLUX’ and ‘NEGATIVE
REGULATION OF LIPID CATABOLIC PROCESS’ (Fig. 2A). Given that enhanced cholesterol
metabolism in tumors is associated with poor prognosis (24), these processes in cluster 1, which
inhibit cholesterol metabolism and reduce the enrichment of
cholesterol metabolites, were linked to a more favorable prognosis.
Conversely, cluster 2 exhibited significant enrichment in processes
such as ‘CELL CYCLE CHECKPOINT SIGNALING’, ‘MEIOTIC CELL CYCLE
PROCESS’, ‘MITOTIC CELL CYCLE CHECKPOINT SIGNALING’, ‘DNA DEPENDENT
DNA REPLICATION’ and ‘DNA RECOMBINATION’, contributing to
accelerated tumor progression (Fig.
2B). These findings were corroborated by Kyoto Encyclopedia of
Genes and Genomes pathway analysis (Fig. 2C and D).
Immune infiltration analysis
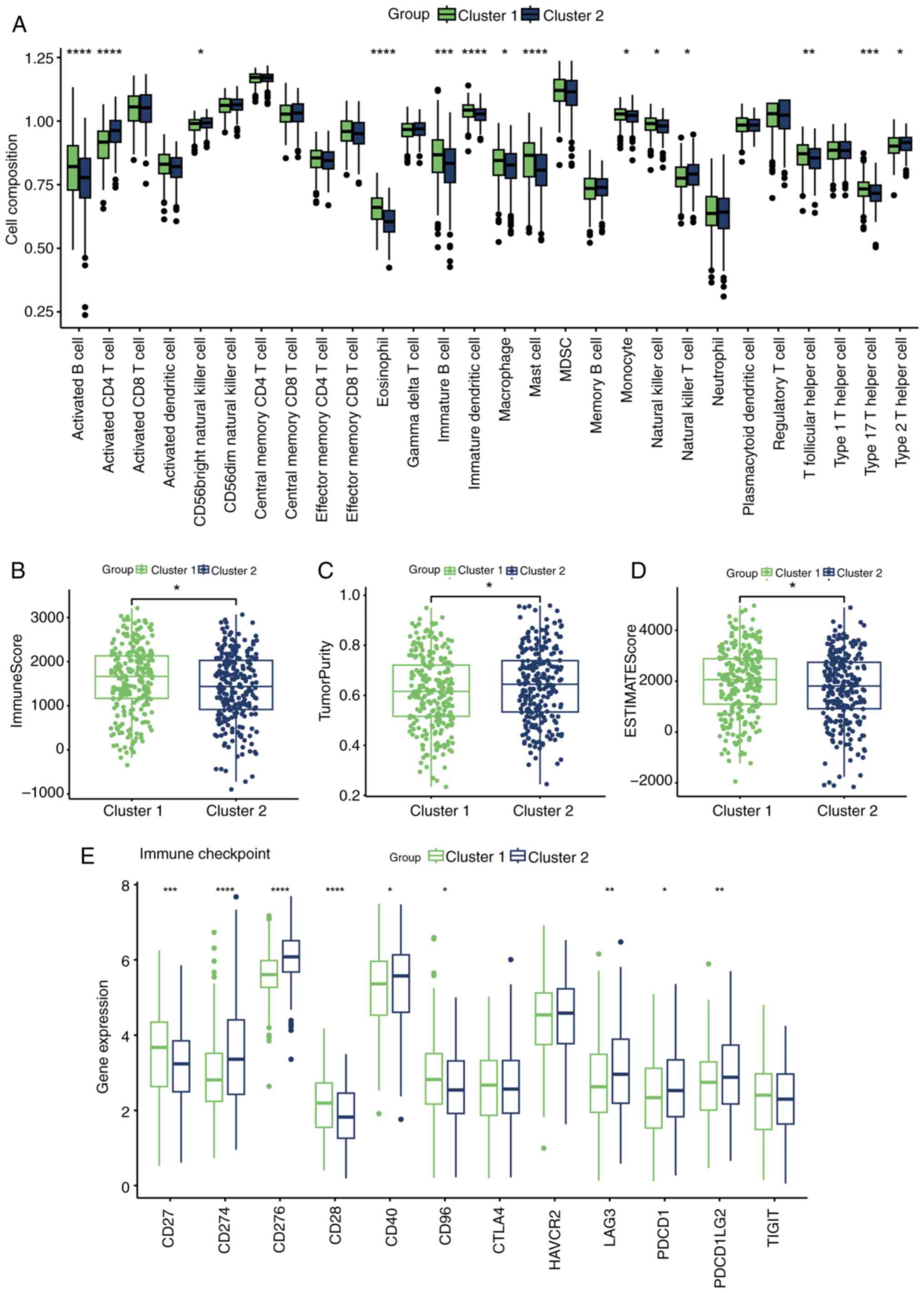
Cholesterol metabolism reprogramming has been shown
to be significantly associated with the formation of an
immunosuppressive microenvironment (6). To investigate this further, the
infiltration of 28 immune cell types was assessed across the
clusters. Cluster 1, with a better prognosis, exhibited more
abundant immune cell infiltration (Fig. 3A), with the majority of these cells
having antitumor effects, such as B cells, macrophages, mast cells,
natural killer cells, T follicular helper cells and type 17 T
helper cells (25). Additionally,
the ESTIMATE algorithm analysis confirmed the higher levels of
immune cell infiltration in cluster 1 (Fig. 3B-D). Furthermore, the expression of
immunosuppressive checkpoints is known to be associated with
immunotherapy responses (26), and
most immunosuppressive checkpoints commonly targeted in clinical
practice were more closely associated with cluster 2, such as
CD274, CD276, LAG3, PDCD1 and PDCD1LG2 (Fig. 3E).
Construction and validation of CMRG
signature
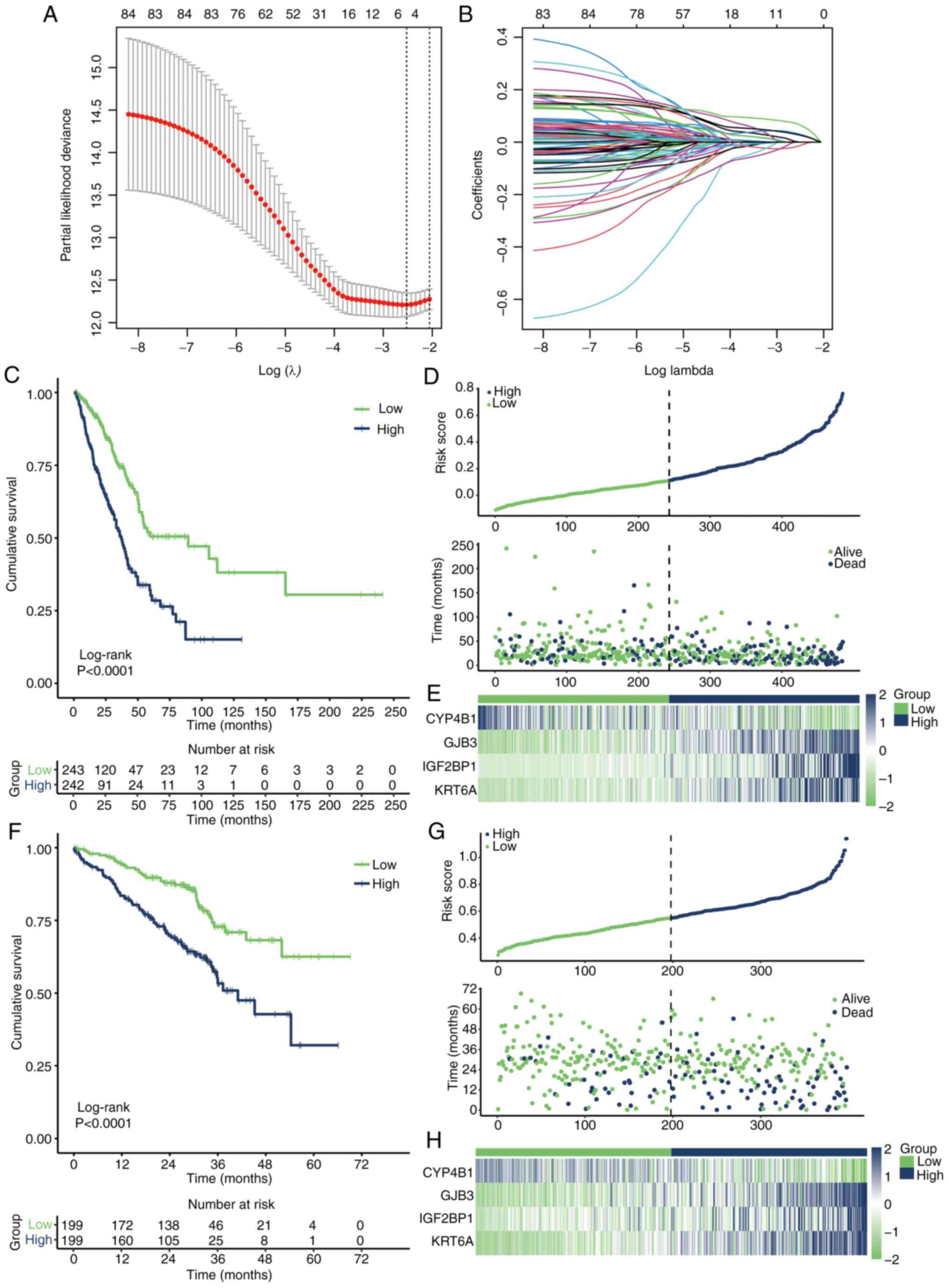
Limma differential analysis identified 124
differentially expressed genes between cluster 1 and cluster 2.
Univariate Cox regression identified 85 genes associated with
prognosis (Table SII). Using the
optimal λ value (Fig. 4A and B),
four key genes: CYP4B1, GJB3, IGF2BP1 and KRT6A, were identified as
significantly influencing the prognosis of the two clusters. The
risk score for each patient was calculated using the following
formula: Risk score=CYP4B1 × −0.01406345 + GJB3 × 0.03109533 +
IGF2BP1 × 0.06671392 + KRT6A × 0.02723521. This model effectively
distinguished patient prognoses in TCGA-LUAD cohort (Fig. 4C). The risk factor distribution
graph demonstrated a higher mortality rate in the high-risk group
(Fig. 4D), and the heatmap
illustrated the distribution of the four key genes across different
patients (Fig. 4E). Applying the
same risk score formula to the GSE72094 dataset yielded similar
results (Fig. 4F-H), further
validating the effectiveness of the prognostic model.
Construction and validation of the
nomogram
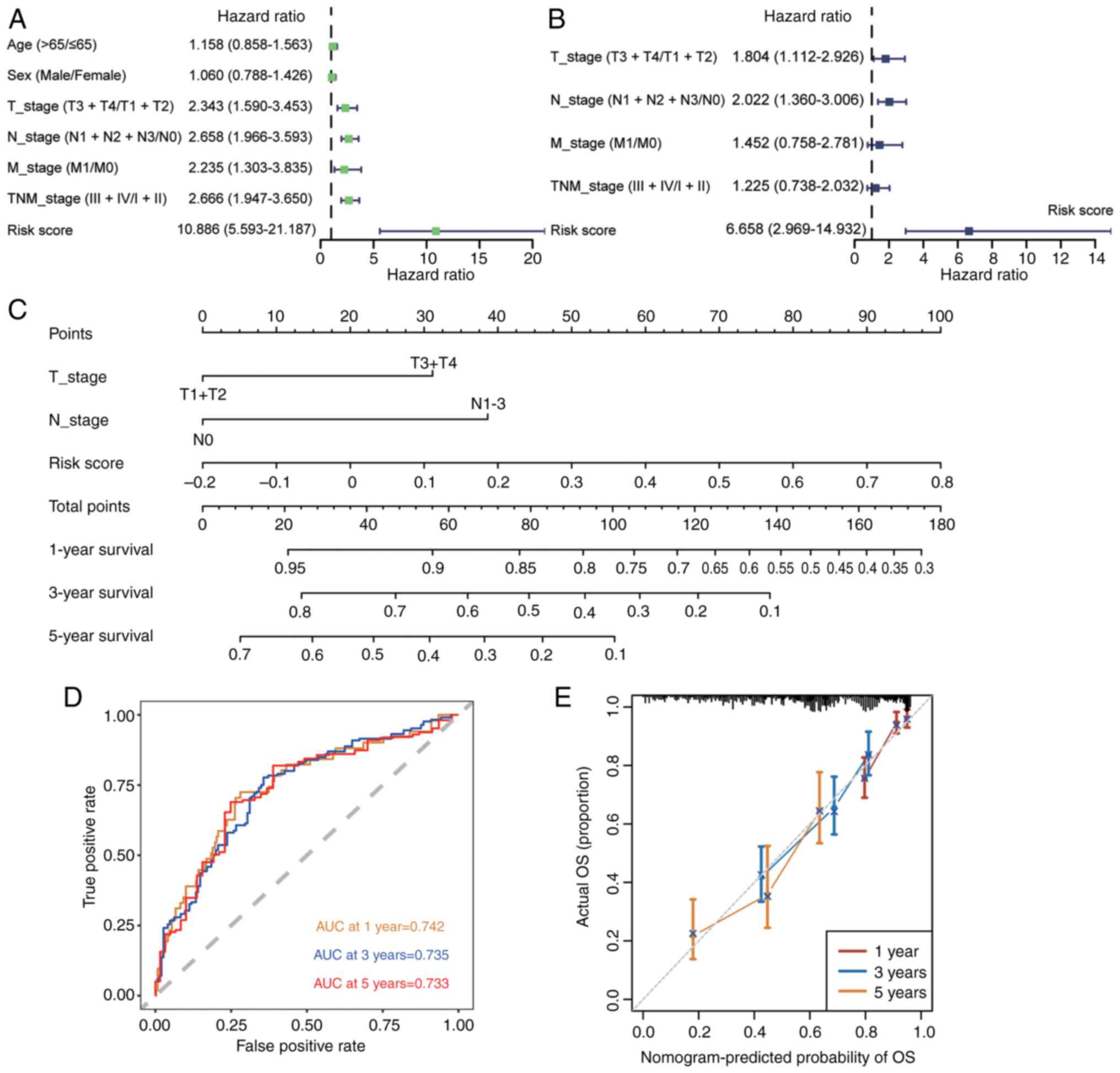
Univariate and multivariate Cox regression analyses
of the key features affecting prognosis found that CMRG risk
signature was an independent prognostic risk factor (Fig. 5A and B). Based on the CMRG risk
signature and clinical characteristics, a nomogram prediction model
was constructed (Fig. 5C). The AUC
values for 1-, 3- and 5-year predictions were 0.742, 0.735 and
0.733, respectively (Fig. 5D). In
addition, the prediction results of the nomogram closely aligned
with the actual outcomes (Fig.
5E).
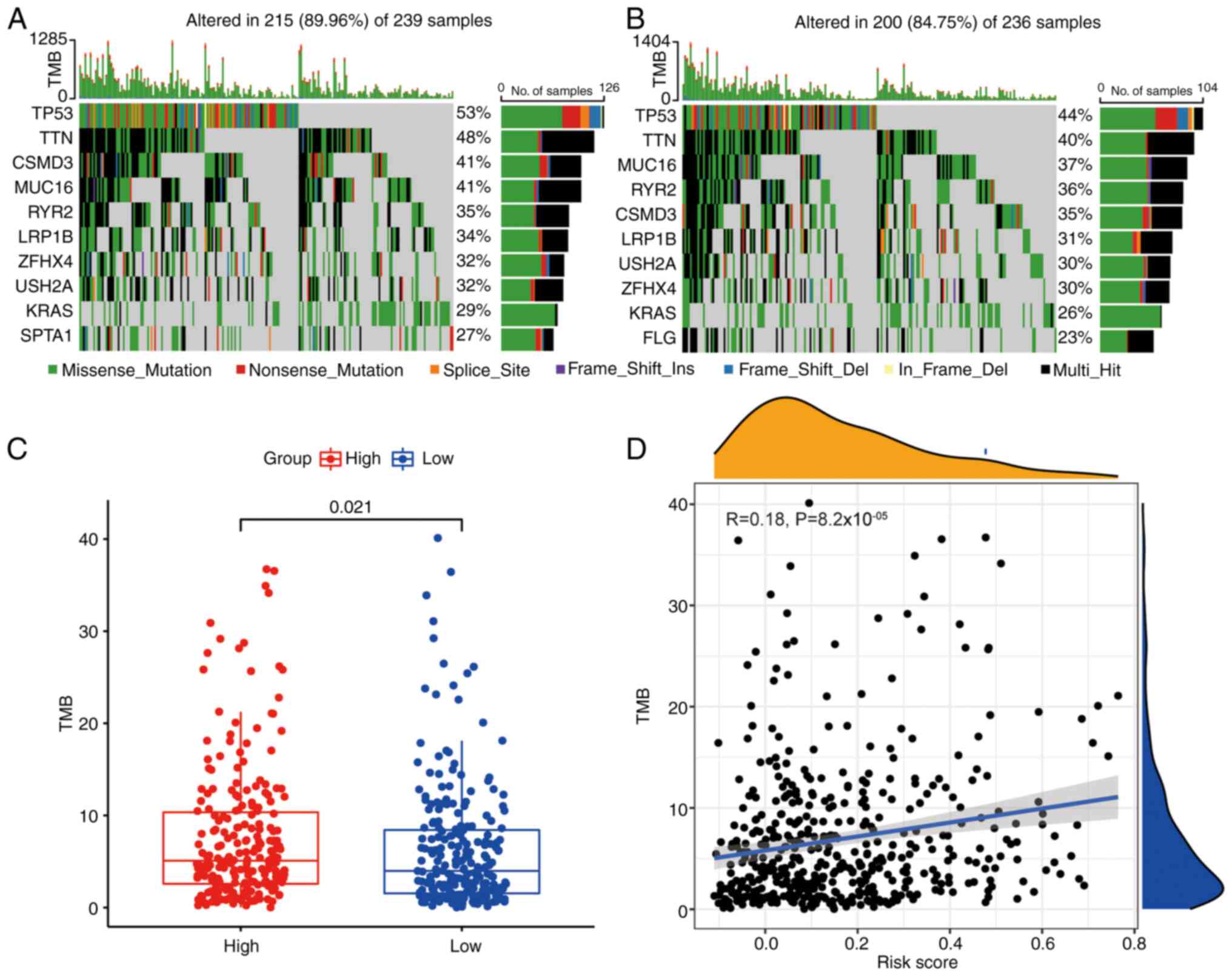
Analysis of TMB
Gene mutations are critical factors influencing
tumor prognosis (27). TMB
analysis of patients in different risk groups showed that most of
the top 10 mutated genes overlapped between the two groups
(Fig. 6A and B), but mutation
incidence was higher in the high-risk group (Fig. 6C). In addition, a low positive
association was confirmed between risk score and TMB (Fig. 6D).
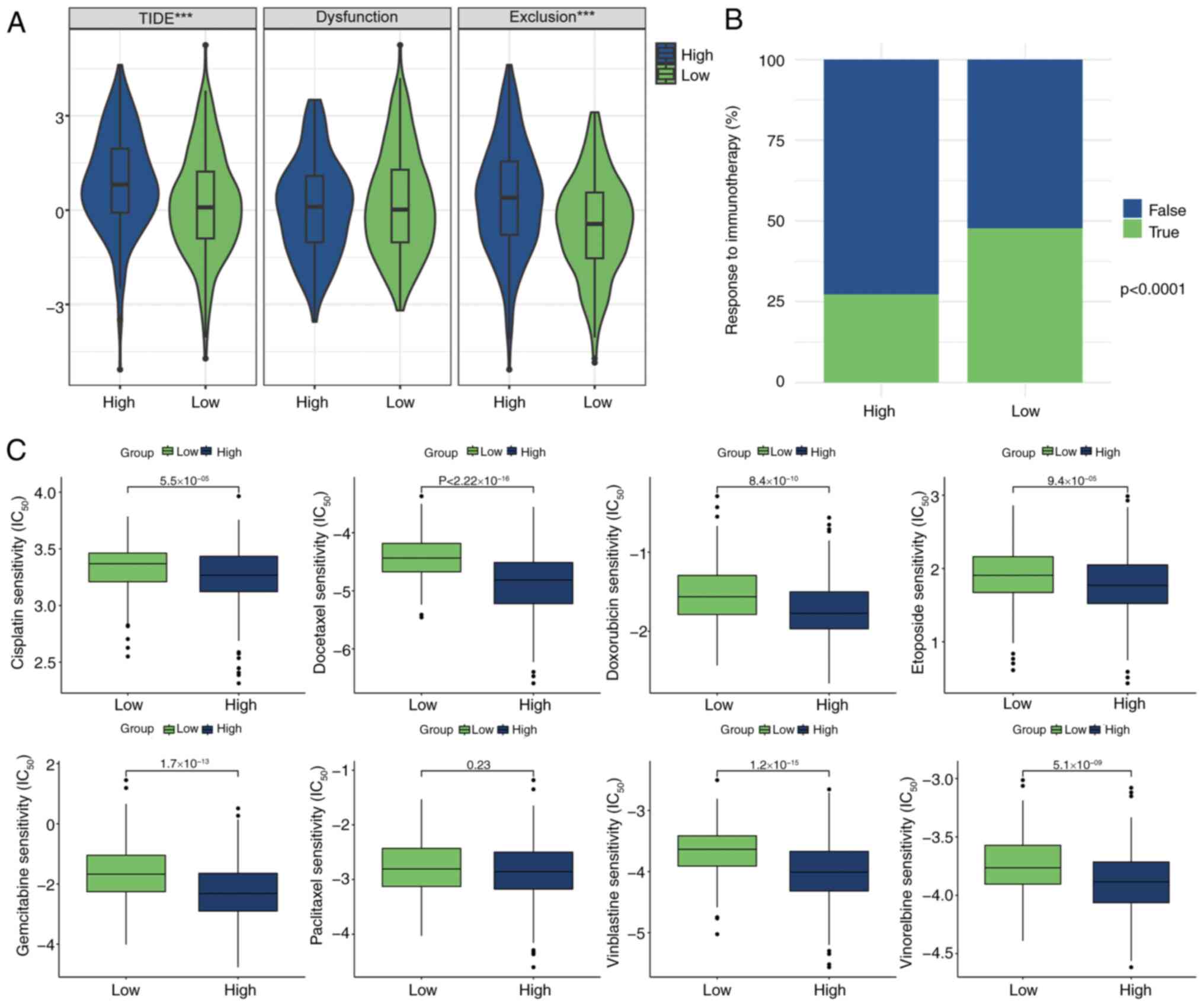
Prediction of immunotherapy and
chemotherapy response
Using the TIDE algorithm, the potential effects of
immunotherapy for patients with LUAD were successfully modeled and
predicted. Notably, high-risk patients exhibited higher exclusion
scores and TIDE values, while there was no significant difference
in dysfunction scores (Fig. 7A),
indicating a greater likelihood of drug resistance during
immunotherapy compared with low-risk patients. Further analysis
revealed that the percentage of low-risk patients responding
positively to immunotherapy was significantly higher than that of
high-risk patients (Fig. 7B),
providing valuable insights for clinical decision-making.
Additionally, the sensitivity of various chemotherapeutic agents
within the patient cohort was assessed. Multiple chemotherapy
drugs, such as cisplatin, docetaxel and doxorubicin, showed lower
half-maximal inhibitory concentration values in the high-risk
group, indicating that high-risk patients were more likely to
benefit from these chemotherapy drugs (Fig. 7C), offering an expanded
understanding of treatment options and providing a scientific
foundation for personalized treatment strategies.
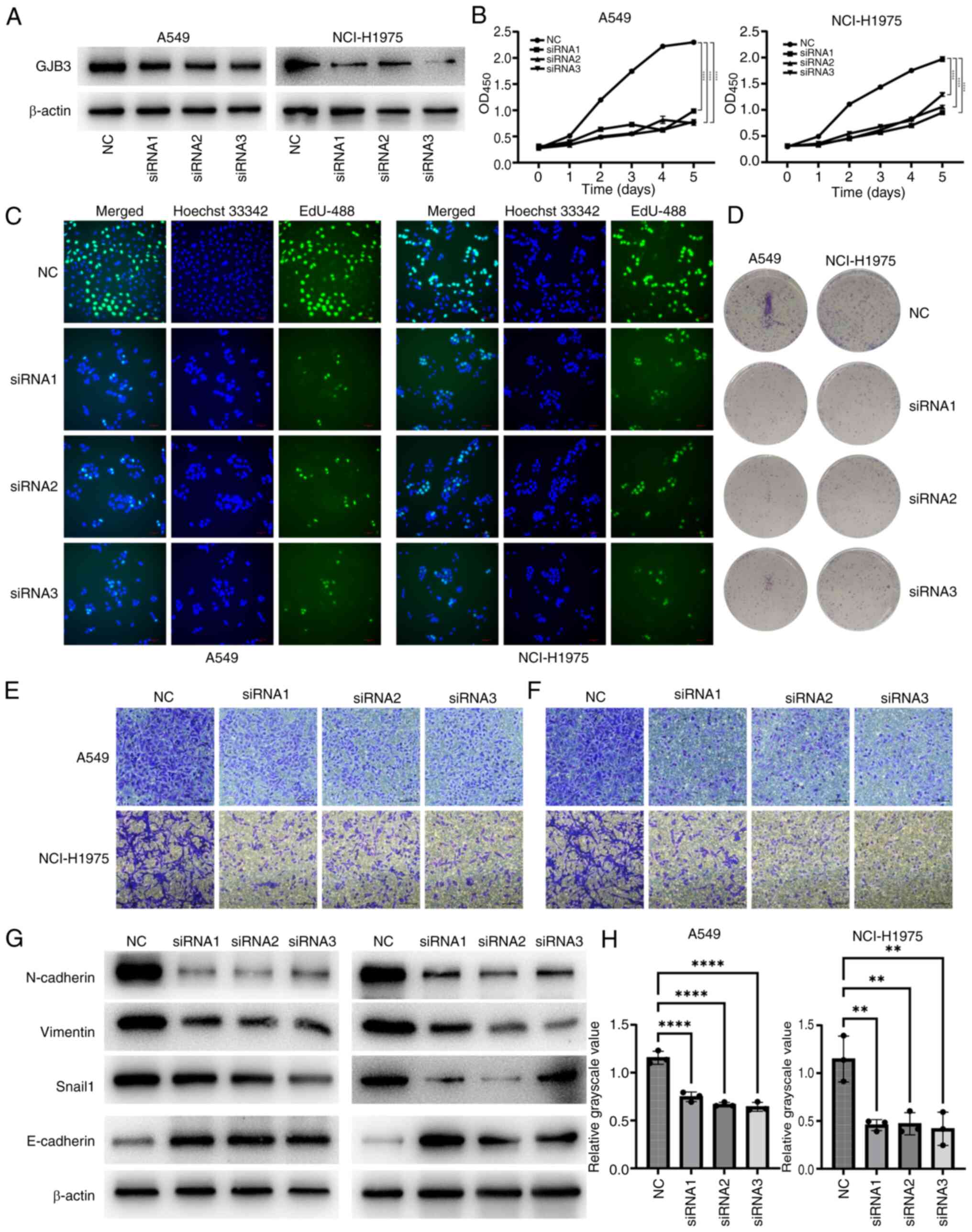
GJB3 promotes the proliferation,
invasion and epithelial-mesenchymal transition (EMT) of LUAD
Bioinformatics analysis revealed that GJB3 was
highly expressed in tumor tissues and functioned as an oncogene
(Table SII). This was confirmed
through in vitro validation. siRNA-mediated knockdown of
GJB3 expression in A549 and NCI-H1975 cells (Fig. 8A and H) resulted in significantly
reduced cell viability (Fig. 8B).
In addition, EdU staining and colony formation assays showed that
GJB3 knockdown decreased the proliferative capacity of both A549
and NCI-H1975 cells (Fig. 8C and
D, and S1). Transwell assays
further demonstrated that GJB3 knockdown inhibited the migration
and invasion of these cells (Fig. 8E
and F). Given that GJB3 encodes a key member of the connexin
gene family (28), its alteration
might impact EMT. Further examination revealed that GJB3 knockdown
downregulated the expression levels of vimentin, Snail1 and
N-cadherin, while upregulating E-cadherin levels in A549 and
NCI-H1975 cells (Fig. 8G), thus
indicating that GJB3 knockdown inhibited the EMT process in these
cell lines.
Discussion
The treatment of NSCLC faces notable challenges,
including treatment resistance and the need for personalized
therapies. Although traditional chemotherapy and radiotherapy
remain essential components of NSCLC management, innovative
treatment strategies have emerged in recent years (29). Immunotherapy, such as
anti-PD-1/PD-L1 therapy, one of the most promising advancements,
has demonstrated marked therapeutic effects and survival benefits
by stimulating the immune system of patients to target and
eliminate cancer cells (30). By
contrast, targeted therapies, such as EGFR-tyrosine kinase
inhibitors (TKIs), ALK-TKIs and entrectinib, offer more tailored
treatment options by focusing on specific molecular markers on
tumor cells, enabling precise intervention. By identifying and
targeting these markers with specific drugs, targeted therapies
enhance treatment efficacy, improve patient quality of life and
minimize side effects (31–33).
Furthermore, genomics-guided therapy represents a new frontier in
personalized treatment for NSCLC. By analyzing the genetic profile
of tumors, this approach provides deeper insights into the genetic
characteristics, mutations and potential resistance mechanisms of
the tumor, enabling the development of more precise and effective
treatment strategies (34). The
combination of immunotherapy and targeted therapies has further
advanced NSCLC treatment, leveraging the synergistic effects of
both approaches to improve therapeutic outcomes while minimizing
adverse effects, thus offering patients greater survival benefits
and a better overall treatment experience (35).
Cholesterol serves a pivotal role in maintaining
cellular homeostasis (36). Due to
the limited availability of nutrients in local tissues, tumors
often experience relative nutrient deficiencies alongside the
accumulation of metabolic waste products (37). In response to these adverse
conditions, tumors undergo metabolic reprogramming to adapt
(38). Preclinical studies have
shown that inhibiting cholesterol synthesis can restrict tumor
progression (39,40), and promising results have been
reported from combining statins with other treatment modalities
(41,42). Identifying key molecules involved
in cholesterol metabolism and developing targeted drugs against
them are critical steps in advancing treatment for LUAD.
Cholesterol metabolism influences both tumor growth
and the immune microenvironment (43), indicating a complex crosstalk
between cholesterol metabolic status and the TME. Two distinct
clusters with different cholesterol metabolism profiles were
identified in the present study, with cluster 1 exhibiting richer
immune cell infiltration, which helps explain the prognostic
differences in patients with LUAD. Given the strong association
between cholesterol metabolism and prognosis in LUAD, a prognostic
model based on cholesterol metabolism was developed, involving
CYP4B1, GJB3, IGF2BP1 and KRT6A. CYP4B1, a member of the cytochrome
P450 enzyme superfamily, serves a key role in cholesterol and
derivative synthesis. Its elevated expression in bladder cancer has
been linked to an increased risk of the disease (44). IGF2BP1 influences tumor development
and therapeutic responses primarily by binding to mRNAs, regulating
their stability and translation (45). Müller et al (46) demonstrated that IGF2BP1 promotes
SRF expression in an m6A-dependent manner, driving tumor growth and
invasion. Additionally, Zhang et al (47) showed that IGF2BP1 overexpression
accelerates the malignant progression of endometrial cancer through
m6A methylation. Together, these studies highlight the importance
of IGF2BP1 in understanding cancer progression. KRT6A is positively
associated with both T-stage and N-stage in LUAD, and its knockdown
inhibits cell proliferation, invasion and the EMT process in LUAD
(48). Furthermore, GJB3 has been
shown to be markedly upregulated in the liver metastases of
pancreatic cancer, with overexpression enhancing the accumulation
of tumor-associated neutrophils and promoting liver metastasis
(49). The current study focused
on GJB3, which is a member of the cell junction protein family.
According to a previous report, high cholesterol metabolism can
affect cell adhesion and promote tumor metastasis (50), and GJB3 may have an important role
in this process. To further investigate its function, in
vitro experiments were conducted, revealing that GJB3 knockdown
inhibited the proliferative ability of LUAD cells, consistent with
the results from GSEA. Additionally, GJB3 knockdown suppressed the
migration and invasion of LUAD cells and reduced the expression of
EMT-related proteins. These findings suggested that GJB3 may serve
a critical role in LUAD progression and could serve as a potential
therapeutic target.
Cholesterol metabolism influences the TME and may
significantly affect the efficacy of immunotherapy (51). The CMRG score constructed in the
current study suggested that low-risk patients exhibited higher
response rates to immune checkpoint therapy, whereas high-risk
patients were predicted to show a better chemotherapy response.
This highlights new perspectives for the precision treatment of
patients with LUAD.
There are some limitations in the present study: The
sequencing data comes from previous patients, and the
characteristics of cholesterol metabolism were only studied in TCGA
and GSE72094 datasets. It is necessary to conduct larger
prospective studies to further confirm the effectiveness of the
prognostic model. Second, the cancer-promoting ability of GJB3 in
LUAD needs further validation in animals. Notably, the mechanism by
which GJB3 promotes the progression of LUAD by affecting
cholesterol metabolism needs to be further studied through
multi-omics, and in vitro and in vivo
experiments.
In conclusion, cholesterol metabolic status is
closely associated with the prognosis, TME and treatment response
in patients with LUAD. The cholesterol metabolism-related
prognostic model developed in the current study may not only
predict patient outcomes but could also provide valuable insights
into personalized treatment strategies.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science
Foundation of Guangdong Province (grant no. 2019A1515011601).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
QY, WC and JW designed and supervised the
experiments. WC and WW performed the bioinformatics analysis. LG
and DL performed the experiments. QY and JY analyzed and
interpreted the data, and wrote the first draft of the manuscript.
QY, JW and WC reviewed the manuscript. LG and WC confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020.
|
|
2
|
Berg CD, Schiller JH, Boffetta P, Cai J,
Connolly C, Kerpel-Fronius A, Kitts AB, Lam DCL, Mohan A, Myers R,
et al: Air pollution and lung cancer: A review by international
association for the study of lung cancer early detection and
screening committee. J Thorac Oncol. 18:1277–1289. 2023. View Article : Google Scholar
|
|
3
|
Akçay S: Deciphering molecular overlaps
between COPD and NSCLC subtypes (LUAD and LUSC): An integrative
bioinformatics study. Medicine (Baltimore). 104:e439062025.
View Article : Google Scholar
|
|
4
|
Thai AA, Solomon BJ, Sequist LV, Gainor JF
and Heist RS: Lung cancer. Lancet. 398:535–554. 2021. View Article : Google Scholar
|
|
5
|
Mamdani H, Matosevic S, Khalid AB, Durm G
and Jalal SI: Immunotherapy in lung cancer: Current landscape and
future directions. Front Immunol. 13:8236182022. View Article : Google Scholar
|
|
6
|
Huang B, Song BL and Xu C: Cholesterol
metabolism in cancer: Mechanisms and therapeutic opportunities. Nat
Metab. 2:132–141. 2020. View Article : Google Scholar
|
|
7
|
Bakiri L, Hamacher R, Graña O,
Guío-Carrión A, Campos-Olivas R, Martinez L, Dienes HP, Thomsen MK,
Hasenfuss SC and Wagner EF: Liver carcinogenesis by FOS-dependent
inflammation and cholesterol dysregulation. J Exp Med.
214:1387–1409. 2017. View Article : Google Scholar
|
|
8
|
Wang X, Huang Z, Wu Q, Prager BC, Mack SC,
Yang K, Kim LJY, Gimple RC, Shi Y, Lai S, et al: MYC-regulated
mevalonate metabolism maintains brain tumor-initiating cells.
Cancer Res. 77:4947–4960. 2017. View Article : Google Scholar
|
|
9
|
Kondo A, Yamamoto S, Nakaki R, Shimamura
T, Hamakubo T, Sakai J, Kodama T, Yoshida T, Aburatani H and Osawa
T: Extracellular acidic pH activates the sterol regulatory
element-binding protein 2 to promote tumor progression. Cell Rep.
18:2228–2242. 2017. View Article : Google Scholar
|
|
10
|
He M, Zhang W, Dong Y, Wang L, Fang T,
Tang W, Lv B, Chen G, Yang B, Huang P and Xia J: Pro-inflammation
NF-κB signaling triggers a positive feedback via enhancing
cholesterol accumulation in liver cancer cells. J Exp Clin Cancer
Res. 36:152017. View Article : Google Scholar
|
|
11
|
Kloudova A, Guengerich FP and Soucek P:
The role of oxysterols in human cancer. Trends Endocrinol Metab.
28:485–496. 2017. View Article : Google Scholar
|
|
12
|
Raccosta L, Fontana R, Maggioni D,
Lanterna C, Villablanca EJ, Paniccia A, Musumeci A, Chiricozzi E,
Trincavelli ML, Daniele S, et al: The oxysterol-CXCR2 axis plays a
key role in the recruitment of tumor-promoting neutrophils. J Exp
Med. 210:1711–1728. 2013. View Article : Google Scholar
|
|
13
|
Goossens P, Rodriguez-Vita J, Etzerodt A,
Masse M, Rastoin O, Gouirand V, Ulas T, Papantonopoulou O, Van Eck
M, Auphan-Anezin N, et al: Membrane cholesterol efflux drives
tumor-associated macrophage reprogramming and tumor progression.
Cell Metab. 29:1376–1389.e4. 2019. View Article : Google Scholar
|
|
14
|
Kopecka J, Trouillas P, Gašparović AČ,
Gazzano E, Assaraf YG and Riganti C: Phospholipids and cholesterol:
Inducers of cancer multidrug resistance and therapeutic targets.
Drug Resist Updat. 49:1006702020. View Article : Google Scholar
|
|
15
|
Schabath MB, Welsh EA, Fulp WJ, Chen L,
Teer JK, Thompson ZJ, Engel BE, Xie M, Berglund AE, Creelan BC, et
al: Differential association of STK11 and TP53 with KRAS
mutation-associated gene expression, proliferation and immune
surveillance in lung adenocarcinoma. Oncogene. 35:3209–3216. 2016.
View Article : Google Scholar
|
|
16
|
Castanza AS, Recla JM, Eby D,
Thorvaldsdottir H, Bult CJ and Mesirov JP: Extending support for
mouse data in the molecular signatures database (MSigDB). Nat
Methods. 20:1619–1620. 2023. View Article : Google Scholar
|
|
17
|
Wilkerson MD and Hayes DN:
ConsensusClusterPlus: A class discovery tool with confidence
assessments and item tracking. Bioinformatics. 26:1572–1573. 2010.
View Article : Google Scholar
|
|
18
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar
|
|
19
|
Barbie DA, Tamayo P, Boehm JS, Kim SY,
Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al:
Systematic RNA interference reveals that oncogenic KRAS-driven
cancers require TBK1. Nature. 462:108–112. 2009. View Article : Google Scholar
|
|
20
|
Yoshihara K, Shahmoradgoli M, Martinez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar
|
|
21
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar
|
|
22
|
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X,
Li Z, Traugh N, Bu X, Li B, et al: Signatures of T cell dysfunction
and exclusion predict cancer immunotherapy response. Nat Med.
24:1550–1558. 2018. View Article : Google Scholar
|
|
23
|
Rami-Porta R, Nishimura KK, Giroux DJ,
Detterbeck F, Cardillo G, Edwards JG, Fong KM, Giuliani M, Huang J,
Kernstine KH Sr, et al: The international association for the study
of lung cancer lung cancer staging project: Proposals for revision
of the TNM stage groups in the forthcoming (ninth) edition of the
TNM classification for lung cancer. J Thorac Oncol. 19:1007–1027.
2024. View Article : Google Scholar
|
|
24
|
Ehmsen S, Pedersen MH, Wang G, Terp MG,
Arslanagic A, Hood BL, Conrads TP, Leth-Larsen R and Ditzel HJ:
Increased cholesterol biosynthesis is a key characteristic of
breast cancer stem cells influencing patient outcome. Cell Rep.
27:3927–3938.e6. 2019. View Article : Google Scholar
|
|
25
|
Turley SJ, Cremasco V and Astarita JL:
Immunological hallmarks of stromal cells in the tumour
microenvironment. Nat Rev Immunol. 15:669–682. 2015. View Article : Google Scholar
|
|
26
|
Zhang Y and Zhang Z: The history and
advances in cancer immunotherapy: Understanding the characteristics
of tumor-infiltrating immune cells and their therapeutic
implications. Cell Mol Immunol. 17:807–821. 2020. View Article : Google Scholar
|
|
27
|
Martínez-Jiménez F, Muiños F, Sentís I,
Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, Mularoni L, Pich O,
Bonet J, Kranas H, et al: A compendium of mutational cancer driver
genes. Nat Rev Cancer. 20:555–572. 2020. View Article : Google Scholar
|
|
28
|
Liu J, Wang X, Jiang W, Azoitei A, Eiseler
T, Eckstein M, Hartmann A, Stilgenbauer S, Elati M, Hohwieler M, et
al: Impairment of α-tubulin and F-actin interactions of GJB3
induces aneuploidy in urothelial cells and promotes bladder cancer
cell invasion. Cell Mol Biol Lett. 29:942024. View Article : Google Scholar
|
|
29
|
Otano I, Ucero AC, Zugazagoitia J and
Paz-Ares L: At the crossroads of immunotherapy for
oncogene-addicted subsets of NSCLC. Nat Rev Clin Oncol. 20:143–159.
2023. View Article : Google Scholar
|
|
30
|
Sui H, Ma N, Wang Y, Li H, Liu X, Su Y and
Yang J: Anti-PD-1/PD-L1 therapy for non-small-cell lung cancer:
Toward personalized medicine and combination strategies. J Immunol
Res. 2018:69849482018. View Article : Google Scholar
|
|
31
|
Santoni-Rugiu E, Melchior LC, Urbanska EM,
Jakobsen JN, Stricker K, Grauslund M and Sørensen JB: Intrinsic
resistance to EGFR-tyrosine kinase inhibitors in EGFR-mutant
non-small cell lung cancer: Differences and similarities with
acquired resistance. Cancers (Basel). 11:9232019. View Article : Google Scholar
|
|
32
|
Shaw AT, Solomon BJ, Besse B, Bauer TM,
Lin CC, Soo RA, Riely GJ, Ou SI, Clancy JS, Li S, et al: ALK
Resistance mutations and efficacy of lorlatinib in advanced
anaplastic lymphoma kinase-positive non-small-cell lung cancer. J
Clin Oncol. 37:1370–1379. 2019. View Article : Google Scholar
|
|
33
|
Drilon A, Siena S, Dziadziuszko R, Barlesi
F, Krebs MG, Shaw AT, de Braud F, Rolfo C, Ahn MJ, Wolf J, et al:
Entrectinib in ROS1 fusion-positive non-small-cell lung cancer:
Integrated analysis of three phase 1–2 trials. Lancet Oncol.
21:261–270. 2020. View Article : Google Scholar
|
|
34
|
Wang Z, Zhang Q, Qi C, Bai Y, Zhao F, Chen
H, Li Z, Wang X, Chen M, Gong J, et al: Combination of AKT1 and
CDH1 mutations predicts primary resistance to immunotherapy in
dMMR/MSI-H gastrointestinal cancer. J Immunother Cancer.
10:e0047032022. View Article : Google Scholar
|
|
35
|
Wang M, Herbst RS and Boshoff C: Toward
personalized treatment approaches for non-small-cell lung cancer.
Nat Med. 27:1345–1356. 2021. View Article : Google Scholar
|
|
36
|
King RJ, Singh PK and Mehla K: The
cholesterol pathway: Impact on immunity and cancer. Trends Immunol.
43:78–92. 2022. View Article : Google Scholar
|
|
37
|
Pavlova NN, Zhu J and Thompson CB: The
hallmarks of cancer metabolism: Still emerging. Cell Metab.
34:355–377. 2022. View Article : Google Scholar
|
|
38
|
Hanahan D: Hallmarks of cancer: New
dimensions. Cancer Discov. 12:31–46. 2022. View Article : Google Scholar
|
|
39
|
Li J, Liu J, Liang Z, He F, Yang L, Li P,
Jiang Y, Wang B, Zhou C, Wang Y, et al: Simvastatin and
Atorvastatin inhibit DNA replication licensing factor MCM7 and
effectively suppress RB-deficient tumors growth. Cell Death Dis.
8:e26732017. View Article : Google Scholar
|
|
40
|
Mullen PJ, Yu R, Longo J, Archer MC and
Penn LZ: The interplay between cell signalling and the mevalonate
pathway in cancer. Nat Rev Cancer. 16:718–731. 2016. View Article : Google Scholar
|
|
41
|
Lee J, Jung KH, Park YS, Ahn JB, Shin SJ,
Im SA, Oh DY, Shin DB, Kim TW, Lee N, et al: Simvastatin plus
irinotecan, 5-fluorouracil, and leucovorin (FOLFIRI) as first-line
chemotherapy in metastatic colorectal patients: A multicenter phase
II study. Cancer Chemother Pharmacol. 64:657–663. 2009. View Article : Google Scholar
|
|
42
|
Liu JC, Hao WR, Hsu YP, Sung LC, Kao PF,
Lin CF, Wu AT, Yuan KS and Wu SY: Statins dose-dependently exert a
significant chemopreventive effect on colon cancer in patients with
chronic obstructive pulmonary disease: A population-based cohort
study. Oncotarget. 7:65270–65283. 2016. View Article : Google Scholar
|
|
43
|
Liu X, Lv M, Zhang W and Zhan Q:
Dysregulation of cholesterol metabolism in cancer progression.
Oncogene. 42:3289–3302. 2023. View Article : Google Scholar
|
|
44
|
Imaoka S, Yoneda Y, Sugimoto T, Hiroi T,
Yamamoto K, Nakatani T and Funae Y: CYP4B1 is a possible risk
factor for bladder cancer in humans. Biochem Biophys Res Commun.
277:776–780. 2000. View Article : Google Scholar
|
|
45
|
Wang T, Kong S, Tao M and Ju S: The
potential role of RNA N6-methyladenosine in cancer progression. Mol
Cancer. 19:882020. View Article : Google Scholar
|
|
46
|
Müller S, Glaß M, Singh AK, Haase J, Bley
N, Fuchs T, Lederer M, Dahl A, Huang H, Chen J, et al: IGF2BP1
promotes SRF-dependent transcription in cancer in a m6A- and
miRNA-dependent manner. Nucleic Acids Res. 47:375–390. 2019.
View Article : Google Scholar
|
|
47
|
Zhang L, Wan Y, Zhang Z, Jiang Y, Gu Z, Ma
X, Nie S, Yang J, Lang J, Cheng W and Zhu L: IGF2BP1 overexpression
stabilizes PEG10 mRNA in an m6A-dependent manner and promotes
endometrial cancer progression. Theranostics. 11:1100–1114. 2021.
View Article : Google Scholar
|
|
48
|
Yang B, Zhang W, Zhang M, Wang X, Peng S
and Zhang R: KRT6A promotes EMT and cancer stem cell transformation
in lung adenocarcinoma. Technol Cancer Res Treat.
19:15330338209212482020. View Article : Google Scholar
|
|
49
|
Huo Y, Zhou Y, Zheng J, Jin G, Tao L, Yao
H, Zhang J, Sun Y, Liu Y and Hu LP: GJB3 promotes pancreatic cancer
liver metastasis by enhancing the polarization and survival of
neutrophil. Front Immunol. 13:9831162022. View Article : Google Scholar
|
|
50
|
Mohammadalipour A, Showalter CA, Muturi
HT, Farnoud AM, Najjar SM and Burdick MM: Cholesterol depletion
decreases adhesion of non-small cell lung cancer cells to
E-selectin. Am J Physiol Cell Physiol. 325:C471–C482. 2023.
View Article : Google Scholar
|
|
51
|
Ngwa VM, Edwards DN, Philip M and Chen J:
Microenvironmental metabolism regulates antitumor immunity. Cancer
Res. 79:4003–4008. 2019. View Article : Google Scholar
|