Lactate, once regarded as a metabolic waste product
of glycolysis, is now considered a multifunctional molecule that
promotes cancer progression. Notably, the Warburg effect is a
hallmark of cancer metabolism; even under oxygen-rich conditions,
tumor cells still preferentially metabolize glucose through
glycolysis and tend to ‘ferment’ glucose into lactic acid, leading
to an accumulation of lactic acid within the tumor microenvironment
(TME) (1). In addition to serving
a role in central carbon metabolism, lactate regulates tumor
immunity, antiviral responses and endoplasmic reticulum
(ER)-mitochondrial magnesium ion dynamics (2), and contributes to pathological
processes such as cancer progression (3). Lactate, as a signaling molecule,
regulates immune evasion, angiogenesis and therapeutic resistance
(4,5).
The discovery of lactylation, a lactate-derived
post-translational modification (PTM) of lysine residues, has
revealed a direct mechanistic link between glycolytic flux and
cellular regulation. Lactate, as the substrate for this
modification, catalyzes the addition of lactate groups to both
histone and non-histone proteins (6). This process links metabolic disorders
to epigenetic reprogramming, alterations in chromatin structure,
transcriptional programs and protein-protein interactions, thereby
promoting tumorigenesis (7). For
example, M1-polarized macrophages rely on aerobic glycolysis to
drive histone lactylation (8),
whereas B-cell adapter for PI3K facilitates their transition to a
reparative macrophage phenotype via lactylation (9). Lactate enhances the activity of
pyruvate kinase M2 (PKM2) by lactylating its K62 residues,
inhibiting the Warburg effect and promoting the transformation of
inflammatory macrophages (iNOS+ CD68+ cells)
into reparative macrophages (ARG1+ CD68+
positive cells) (10). In
non-small cell lung cancer (NSCLC), hypoxia-induced long noncoding
RNA-AC020978 amplifies glycolysis by promoting PKM2 nuclear
translocation and hypoxia-inducible factor-1α (HIF-1α) activation
(11). Furthermore, lactate
directly inhibits CD8+ T cells, natural killer T (NKT)
cells, dendritic cells and macrophages, while enhancing regulatory
T (Treg) cell stability and function, promoting immunosuppression
(12).
Researchers have also demonstrated the notable role
of lactylation in various types of cancer via numerous mechanisms
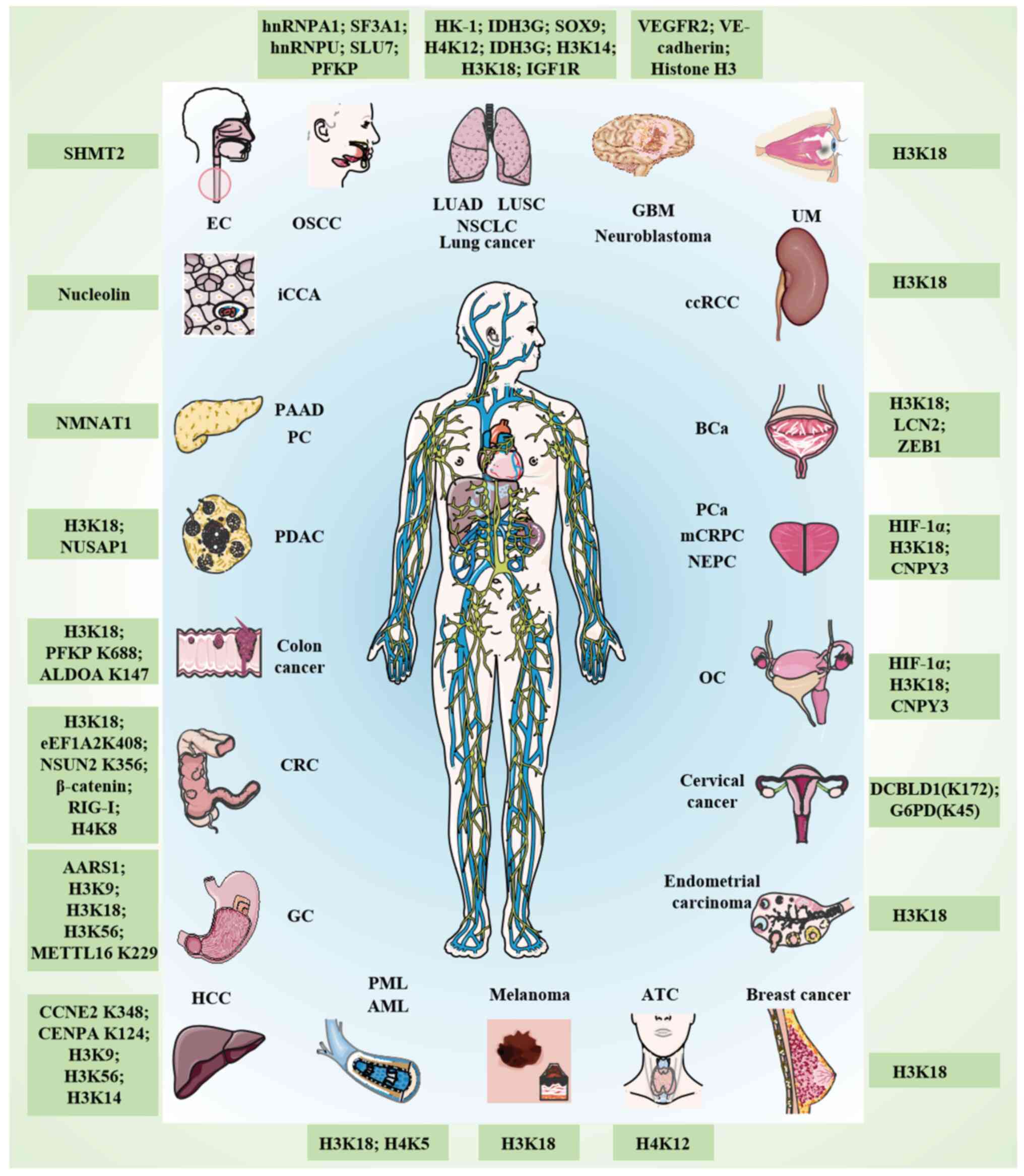
(Table I; Fig. 1). For example, pan-cancer analyses
have revealed upregulated lactylation-related genes (such as CREBBP
and EP300) and their association with kidney renal papillary cell
carcinoma and hepatocellular carcinoma (HCC) (13). Furthermore,
hypoxia-glycolysis-lactylation-related genes predict gastric cancer
(GC) progression (14), whereas
lactate-driven lactylation in colorectal cancer (CRC) is associated
with proliferation, metastasis and immune evasion (15). Lactylation serves a role in
tumorigenesis, including in tumor cell proliferation, invasion,
metastasis, DNA damage repair and immune cell killing, and also
promotes therapeutic resistance through autophagy activation, drug
efflux pumps and DNA damage repair (16).
The present review discusses the latest progress in
lactic acid biology, highlighting its dual role as a driving factor
in the pathogenesis of cancer and in therapeutic sensitivity. The
review also explores the impact of lactylation on metabolic
reprogramming, immune evasion and therapeutic resistance, and
discusses new strategies for precision oncology using this
pathway.
Lactylation critically regulates glycolytic flux in
cancer. Digestive system cancers account for >25% of global
cancer cases (17), with
lactylation emerging as a key regulator of immunosuppression and
metabolic reprogramming in the TME. Yang et al (18) identified 9,256 non-histone lysine
lactylation (Kla) sites in hepatitis B virus-associated HCC,
including adenylate kinase 2 lactylation at K28, which may drive
metastasis by disrupting metabolic regulation.
Abundant immune cell infiltration, particularly
macrophages, and increased genetic instability are traits
associated with high lactylation scores. Lactylation score can be
used to predict the malignant progression of GC and immune escape
(19). Hypoxia-induced
mitochondrial alanyl-tRNA synthetase AARS2 drives lactylation of
pyruvate dehydrogenase (PDH) complex components (PDHA1-K336 and
CPT2-K457/8), suppressing oxidative phosphorylation (OXPHOS) by
limiting pyruvate and acetyl-CoA influx (20). AARS1, upregulated in GC, catalyzes
lactate-AMP formation, promoting lactylation of p53 at lysine
120/139. This disrupts the DNA binding and transcriptional activity
of p53, coupling metabolic rewiring to proteomic changes that fuel
tumorigenesis (21). AARS1 also
activates the YAP-TEAD complex via nuclear lactylation, forming a
Hippo pathway feedback loop that accelerates GC progression and is
associated with poor prognosis (22). In addition, G-protein-coupled
receptor 37 (GPR37) activates the Hippo pathway, upregulating
lactate dehydrogenase A (LDHA) to enhance glycolysis and H3K18
lactylation (H3K18la). This promotes CXCL1/CXCL5 secretion,
remodeling the TME to favor metastasis. By contrast, GPR37
depletion suppresses the progression of liver metastasis in CRC
(23). Lactate also stabilizes
β-catenin via hypoxia-induced lactylation, activating Wnt signaling
and CRC proliferation (24).
Nucleolin lactylation promotes MAPK-activated death
domain protein translation and ERK activation, which is associated
with poor intrahepatic cholangiocarcinoma (iCCA) prognosis
(25). Non-histone lactylation
sites in oral squamous cell carcinoma (OSCC) cells, such as
hnRNPA1, SF3A1, hnRNPU and SLU7, are linked to glycolytic
dysregulation and tumorigenesis, highlighting the role of Kla in
pathogenesis (26). In lung
adenocarcinoma (LUAD), basic leucine zipper and W2 domains 2 (BZW2)
enhances glycolysis and isocitrate dehydrogenase 3 (IDH3G)
lactylation. Combined inhibition of BZW2 and glycolysis [such as
2-deoxy-D-glucose (2-DG)] suppresses tumor growth (27).
CircXRN2 is downregulated in bladder cancer (BCa)
tissues and suppresses tumor growth by binding to the speckle-type
POZ protein (SPOP) degron. This interaction blocks SPOP-mediated
degradation of large tumor suppressor kinase 1, activating the
Hippo signaling pathway to suppress tumor progression driven by
H3K18la (28). Upregulation of
zinc finger E-box binding homeobox 1 (ZEB1) in BCa is counteracted
by phosphofructokinase-1 (PFK-1), and PFK-1 inhibits glycolysis
through the lactylation of ZEB1 and suppresses its malignant
effects, including cell proliferation, migration and invasion
(29). During neuroendocrine
prostate cancer (NEPC) development, ZEB1 transcriptionally
regulates the expression of several key glycolytic enzymes, thereby
predisposing tumor cells to utilize glycolysis for energy
metabolism. Lactate accumulation enhances histone lactylation,
increasing chromatin accessibility and cellular plasticity
(including neural gene expression), thereby promoting NEPC
progression (30).
In BCa, the enhanced aerobic glycolysis rate
supports c-Myc expression through histone lactylation at the
promoter level. c-Myc further upregulates serine/arginine splicing
factor 10 (SRSF10) through transcription to drive the selective
splicing of MDM4 and Bcl-x in BCa cells. Restricting the activity
of key glycolytic enzymes may affect the c-Myc/SRSF10 axis to
reduce the proliferation of BCa cells (31). Potassium channel subfamily K member
1, which is upregulated in BCa, activates LDHA to enhance
glycolysis and H3K18la. LDHA upregulation creates a feed-forward
loop, reducing tumor cell adhesion and promoting
invasion/metastasis (32).
Deficiency in the Numb/Parkin pathway in prostate cancer (PCa) or
LUAD induces metabolic reprogramming, resulting in a significant
increase in the production of lactate acid, which subsequently
leads to the upregulation of histone lactylation and transcription
of neuroendocrine-associated genes (33). Aerobic glycolysis in anaplastic
thyroid cancer (ATC) increases overall protein lactylation by
increasing cellular lactate availability, and H4K12la activates the
expression of multiple genes necessary for ATC proliferation.
Furthermore, the oncogene BRAFV600E enhances glycolysis to
reprogram the cellular lactylation landscape, resulting in
H4K12la-driven gene transcription and cell cycle dysregulation.
Notably, treatment with a BRAFV600E inhibitor, combined with
blocking cell emulsification, can inhibit ATC progression in an
8505c xenograft tumor mouse model (34).
Autophagy and glycolysis are highly conserved
biological processes involving physiological and pathological
cellular activities. A number of core autophagy proteins undergo
lactylation during cancer (35).
Under nutrient deprivation, lactate-mediated lactylation of
autophagy proteins (such as VPS34, ULK1) enhances autophagic flux,
promoting tumor cell survival (36). In lung cancer and GC, lactate
mediates the lactylation of PIK3C3/VPS34 at lysine 356 and lysine
781 through acyltransferase KAT5/TIP60. The lactylation of
PIK3C3/VPS34 enhances the binding of PIK3C3/VPS34 to beclin 1,
ATG14 and UVRAG, increases the lipid kinase activity of
PIK3C3/VPS34, promotes macroautophagy/autophagy and advances the
endolysome degradation pathway (37).
In the TME, tumor cells export lactate via
monocarboxylate transporter 4 (MCT4), while oxidative tumor cells
import it through MCT1 to fuel OXPHOS (38). Although glycolysis dominates in
most types of cancer, OXPHOS remains active in malignancies such as
leukemia and lymphoma, enabling metabolic flexibility (39). Glucose fuels glycolysis, which
feeds into the TCA cycle and OXPHOS in oxidative cells, or sustains
lactate fermentation in hypoxic regions (40). In SW480 colon cancer cells,
lactylation targets glycolytic enzymes (such as PFKP-K688 and
ALDOA-K147), potentially establishing negative feedback loops to
modulate glycolysis (41). In
pancreatic adenocarcinoma cells, under glucose deprivation, lactate
enhances glutaminase 1-mediated glutamine metabolism and NMNAT1
lactylation, suppressing p38 MAPK and promoting survival (42). Solute carrier family 16 member 1
lactylation is critical for pancreatic ductal adenocarcinoma (PDAC)
progression, and targeting this axis may inhibit tumor growth
(43). In NSCLC, metabolic
dysregulation drives lactate accumulation, which replenishes the
TCA cycle while suppressing glucose uptake and glycolysis. Lactate
downregulates glycolytic enzymes [such as hexokinase (HK)-1 and
PKM] and upregulates TCA enzymes (including SDHA and IDH3G) via
histone lactylation at their promoters, enhancing NSCLC
proliferation and migration (44).
The hypoxic microenvironment resulting from reduced
local blood flow in pancreatic cancer (PC) triggers a shift in
glycolytic-dependent energy metabolism by stabilizing HIF-1α
(51). Elevated H3K18la in PDAC
activates TTK and BUB1B transcription, upregulating P300 to enhance
glycolysis. TTK phosphorylates LDHA at Y239, amplifying lactate and
H3K18la levels (52). Nuclear and
spindle-associated protein 1 (NUSAP1) binds c-Myc and HIF-1α to
upregulate LDHA. Lactate stabilizes NUSAP1 via Kla, forming a
feed-forward loop (NUSAP1-LDHA-lactate-NUSAP1) that drives
metastasis (53). In esophageal
cancer (EC), hypoxia not only enhances the expression of serine
hydroxymethyltransferase 2 (SHMT2) protein but also initiates the
lactylation of SHMT2 protein and enhances its stability, thus
accelerating the malignant progression of EC (54).
Angiogenic mimicry provides tumor cells with a blood
supply independent of endothelial cells (55). MAPK 6 pseudogene 4 stabilizes
Krüppel-like factor 15, which transcriptionally activates LDHA.
LDHA-mediated lactylation of vascular endothelial growth factor
receptor 2 (VEGFR2) and VE-cadherin enhances their expression,
fostering GBM cell proliferation, migration and angiogenic mimicry.
However, the specific lactylation sites in VEGFR2 and VE-cadherin
involved in GBM still require further exploration (56). FK506-binding protein 10 (FKBP10)
binds LDHA, enhancing LDHA-Y10 phosphorylation to amplify the
Warburg effect and histone lactylation. By contrast, HIF-2α, a
driver of angiogenesis and redox balance, suppresses FKBP10
transcription. Combining HIF-2α inhibitors (such as PT2385) with
FKBP10 targeting enhances antitumor efficacy, particularly in
low-FKBP10 ccRCC (57). Lactate
import via MCT1 stabilizes HIF-1α under normoxia through HIF-1α
lactylation, perpetuating KIAA1199 expression. By contrast,
silencing KIAA1199 restores 3A semaphoring (sema)3A and inhibits
angiogenesis (58). Lactate also
stabilizes HIF-1α, promoting discoidin, CUB, and LCCL
domain-containing protein 1 (DCBLD1) transcription. DCBLD1-K172
lactylation inhibits ubiquitination, blocking autophagy-mediated
degradation of G6PD to fuel the pentose phosphate pathway (PPP).
This activates the PPP, fueling cervical cancer cell migration,
invasion and growth (59).
G6PD K45 is lactylated during G6PD-mediated
antioxidant stress. In primary human keratinocytes and HPV-negative
cervical cancer C33A cell lines ectopically expressing HPV16 E6,
G6PD K45A is not lactylated, and the transduction of G6PD K45A has
been shown to increase the levels of glutathione and NADPH, and can
accordingly decrease the levels of reactive oxygen species. In
vivo, 6-aminonicotinamide inhibits the activity of the G6PD
enzyme or the re-expression of G6PD K45T, thereby suppressing tumor
proliferation (60). The
non-metabolizable glucose analog 2-DG and oxamate treatment have
been reported to decrease the levels of lactylation to inhibit
proliferation and migration, induce apoptosis and arrest the cell
cycle of EC cells. In addition, a study on EC xenograft tumor model
mice and EC cells confirmed that H3K18la may upregulate the
deubiquitinase ubiquitin-specific protease (USP)39, stabilizing
phosphoglycerate kinase 1 to activate the PI3K/AKT/HIF-1α axis and
to enhance glycolysis (61).
Lactylation orchestrates the progression of cancer
through extensive epigenetic reprogramming, modulating both histone
and non-histone targets to alter gene expression, RNA processing
and cellular phenotypes.
Histone lactylation directly modulates chromatin
dynamics. In GC, H3K18la activates vascular cell adhesion molecule
1, promoting GC cell proliferation and migration via AKT/mTOR
signaling, and recruits immunosuppressive mesenchymal stem cells
through CXCL1 upregulation, thereby promoting immune suppression
and tumor metastasis (62). In
CRC, tumor-derived lactate inhibits retinoic acid receptor γ in
macrophages, elevating H3K18la and IL-6/STAT3 signaling to polarize
macrophages toward a pro-tumorigenic state (63). Histone lactylation (such as H4K8la,
H4K16la) is suppressed by liver kinase B1, which induces senescence
by inhibiting telomerase reverse transcriptase (64). Furthermore, lactate in the TME of
LUAD reduces the transcription of solute carrier family 25, member
29 (SLC25A29) in endothelial cells. Specifically, the increase of
H3K14la and H3K18la in the SLC25A29 promoter region reduces the
transcription of SLC25A29, which affects the proliferation,
migration and apoptosis of endothelial cells (65). The NF-κB pathway promotes the
Warburg effect, thereby inducing the lactylation of H3 histone and
increasing the expression of LINC01127. The enhanced expression of
LINC01127 promotes the self-renewal of GBM cells and directly
guides POLR2A to the MAP4K4 promoter region to regulate the
expression of MAP4K4, thereby activating the JNK pathway and
ultimately regulating the self-renewal of GBM cells (66). Inactivation of the von
Hippel-Lindau (VHL) tumor suppressor triggers histone lactylation,
activating platelet-derived growth factor receptor β (PDGFRβ)
transcription; PDGFRβ signaling reciprocally enhances histone
lactylation, creating an oncogenic feedback loop (67).
The lactylation and delactylation of non-histone
proteins are closely related to the pathogenesis of HCC. Sirtuin
(SIRT)3 deficiency in HCC permits the accumulation of cyclin
(CCN)E2 lactylation, whereas honokiol-mediated SIRT3 reactivation
delactylates CCNE2-K348 to suppress HCC cell proliferation
(68). Furthermore, CENPA-K124
lactylation enhances the interaction between CENPA and the
transcription factor YY1, driving CCND1 and neuropilin 2 expression
to promote HCC progression (69).
In CRC, KAT8-catalyzed lactylation of eEF1A2K408 has been shown to
result in boosted translation elongation and enhanced protein
synthesis, which contribute to tumorigenesis (70). Furthermore, H3K18la has been
reported to increase METTL3 expression in tumor-infiltrating
myeloid cells. Notably, METTL3 mediates m6A modification on Jak1
mRNA in tumor-infiltrating myeloid cells, and the m6A-YTHDF1 axis
enhances JAK1 protein translation efficiency, subsequent
phosphorylation of STAT3 and immunosuppression (71). In ocular melanoma, histone
lactylation promotes tumorigenesis by increasing the expression of
YTHDF2. YTHDF2 recognizes m6A-modified PER1 and TP53 mRNA, promotes
their degradation and accelerates the occurrence of ocular melanoma
(72). Histone lactylation
enhances the expression of AlkB homolog 3 (ALKBH3) by removing the
m1A methylation of SP100A. ALKBH3 lactylation facilitates
promyelocytic leukemia protein nuclear condensate formation,
bridging m1A modification to metabolic reprogramming (73). Elevated NOP2/Sun RNA
methyltransferase family member 2 (NSUN2), an m5C
methyltransferase, and Y-box binding protein 1 (YBX1), an m5C
methylation recognition enzyme, drive m5C modification of ENO1 mRNA
in CRC, promoting lactate production. This lactate then induces
NSUN2 expression via histone H3K18 lactylation and enhances RNA
binding of NSUN2 through its own lactylation (K356), creating a
feed-forward loop linking metabolism and epigenetics (74). Furthermore, the chemotherapeutic
agent gemcitabine (GEM) increases histone acetylation but reduces
histone lactylation, suggesting cross-talk between PTMs in the
mechanism of GEM (75). These
mechanisms collectively establish lactylation as a master regulator
of cancer epigenetics, influencing angiogenesis, therapy resistance
and metabolic adaptation.
Lactylation orchestrates immunosuppression within
the TME by reprogramming immune cell function and recruitment. In
HCC, dysregulation of histone acetyltransferase EP300 and histone
deacetylase (HDAC)1-3 alters immune cell infiltration (including B
cells) and predicts poor prognosis when HDAC1/2 are upregulated
(76). Glucose transporter 3
(GLUT3) levels are significantly increased in GC tissues. By
contrast, after knocking down GLUT3, the levels of LDHA,
L-lactylation, H3K9, H3K18 and H3K56 are markedly reduced. Notably,
in GLUT3-knockdown cell lines, upregulation of LDHA reverses the
lactylation and epithelial-mesenchymal transition functional
phenotypes, while Kla is enriched in GC tissues and predicts poor
survival, underscoring its role as a prognostic biomarker (77,78).
Claudin-9 promotes glycolytic metabolism in GC cells by activating
the PI3K/AKT/HIF-1α signaling pathway, resulting in increased
lactate production. Lactate, as a glycolytic metabolite, can
enhance the lactylation of programmed death ligand 1 (PD-L1) and
improve its stability. This modified PD-L1 will inhibit the
antitumor immune response of CD8+ T cells, thereby
enhancing GC cell immune evasion and promoting the progression of
GC (79). STAT5-induced lactate
accumulation promotes nuclear translocation of E3 binding protein,
increases lactylation of the PD-L1 promoter, and subsequently
induces PD-L1 transcription, driving immunosuppression in acute
myeloid leukemia (80).
The gut microbiome further amplifies
immunosuppression through lactylation. In CRC, RIG-I lactylation
induced by Escherichia coli inhibits the recruitment of
NF-κB to the NLRP3 promoter, suppresses inflammasome activity,
impairs CD8+ T-cell function and simultaneously enhances
Treg cell-mediated tolerance (81). Similarly, gram-negative bacterial
lipopolysaccharide upregulates the expression of LINC00152 and
promotes the invasion of CRC cells by introducing histone
lactylation on its promoter, reducing the binding efficiency of
inhibitory factor YY1 to it (82).
In lung squamous cell carcinoma, solute carrier
family 2 member 1 (SLC2A1) upregulation is associated with elevated
protein lactylation and SPP1+ macrophage abundance,
driving therapy resistance (83).
In advanced GBM, monocyte-derived macrophages dominate the TME and
secrete IL-10 via PERK-driven histone lactylation, inhibiting
T-cell activity. PERK deletion abolishes lactylation, restores
T-cell function and delays tumor growth (84). Furthermore, single-cell RNA
sequencing has confirmed lactylation-related gene expression across
glioma-infiltrating immune cells (monocytes, macrophages,
CD8+ T cells), and lactylation scores have been reported
to be associated with lipogenesis, DNA repair and mTORC1 signaling
(85). HK-3 indirectly affects
neuroblastoma progression by recruiting and polarizing M2-like
macrophages through the PI3K/AKT/CXCL14 axis. After knocking down
the expression of HK3, the levels of histone lysine lactylation in
neuroblastoma cell lines SK-N-SH and SK-N-BE (2) are significantly reduced, and the
lactate concentration in the supernatant of SK-N-SH/SK-N-BE
(2) cell cultures is significantly
reduced. These findings indicate that in neuroblastoma HK3 affects
lactate secretion in the microenvironment and regulates histone
lactylation (86). In the TME of
malignant pleural effusion (MPE), the antitumor activity of
CD8+ T cells and NKT-like cells is inhibited, and the
function of Treg cells is enhanced. The glycolytic pathway and
pyruvate metabolism are highly activated in a distinct
subpopulation of NKT-like cells expressing FOXP3 in MPE. Notably, a
small molecule inhibitor of MCT1, 7ACC2, has been shown to reduce
FOXP3 expression and histone lactylation levels in NKT-like cells
(87).
In addition to regulating metabolic reprogramming,
and epigenetic and immune pathways, lactylation drives
multi-faceted treatment resistance in cancer. Previous proteomic
profiling identified 1,438 lactylation sites across 772 proteins in
HCC tissues, implicating lactylation in amino acid metabolism,
ribosomal function and fatty acid metabolism (88). Lactylation of USP14 and ATP-binding
cassette (ABC) subfamily F member 1 is associated with drug
sensitivity. Notably, patients with high-risk HCC respond better to
sorafenib, whereas low-risk patients exhibit elevated Tumor Immune
Dysfunction and Exclusion scores and benefit from immunotherapy
(89). Lactylation also activates
oncogenic pathways (Wnt, MAPK, mTOR, NOTCH) via nuclear receptor
6A1, oxysterol-binding protein 2 and UNC119B, linking Kla to immune
evasion and therapy resistance (90). Apicidin treatment reduces
lactylation, suppressing HCC migration and proliferation, although
clinical validation and mechanistic crosstalk with other PTMs
require further study (91).
In GC, copper stress lactylates METTL16 at K229,
enhancing copper ionophore (elesclomol) efficacy. Combining
elesclomol with the SIRT2 inhibitor AGK2 amplifies copper toxicity,
suggesting a therapeutic strategy to overcome chemoresistance
(92,93). In CRC, H3K18la upregulation in
bevacizumab-resistant tumors, which upregulates autophagy protein
RUBCNL/Pacer through BECN1 interaction, enhances autophagosome
maturation and survival (94). In
diapause-like CRC, SMC4 and phosphoglycerate mutase 1 loss disrupts
F-actin assembly and upregulates ABC transporters via histone
lactylation, reducing chemotherapy sensitivity (95). Aldolase B-activated PDH kinase 1
drives lactylation of circulating carcinoembryonic antigen,
activating CEACAM6 to promote proliferation and chemoresistance in
CRC (96).
Hypoxia in the TME amplifies lactate production,
global lactylation and immunosuppression (97,98).
Hypoxia upregulates SOX9 lactylation, enhancing stemness and
invasion in NSCLC (99). AKR1B10
promotes LDHA-driven lactate accumulation, stimulating H4K12la to
activate CCNB1 and accelerate DNA replication, conferring
pemetrexed resistance in lung cancer brain metastasis (100). This aligns with findings that
tumor resistance stems from heterogeneity and immunosuppressive TME
interactions (101).
In BCa, H3K18la drives the key transcription factors
YBX1 and YY1 associated with cisplatin resistance, thereby
promoting cisplatin resistance (102). Sema3A suppresses VEGFA-induced
colony formation, proliferation and PD-L1 expression in PCa.
Sema3A, along with sema3B/C/E, predicts biochemical recurrence in
low- to intermediate-risk PCa post-prostatectomy (103). Prognostic models combining the
lactylation-related genes ALDOA, DDX39A, H2AX, KIF2C and RACGAP1
has been shown to predict disease-free survival and treatment
response in PCa. These genes are highly expressed in
castration-resistant tumors, underscoring the clinical relevance of
lactylation (104). In
PTEN-deficient metastatic castration-resistant PCa, a PI3K
inhibitor (PI3Ki) has been reported to reduce histone lactylation
within tumor-associated macrophages (TAMs), resulting in their
anticancer phagocytic activation, which is augmented by ADT/aPD-1
treatment and abrogated by feedback activation of the Wnt/β-catenin
pathway. Furthermore, co-targeting Wnt/β-catenin signaling with
LGK-974 in combination with PI3Ki, has previously demonstrated
durable tumor control in 100% of mice via H3K18lac suppression and
complete TAM activation (105,106). Gambogic acid disrupts this cycle
by recruiting SIRT1 to delactylate chaperone protein CNPY3,
inducing lysosomal rupture and pyroptotic cell death (107). Lactylation-related genes
influence BCa growth, immune microenvironment remodeling and
therapy resistance by modulating lactate transport within the TME
(108). In addition, H4K12la
contributes to chemoresistance and adverse immune responses in
breast cancer (109).
Lactylation modulation represents an emerging
frontier for targeted cancer therapy, with implications for
treatment response prediction and combination strategies. Yu et
al (110) identified 14
lactylation-related genes in ovarian cancer (OC) using The Cancer
Genome Atlas database, stratifying patients into low- and high-risk
groups. The low-risk profile was associated with metabolic
processes (thermogenesis and OXPHOS) and immune responses
(neutrophil extracellular traps and IL-17 signaling). High-risk
features were associated with cell adhesion (proteoglycans and
adhesive plaques) and carcinogenic signaling (Wnt and extracellular
matrix-receptor interactions). Notably, lactylation-related genes
were shown to be closely related to tumor classification and
immunity, and OC based on lactylation-related characteristics was
indicated to have a good prognostic performance.
Similarly, in skin cutaneous melanoma (CM),
Kaplan-Meier survival analysis revealed that the lactylation
low/TME high group had the highest overall survival (OS) rate,
whereas the lactylation high/TME low group had the lowest OS rate.
In addition, CM immunotherapy was revealed to be more suitable for
patients in the lactylation low/TME high group (111). Notably, calmodulin-like protein 5
is a core lactylation-associated gene in CM, which is associated
with patient survival and immune infiltration (112).
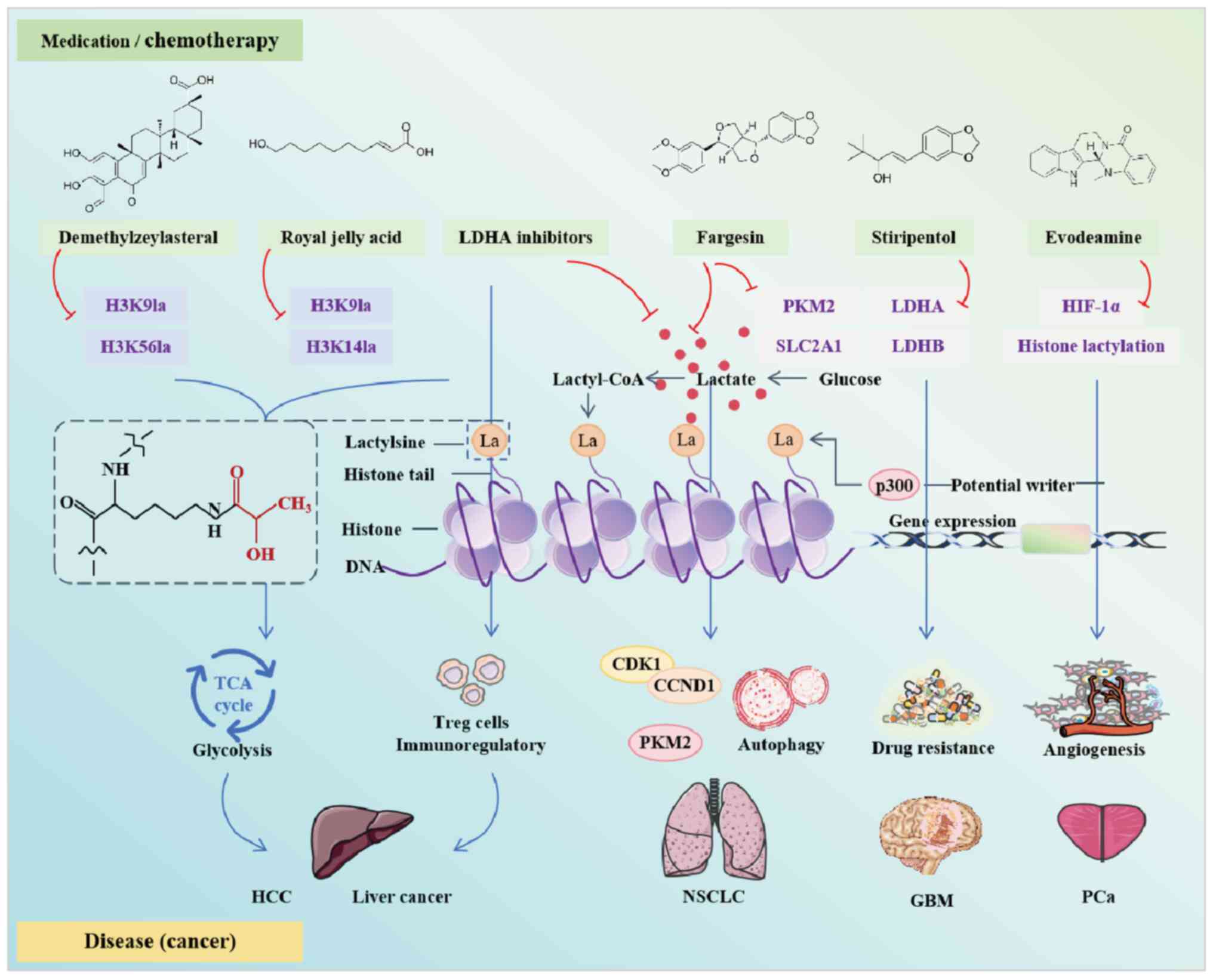
Lactate accumulation in the TME promotes immune
evasion and tumor survival, but lactylation also offers therapeutic
opportunities. Preclinical studies have highlighted strategies such
as inhibiting lactate production (for example, via LDHA inhibitors)
or disrupting histone lactylation [such as with demethylzeylasteral
(DML), royal jelly acid (RJA) or fargesin] to reduce tumor energy
supply or enhance immunotherapy efficacy (Fig. 2). However, these findings are
limited by preclinical risks, as results from homogeneous mouse
models may overestimate efficacy in humans, where TME heterogeneity
remains unresolved.
In a mouse liver cancer cell model (Hepa1-6), LDHA
inhibitors have been shown to suppress the immunoregulatory effects
of Treg cells in the TME by reducing lactate concentration.
Notably, combined treatment with anti-PD-1 and LDHA inhibitors has
a stronger antitumor effect than anti-PD-1 alone (118). The anti-epileptic drug,
stiripentol enhances the sensitivity of cancer cells to
temozolomide by inhibiting lactylation. Lactylation is upregulated
in relapsed GBM tissues and temozolomide-resistant cells, mainly
since H3K9la confers temozolomide resistance in GBM through
Luc7L2-mediated intron-7 retention of MLH1. Stiripentol enhances
temozolomide sensitivity in GBM by inhibiting LDH activity and
H3K9la-mediated resistance (119). Oxamate promotes immune activation
of chimeric antigen receptor (CAR)-T cells infiltrating tumors. In
a GBM mouse model, oxamate has been shown to promote the immune
activation of tumor-infiltrating CAR-T cells through altering the
phenotypes of immune molecules and increasing Treg-cell
infiltration (120). However,
murine models cannot fully recapitulate human metabolic plasticity.
PD-1/PD-L1 is a major inhibitory checkpoint pathway that regulates
immune escape in patients with cancer, and its participation and
inhibition notably reshape the pattern of tumor clearance. Immune
checkpoint inhibition targeting PD-1/PD-L1 is a reliable tumor
therapy (121). Notably, lactate
treatment in PCa cells can increase the expression of HIF-1α and
PD-L1, while restricting the expression of sema3A, whereas
silencing the expression of MCT4 reverses this process. Evodiamine
blocks lactate-induced angiogenesis by restricting the histone
lactylation and expression of HIF-1α in PCa, further enhances
Sema3A transcription, inhibits PD-L1 transcription, and induces
ferroptosis by decreasing the expression of low GSH peroxidase 4
(122).
Proteomics analysis has shown that H4K12la is
elevated in triple-negative breast cancer (TNBC) tissues, and
H4K12la has been shown to be positively associated with the
proliferative marker Ki-67 and negatively associated with OS rate
(123). However, it is still
necessary to validate H4K12la as a biomarker for TNBC
resistance.
While lactylation is a promising therapeutic target,
integrating preclinical findings with clinical practice requires:
i) Interdisciplinary collaboration; for example, integrating
metabolomics, epigenetics and immunology to address the complexity
of the underlying mechanism. ii) Robust clinical validation, with
staged trials to evaluate safety, efficacy and related biomarkers.
iii) A patient-centered approach to develop guidelines for targeted
lactylation therapy based on tumor subtypes, TME background and
metabolic vulnerability. By addressing these challenges, lactate
regulation could become a cornerstone of precision oncology.
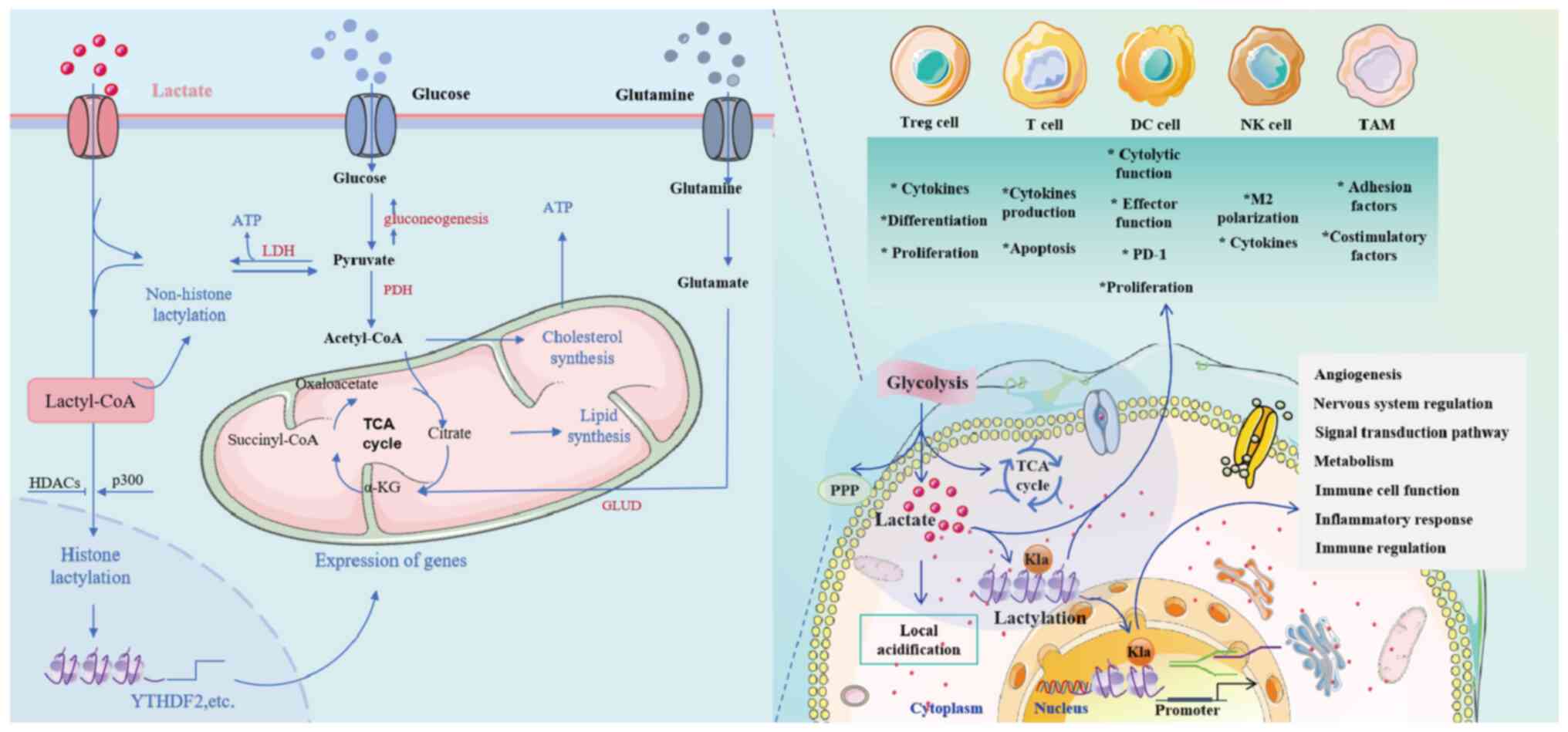
Tumor cells produce lactate through glycolysis,
resulting in local acidification and altering the pH value of the
TME. The acidified environment affects the cellular components of
the TME, and affects tumor growth and spread. Lactate acts as a
signaling molecule to regulate tumor growth, invasion, metastasis
and therapeutic response. Furthermore, lactylation is involved in
critical biological processes such as glycolytic cell function,
macrophage polarization, angiogenesis, mitochondrial activity and
nervous system regulation. Lactylation also modulates tumor
behavior by influencing signal transduction pathways. The interplay
between lactylation and the immune cycle is complex, involving
metabolism, immune cell function, inflammatory responses and immune
regulation (Fig. 3). Therefore,
intervening in lactylation may be a new target for tumor
therapy.
The structural complexity and functional diversity
of histones necessitate further exploration of their lactylation in
cancer to elucidate its role and mechanism in tumor development.
H3K18la is associated with shifts in cellular states, with
quantitative lactylation changes at promoters and enhancers
potentially driving these transitions (124). H3K9la levels are upregulated in
endothelial cells in response to VEGF stimulation, and
hyperlactylation of H3K9 inhibits expression of the lactylation
eraser HDAC2, whereas upregulation of HDAC2 decreases H3K9la and
suppresses angiogenesis (125,126). Elevated H3K9la levels in LCSCs,
HCC and GBM are associated with tumorigenicity, drug resistance and
immunosuppression (127). Histone
lactylation thus influences gene expression, tumor progression and
immune evasion, underscoring the need to determine its underlying
mechanisms for developing novel therapies.
In GC, lactate-driven lactylation of NBS1 promotes
homologous recombination (HR)-mediated DNA repair, fostering
chemoresistance, while inhibition of lactate production has emerged
as a promising strategy (128).
Similarly, MRE11 lactylation at K673, mediated by cyclic-AMP
response binding protein-binding protein (CBP), enhances HR repair,
whereas targeting CBP or LDH sensitizes tumors to chemotherapy in
preclinical models (129).
KRAS-driven lactylation activates circATXN7, which sequesters NF-κB
p65 in the cytoplasm, promoting immune evasion (130). However, the potential for
resistance to lactylation-targeted therapies remains a concern, as
tumors may exploit alternative DNA repair pathways or upregulate
compensatory PTMs, highlighting the need for combination
therapies.
While preclinical studies have demonstrated the
potential of lactylation inhibition (such as by targeting LDH or
CBP) to enhance chemosensitivity and reduce immune evasion,
clinical translation faces notable hurdles (131,132). Small molecule inhibitors and
antibodies targeting lactylation-related proteins have shown
promising prospects in preclinical models, and targeted lactylation
is expected to improve the immunosuppressive TME. For example, it
has been shown that activating the SIRT3 lactase function can
restore the anti-leukemia activity of NK cells (133). In addition, irinotecan has been
found to possess new functions of inhibiting lactylation. Their
safety is known and they can enter clinical trials more quickly
(134). A number of
small-molecule inhibitors also exhibit synergistic effects when
used in combination with existing chemotherapy drugs or
immunotherapies. Furthermore, lactylation levels vary among
different tumor types and individuals Therefore, specific
lactylation sites may be considered new and effective targets for
tumor diagnosis and treatment. Furthermore, developing highly
specific inhibitors that can precisely target specific lactylated
proteins or sites is currently a challenge, and it is necessary to
avoid interfering with other similar post-translational
modifications (such as acetylation). How to efficiently deliver
drugs to tumor tissues and penetrate into the interior of tumor
cells is a common problem faced by clinical application. New
delivery systems such as nano-strategies may be the solution. In
addition, the lactylation regulatory network is complex, and tumor
cells may develop drug resistance through other compensatory
pathways; thus, it is necessary to continuously explore the
mechanism and develop combined strategies (135,136).
Lactylation detection remains technically
challenging. While high-performance LC-MS/MS, LC/MS and anti-Kla
immunoblotting are widely used, improved precision is needed to
distinguish lactylation from other PTMs, particularly given the
overlapping molecular weights of histones H1, H3 and H4 (114). Advanced proteomics analysis and
site-specific antibodies could address these limitations.
Clinically, interventions must account for tissue-specific oxygen
gradients and lactylation heterogeneity. Although lactylation
biomarkers could monitor therapeutic response, validation in human
cohorts is lacking. Early-phase trials of lactylation inhibitors
are needed to assess safety, efficacy and optimal dosing.
As understanding of the role of lactylation in tumor
biology deepens, targeting this pathway is expected to address
treatment resistance and immunosuppression in some of the most
aggressive malignant tumors, especially those driven by glycolytic
metabolism and immune rejection. Future success will depend on a
careful balance of efficacy and toxicity, the intelligent design of
combination regimens, and the development of diagnostic tools to
identify patients most likely to benefit from lactylation-directed
therapy.
Not applicable.
This work was supported by Research Funds of Center for Xin'an
Medicine and Modernization of Traditional Chinese Medicine of
institute of Health and Medicine, Hefei Comprehensive National
Science Center (grant no. 2023CXMMTCM025), the Anhui Education
Department (grant no. gxgwfx2022019), the Anhui University of
Chinese Medicine (grant no. 2022BHTNXA05, DT2400001288) and the
Anhui Administration of Traditional Chinese Medicine (grant no.
2022CCYB10).
Not applicable.
XW, JC and BW collected and reviewed the literature,
and wrote the review. YL, XZ and YS collected and reviewed the
literature. CM made substantial contributions to the conception and
design of the work. YH is responsible for the design of this work,
the final approval of the version to be published, and for all
aspects of the work in ensuring that questions related to the
accuracy or integrity of any part of the work are appropriately
investigated and resolved. Data authentication is not applicable.
All authors read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Zhu W, Fan C, Hou Y and Zhang Y:
Lactylation in tumor microenvironment and immunotherapy resistance:
New mechanisms and challenges. Cancer Lett. 627:2178352025.
View Article : Google Scholar
|
|
2
|
Brooks GA, Curl CC, Leija RG, Osmond AD,
Duong JJ and Arevalo JA: Tracing the lactate shuttle to the
mitochondrial reticulum. Exp Mol Med. 54:1332–1347. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fan H, Yang F, Xiao Z, Luo H, Chen H, Chen
Z, Liu Q and Xiao Y: Lactylation: Novel epigenetic regulatory and
therapeutic opportunities. Am J Physiol Endocrinol Metab.
324:E330–E338. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lv X, Lv Y and Dai X: Lactate, histone
lactylation and cancer hallmarks. Expert Rev Mol Med. 25:e72023.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen L, Huang L, Gu Y, Cang W, Sun P and
Xiang Y: Lactate-lactylation hands between metabolic reprogramming
and immunosuppression. Int J Mol Sci. 23:119432022. View Article : Google Scholar
|
|
6
|
Li S, Dong L and Wang K: Current and
future perspectives of lysine lactylation in cancer. Trends Cell
Biol. 35:190–193. 2025. View Article : Google Scholar
|
|
7
|
Yu X, Yang J, Xu J, Pan H, Wang W, Yu X
and Shi S: Histone lactylation: From tumor lactate metabolism to
epigenetic regulation. Int J Biol Sci. 20:1833–1854. 2024.
View Article : Google Scholar
|
|
8
|
Zhang D, Tang Z, Huang H, Zhou G, Cui C,
Weng Y, Liu W, Kim S, Lee S, Perez-Neut M, et al: Metabolic
regulation of gene expression by histone lactylation. Nature.
574:575–580. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Irizarry-Caro RA, McDaniel MM, Overcast
GR, Jain VG, Troutman TD and Pasare C: TLR signaling adapter BCAP
regulates inflammatory to reparatory macrophage transition by
promoting histone lactylation. Proc Natl Acad Sci USA.
117:30628–30638. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang J, Yang P, Yu T, Gao M, Liu D, Zhang
J, Lu C, Chen X, Zhang X and Liu Y: Lactylation of PKM2 suppresses
inflammatory metabolic adaptation in pro-inflammatory macrophages.
Int J Biol Sci. 18:6210–6225. 2022. View Article : Google Scholar
|
|
11
|
Hua Q, Mi B, Xu F, Wen J, Zhao L, Liu J
and Huang G: Hypoxia-induced lncRNA-AC020978 promotes proliferation
and glycolytic metabolism of non-small cell lung cancer by
regulating PKM2/HIF-1α axis. Theranostics. 10:4762–4778. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Apostolova P and Pearce EL: Lactic acid
and lactate: Revisiting the physiological roles in the tumor
microenvironment. Trends Immunol. 43:969–977. 2022. View Article : Google Scholar
|
|
13
|
Wu Z, Wu H, Dai Y, Wang Z, Han H, Shen Y,
Zhang R and Wang X: A pan-cancer multi-omics analysis of
lactylation genes associated with tumor microenvironment and cancer
development. Heliyon. 10:e274652024. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang X, Li Y and Chen Y: Development of a
comprehensive gene signature linking hypoxia, glycolysis,
lactylation, and metabolomic insights in gastric cancer through the
integration of bulk and single-cell RNA-Seq data. Biomedicines.
11:29482023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang H, Chen K, Zhu Y, Hu Z, Wang Y, Chen
J, Li Y, Li D and Wei P: A multi-dimensional approach to unravel
the intricacies of lactylation related signature for prognostic and
therapeutic insight in colorectal cancer. J Transl Med. 22:2112024.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun Y, Wang H, Cui Z, Yu T, Song Y, Gao H,
Tang R, Wang X, Li B, Li W and Wang Z: Lactylation in cancer
progression and drug resistance. Drug Resist Updat. 81:1012482025.
View Article : Google Scholar
|
|
17
|
Zeng Y, Yu T, Lou Z, Chen L, Pan L and
Ruan B: Emerging function of main RNA methylation modifications in
the immune microenvironment of digestive system tumors. Pathol Res
Pract. 256:1552682024. View Article : Google Scholar
|
|
18
|
Yang Z, Yan C, Ma J, Peng P, Ren X, Cai S,
Shen X, Wu Y, Zhang S, Wang X, et al: Lactylome analysis suggests
lactylation-dependent mechanisms of metabolic adaptation in
hepatocellular carcinoma. Nat Metab. 5:61–79. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang H, Zou X, Yang S, Zhang A, Li N and
Ma Z: Identification of lactylation related model to predict
prognostic, tumor infiltrating immunocytes and response of
immunotherapy in gastric cancer. Front Immunol. 14:11499892023.
View Article : Google Scholar
|
|
20
|
Mao Y, Zhang J, Zhou Q, He X, Zheng Z, Wei
Y, Zhou K, Lin Y, Yu H, Zhang H, et al: Hypoxia induces
mitochondrial protein lactylation to limit oxidative
phosphorylation. Cell Res. 34:13–30. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zong Z, Xie F, Wang S, Wu X, Zhang Z, Yang
B and Zhou F: Alanyl-tRNA synthetase, AARS1, is a lactate sensor
and lactyltransferase that lactylates p53 and contributes to
tumorigenesis. Cell. 187:2375–2392.e33. 2024. View Article : Google Scholar
|
|
22
|
Ju J, Zhang H, Lin M, Yan Z, An L, Cao Z,
Geng D, Yue J, Tang Y, Tian L, et al: The alanyl-tRNA synthetase
AARS1 moonlights as a lactyltransferase to promote YAP signaling in
gastric cancer. J Clin Invest. 134:e1745872024. View Article : Google Scholar
|
|
23
|
Zhou J, Xu W, Wu Y, Wang M, Zhang N, Wang
L, Feng Y, Zhang T, Wang L and Mao A: GPR37 promotes colorectal
cancer liver metastases by enhancing the glycolysis and histone
lactylation via Hippo pathway. Oncogene. 42:3319–3330. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Miao Z, Zhao X and Liu X: Hypoxia induced
β-catenin lactylation promotes the cell proliferation and stemness
of colorectal cancer through the wnt signaling pathway. Exp Cell
Res. 422:1134392023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang L, Niu K, Wang J, Shen W, Jiang R,
Liu L, Song W, Wang X, Zhang X, Zhang R, et al: Nucleolin
lactylation contributes to intrahepatic cholangiocarcinoma
pathogenesis via RNA splicing regulation of MADD. J Hepatol.
81:651–666. 2024. View Article : Google Scholar
|
|
26
|
Song F, Hou C, Huang Y, Liang J, Cai H,
Tian G, Jiang Y, Wang Z and Hou J: Lactylome analyses suggest
systematic lysine-lactylated substrates in oral squamous cell
carcinoma under normoxia and hypoxia. Cell Signal. 120:1112282024.
View Article : Google Scholar
|
|
27
|
Wang M, He T, Meng D, Lv W, Ye J, Cheng L
and Hu J: BZW2 modulates lung adenocarcinoma progression through
glycolysis-mediated IDH3G lactylation modification. J Proteome Res.
22:3854–3865. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xie B, Lin J, Chen X, Zhou X, Zhang Y, Fan
M, Xiang J, He N, Hu Z and Wang F: CircXRN2 suppresses tumor
progression driven by histone lactylation through activating the
Hippo pathway in human bladder cancer. Mol Cancer. 22:1512023.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang R, Xu F, Yang Z, Cao J, Hu L and She
Y: The mechanism of PFK-1 in the occurrence and development of
bladder cancer by regulating ZEB1 lactylation. BMC Urol. 24:592024.
View Article : Google Scholar
|
|
30
|
Wang D, Du G, Chen X, Wang J, Liu K, Zhao
H, Cheng C, He Y, Jing N, Xu P, et al: Zeb1-controlled metabolic
plasticity enables remodeling of chromatin accessibility in the
development of neuroendocrine prostate cancer. Cell Death Differ.
31:779–791. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pandkar MR, Sinha S, Samaiya A and Shukla
S: Oncometabolite lactate enhances breast cancer progression by
orchestrating histone lactylation-dependent c-Myc expression.
Transl Oncol. 37:1017582023. View Article : Google Scholar
|
|
32
|
Hou X, Ouyang J, Tang L, Wu P, Deng X, Yan
Q, Shi L, Fan S, Fan C, Guo C, et al: KCNK1 promotes proliferation
and metastasis of breast cancer cells by activating lactate
dehydrogenase A (LDHA) and up-regulating H3K18 lactylation. PLoS
Biol. 22:e30026662024. View Article : Google Scholar
|
|
33
|
He Y, Ji Z, Gong Y, Fan L, Xu P, Chen X,
Miao J, Zhang K, Zhang W, Ma P, et al: Numb/Parkin-directed
mitochondrial fitness governs cancer cell fate via metabolic
regulation of histone lactylation. Cell Rep. 42:1120332023.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang X, Ying T, Yuan J, Wang Y, Su X, Chen
S, Zhao Y, Zhao Y, Sheng J, Teng L, et al: BRAFV600E restructures
cellular lactylation to promote anaplastic thyroid cancer
proliferation. Endocr Relat Cancer. 30:e2203442023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Marcucci F and Rumio C: Tumor cell
glycolysis-at the crossroad of epithelial-mesenchymal transition
and autophagy. Cells. 11:10412022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jia M, Yue X, Sun W, Zhou Q, Chang C, Gong
W, Feng J, Li X, Zhan R, Mo K, et al: ULK1-mediated metabolic
reprogramming regulates Vps34 lipid kinase activity by its
lactylation. Sci Adv. 9:eadg49932023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun W, Jia M, Feng Y and Cheng X: Lactate
is a bridge linking glycolysis and autophagy through lactylation.
Autophagy. 19:3240–3241. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ashton TM, McKenna WG, Kunz-Schughart LA
and Higgins GS: Oxidative phosphorylation as an emerging target in
cancer therapy. Clin Cancer Res. 24:2482–2490. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sancho P, Barneda D and Heeschen C:
Hallmarks of cancer stem cell metabolism. Br J Cancer.
114:1305–1312. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Payen VL, Mina E, Van Hée VF, Porporato PE
and Sonveaux P: Monocarboxylate transporters in cancer. Mol Metab.
33:48–66. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cheng Z, Huang H, Li M and Chen Y:
Proteomic analysis identifies PFKP lactylation in SW480 colon
cancer cells. iScience. 27:1086452023. View Article : Google Scholar
|
|
42
|
Huang H, Wang S, Xia H, Zhao X, Chen K,
Jin G, Zhou S, Lu Z, Chen T, Yu H, et al: Lactate enhances NMNAT1
lactylation to sustain nuclear NAD+ salvage pathway and
promote survival of pancreatic adenocarcinoma cells under
glucose-deprived conditions. Cancer Lett. 588:2168062024.
View Article : Google Scholar
|
|
43
|
Peng T, Sun F, Yang JC, Cai MH, Huai MX,
Pan JX, Zhang FY and Xu LM: Novel lactylation-related signature to
predict prognosis for pancreatic adenocarcinoma. World J
Gastroenterol. 30:2575–2602. 2024. View Article : Google Scholar
|
|
44
|
Jiang J, Huang D, Jiang Y, Hou J, Tian M,
Li J, Sun L, Zhang Y, Zhang T, Li Z, et al: Lactate modulates
cellular metabolism through histone lactylation-mediated gene
expression in non-small cell lung cancer. Front Oncol.
11:6475592021. View Article : Google Scholar
|
|
45
|
Daw CC, Ramachandran K, Enslow BT, Maity
S, Bursic B, Novello MJ, Rubannelsonkumar CS, Mashal AH,
Ravichandran J, Bakewell TM, et al: Lactate elicits
ER-mitochondrial Mg2+ dynamics to integrate cellular
metabolism. Cell. 183:474–489.e17. 2020. View Article : Google Scholar
|
|
46
|
Zhang R, Li L and Yu J: Lactate-induced
IGF1R protein lactylation promotes proliferation and metabolic
reprogramming of lung cancer cells. Open Life Sci. 19:202208742024.
View Article : Google Scholar
|
|
47
|
Seo SH, Hwang SY, Hwang S, Han S, Park H,
Lee YS, Rho SB and Kwon Y: Hypoxia-induced ELF3 promotes tumor
angiogenesis through IGF1/IGF1R. EMBO Rep. 23:e529772022.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Longhitano L, Vicario N, Tibullo D,
Giallongo C, Broggi G, Caltabiano R, Barbagallo GMV, Altieri R,
Baghini M, Di Rosa M, et al: Lactate induces the expressions of
MCT1 and HCAR1 to promote tumor growth and progression in
glioblastoma. Front Oncol. 12:8717982022. View Article : Google Scholar
|
|
49
|
Longhitano L, Giallongo S, Orlando L,
Broggi G, Longo A, Russo A, Caltabiano R, Giallongo C, Barbagallo
I, Di Rosa M, et al: Lactate rewrites the metabolic reprogramming
of uveal melanoma cells and induces quiescence phenotype. Int J Mol
Sci. 24:242022. View Article : Google Scholar
|
|
50
|
Yang L, Wang X, Liu J, Liu X, Li S, Zheng
F, Dong Q, Xu S, Xiong J and Fu B: Prognostic and tumor
microenvironmental feature of clear cell renal cell carcinoma
revealed by m6A and lactylation modification-related genes. Front
Immunol. 14:12250232023. View Article : Google Scholar
|
|
51
|
Qu J, Li P and Sun Z: Histone lactylation
regulates cancer progression by reshaping the tumor
microenvironment. Front Immunol. 14:12843442023. View Article : Google Scholar
|
|
52
|
Li F, Si W, Xia L, Yin D, Wei T, Tao M,
Cui X, Yang J, Hong T and Wei R: Positive feedback regulation
between glycolysis and histone lactylation drives oncogenesis in
pancreatic ductal adenocarcinoma. Mol Cancer. 23:902024. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen M, Cen K, Song Y, Zhang X, Liou YC,
Liu P, Huang J, Ruan J, He J, Ye W, et al:
NUSAP1-LDHA-glycolysis-lactate feedforward loop promotes Warburg
effect and metastasis in pancreatic ductal adenocarcinoma. Cancer
Lett. 567:2162852023. View Article : Google Scholar
|
|
54
|
Qiao Z, Li Y, Li S, Liu S and Cheng Y:
Hypoxia-induced SHMT2 protein lactylation facilitates glycolysis
and stemness of esophageal cancer cells. Mol Cell Biochem.
479:3063–3076. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wei X, Chen Y, Jiang X, Peng M, Liu Y, Mo
Y, Ren D, Hua Y, Yu B, Zhou Y, et al: Mechanisms of vasculogenic
mimicry in hypoxic tumor microenvironments. Mol Cancer. 20:72021.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang M, Zhao Y, Liu X, Ruan X, Wang P,
Liu L, Wang D, Dong W, Yang C and Xue Y: Pseudogene MAPK6P4-encoded
functional peptide promotes glioblastoma vasculogenic mimicry
development. Commun Biol. 6:10592023. View Article : Google Scholar
|
|
57
|
Liu R, Zou Z, Chen L, Feng Y, Ye J, Deng
Y, Zhu X, Zhang Y, Lin J, Cai S, et al: FKBP10 promotes clear cell
renal cell carcinoma progression and regulates sensitivity to the
HIF2α blockade by facilitating LDHA phosphorylation. Cell Death
Dis. 15:642024. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Luo Y, Yang Z, Yu Y and Zhang P: HIF1α
lactylation enhances KIAA1199 transcription to promote angiogenesis
and vasculogenic mimicry in prostate cancer. Int J Biol Macromol.
222:2225–2243. 2022. View Article : Google Scholar
|
|
59
|
Meng Q, Sun H, Zhang Y, Yang X, Hao S, Liu
B, Zhou H, Xu ZX and Wang Y: Lactylation stabilizes DCBLD1
activating the pentose phosphate pathway to promote cervical cancer
progression. J Exp Clin Cancer Res. 43:362024. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Meng Q, Zhang Y, Sun H, Yang X, Hao S, Liu
B, Zhou H, Wang Y and Xu ZX: Human papillomavirus-16 E6 activates
the pentose phosphate pathway to promote cervical cancer cell
proliferation by inhibiting G6PD lactylation. Redox Biol.
71:1031082024. View Article : Google Scholar
|
|
61
|
Wei S, Zhang J, Zhao R, Shi R, An L, Yu Z,
Zhang Q, Zhang J, Yao Y, Li H and Wang H: Histone lactylation
promotes malignant progression by facilitating USP39 expression to
target PI3K/AKT/HIF-1α signal pathway in endometrial carcinoma.
Cell Death Discov. 10:1212024. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhao Y, Jiang J, Zhou P, Deng K, Liu Z,
Yang M, Yang X, Li J, Li R and Xia J: H3K18 lactylation-mediated
VCAM1 expression promotes gastric cancer progression and metastasis
via AKT-mTOR-CXCL1 axis. Biochem Pharmacol. 222:1161202024.
View Article : Google Scholar
|
|
63
|
Li XM, Yang Y, Jiang FQ, Hu G, Wan S, Yan
WY, He XS, Xiao F, Yang XM, Guo X, et al: Histone lactylation
inhibits RARγ expression in macrophages to promote colorectal
tumorigenesis through activation of TRAF6-IL-6-STAT3 signaling.
Cell Rep. 43:1136882024. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Liu M, Gu L, Zhang Y, Li Y, Zhang L, Xin
Y, Wang Y and Xu ZX: LKB1 inhibits telomerase activity resulting in
cellular senescence through histone lactylation in lung
adenocarcinoma. Cancer Lett. 595:2170252024. View Article : Google Scholar
|
|
65
|
Zheng P, Mao Z, Luo M, Zhou L, Wang L, Liu
H, Liu W and Wei S: Comprehensive bioinformatics analysis of the
solute carrier family and preliminary exploration of SLC25A29 in
lung adenocarcinoma. Cancer Cell Int. 23:2222023. View Article : Google Scholar
|
|
66
|
Li L, Li Z, Meng X, Wang X, Song D, Liu Y,
Xu T, Qin J, Sun N, Tian K, et al: Histone lactylation-derived
LINC01127 promotes the self-renewal of glioblastoma stem cells via
the cis-regulating the MAP4K4 to activate JNK pathway. Cancer Lett.
579:2164672023. View Article : Google Scholar
|
|
67
|
Yang J, Luo L, Zhao C, Li X, Wang Z, Zeng
Z, Yang X, Zheng X, Jie H, Kang L, et al: A positive feedback loop
between inactive VHL-triggered histone lactylation and PDGFRβ
signaling drives clear cell renal cell carcinoma progression. Int J
Biol Sci. 18:3470–3483. 2022. View Article : Google Scholar
|
|
68
|
Jin J, Bai L, Wang D, Ding W, Cao Z, Yan
P, Li Y, Xi L, Wang Y, Zheng X, et al: SIRT3-dependent
delactylation of cyclin E2 prevents hepatocellular carcinoma
growth. EMBO Rep. 24:e560522023. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Liao J, Chen Z, Chang R, Yuan T, Li G, Zhu
C, Wen J, Wei Y, Huang Z, Ding Z, et al: CENPA functions as a
transcriptional regulator to promote hepatocellular carcinoma
progression via cooperating with YY1. Int J Biol Sci. 19:5218–5232.
2023. View Article : Google Scholar
|
|
70
|
Xie B, Zhang M, Li J, Cui J, Zhang P, Liu
F, Wu Y, Deng W, Ma J, Li X, et al: KAT8-catalyzed lactylation
promotes eEF1A2-mediated protein synthesis and colorectal
carcinogenesis. Proc Natl Acad Sci USA. 121:e23141281212024.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Xiong J, He J, Zhu J, Pan J, Liao W, Ye H,
Wang H, Song Y, Du Y, Cui B, et al: Lactylation-driven
METTL3-mediated RNA m6A modification promotes
immunosuppression of tumor-infiltrating myeloid cells. Mol Cell.
82:1660–1677.e10. 2022. View Article : Google Scholar
|
|
72
|
Yu J, Chai P, Xie M, Ge S, Ruan J, Fan X
and Jia R: Histone lactylation drives oncogenesis by facilitating
m6A reader protein YTHDF2 expression in ocular melanoma.
Genome Biol. 22:852021. View Article : Google Scholar
|
|
73
|
Gu X, Zhuang A, Yu J, Yang L, Ge S, Ruan
J, Jia R, Fan X and Chai P: Histone lactylation-boosted ALKBH3
potentiates tumor progression and diminished promyelocytic leukemia
protein nuclear condensates by m1A demethylation of SP100A. Nucleic
Acids Res. 52:2273–2289. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chen B, Deng Y, Hong Y, Fan L, Zhai X, Hu
H, Yin S, Chen Q, Xie X, Ren X, et al: Metabolic recoding of
NSUN2-mediated m5c modification promotes the progression
of colorectal cancer via the NSUN2/YBX1/m5C-ENO1
positive feedback loop. Adv Sci (Weinh). 11:e23098402024.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Takata T, Nakamura A, Yasuda H, Miyake H,
Sogame Y, Sawai Y, Hayakawa M, Mochizuki K, Nakao R, Ogata T, et
al: Pathophysiological implications of protein lactylation in
pancreatic epithelial tumors. Acta Histochem Cytochem. 57:57–66.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Cai D, Yuan X, Cai DQ, Li A, Yang S, Yang
W, Duan J, Zhuo W, Min J, Peng L and Wei J: Integrative analysis of
lactylation-related genes and establishment of a novel prognostic
signature for hepatocellular carcinoma. J Cancer Res Clin Oncol.
149:11517–11530. 2023. View Article : Google Scholar
|
|
77
|
Yang H, Yang S, He J, Li W, Zhang A, Li N,
Zhou G and Sun B: Glucose transporter 3 (GLUT3) promotes
lactylation modifications by regulating lactate dehydrogenase A
(LDHA) in gastric cancer. Cancer Cell Int. 23:3032023. View Article : Google Scholar
|
|
78
|
Yang D, Yin J, Shan L, Yi X, Zhang W and
Ding Y: Identification of lysine-lactylated substrates in gastric
cancer cells. iScience. 25:1046302022. View Article : Google Scholar
|
|
79
|
Hu X, Ouyang W, Chen H, Liu Z, Lai Z and
Yao H: Claudin-9 (CLDN9) promotes gastric cancer progression by
enhancing the glycolysis pathway and facilitating PD-L1 lactylation
to suppress CD8+ T cell anti-tumor immunity. Cancer
Pathog Ther. 3:253–266. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Huang ZW, Zhang XN, Zhang L, Liu LL, Zhang
JW, Sun YX, Xu JQ, Liu Q and Long ZJ: STAT5 promotes PD-L1
expression by facilitating histone lactylation to drive
immunosuppression in acute myeloid leukemia. Signal Transduct
Target Ther. 8:3912023. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Gu J, Xu X, Li X, Yue L, Zhu X, Chen Q,
Gao J, Takashi M, Zhao W, Zhao B, et al: Tumor-resident microbiota
contributes to colorectal cancer liver metastasis by lactylation
and immune modulation. Oncogene. 43:2389–2404. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang J, Liu Z, Xu Y, Wang Y, Wang F, Zhang
Q, Ni C, Zhen Y, Xu R, Liu Q, et al: Enterobacterial LPS-inducible
LINC00152 is regulated by histone lactylation and promotes cancer
cells invasion and migration. Front Cell Infect Microbiol.
12:9138152022. View Article : Google Scholar
|
|
83
|
Hao B, Dong H, Xiong R, Song C, Xu C, Li N
and Geng Q: Identification of SLC2A1 as a predictive biomarker for
survival and response to immunotherapy in lung squamous cell
carcinoma. Comput Biol Med. 171:1081832024. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
De Leo A, Ugolini A, Yu X, Scirocchi F,
Scocozza D, Peixoto B, Pace A, D'Angelo L, Liu JKC, Etame AB, et
al: Glucose-driven histone lactylation promotes the
immunosuppressive activity of monocyte-derived macrophages in
glioblastoma. Immunity. 57:1105–1123.e8. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Lu X, Zhou Z, Qiu P and Xin T: Integrated
single-cell and bulk RNA-sequencing data reveal molecular subtypes
based on lactylation-related genes and prognosis and therapeutic
response in glioma. Heliyon. 10:e307262024. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Wu X, Mi T, Jin L, Ren C, Wang J, Zhang Z,
Liu J, Wang Z, Guo P and He D: Dual roles of HK3 in regulating the
network between tumor cells and tumor-associated macrophages in
neuroblastoma. Cancer Immunol Immunother. 73:1222024. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wang ZH, Zhang P, Peng WB, Ye LL, Xiang X,
Wei XS, Niu YR, Zhang SY, Xue QQ, Wang HL and Zhou Q: Altered
phenotypic and metabolic characteristics of
FOXP3+CD3+CD56+ natural killer T
(NKT)-like cells in human malignant pleural effusion.
Oncoimmunology. 12:21605582022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Hong H, Chen X, Wang H, Gu X, Yuan Y and
Zhang Z: Global profiling of protein lysine lactylation and
potential target modified protein analysis in hepatocellular
carcinoma. Proteomics. 23:e22004322023. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Cheng Z, Huang H, Li M, Liang X, Tan Y and
Chen Y: Lactylation-related gene signature effectively predicts
prognosis and treatment responsiveness in hepatocellular carcinoma.
Pharmaceuticals (Basel). 16:6442023. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Wu Q, Li X, Long M, Xie X and Liu Q:
Integrated analysis of histone lysine lactylation (Kla)-specific
genes suggests that NR6A1, OSBP2 and UNC119B are novel therapeutic
targets for hepatocellular carcinoma. Sci Rep. 13:186422023.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Lai JP, Sandhu DS, Moser CD, Cazanave SC,
Oseini AM, Shire AM, Shridhar V, Sanderson SO and Roberts LR:
Additive effect of apicidin and doxorubicin in sulfatase 1
expressing hepatocellular carcinoma in vitro and in vivo. J
Hepatol. 50:1112–1121. 2009. View Article : Google Scholar
|
|
92
|
Wang Y, Pei P, Yang K, Guo L and Li Y:
Copper in colorectal cancer: From copper-related mechanisms to
clinical cancer therapies. Clin Transl Med. 14:e17242024.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Sun L, Zhang Y, Yang B, Sun S, Zhang P,
Luo Z, Feng T, Cui Z, Zhu T, Li Y, et al: Lactylation of METTL16
promotes cuproptosis via m6A-modification on FDX1 mRNA
in gastric cancer. Nat Commun. 14:65232023. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Li W, Zhou C, Yu L, Hou Z, Liu H, Kong L,
Xu Y, He J, Lan J, Ou Q, et al: Tumor-derived lactate promotes
resistance to bevacizumab treatment by facilitating autophagy
enhancer protein RUBCNL expression through histone H3 lysine 18
lactylation (H3K18la) in colorectal cancer. Autophagy. 20:114–130.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Sun X, He L, Liu H, Thorne RF, Zeng T, Liu
L, Zhang B, He M, Huang Y, Li M, et al: The diapause-like
colorectal cancer cells induced by SMC4 attenuation are
characterized by low proliferation and chemotherapy insensitivity.
Cell Metab. 35:1563–1579.e8. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Chu YD, Cheng LC, Lim SN, Lai MW, Yeh CT
and Lin WR: Aldolase B-driven lactagenesis and CEACAM6 activation
promote cell renewal and chemoresistance in colorectal cancer
through the Warburg effect. Cell Death Dis. 14:6602023. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Brown TP and Ganapathy V: Lactate/GPR81
signaling and proton motive force in cancer: Role in angiogenesis,
immune escape, nutrition, and Warburg phenomenon. Pharmacol Ther.
206:1074512020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Xia L, Oyang L, Lin J, Tan S, Han Y, Wu N,
Yi P, Tang L, Pan Q, Rao S, et al: The cancer metabolic
reprogramming and immune response. Mol Cancer. 20:282021.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Yan F, Teng Y, Li X, Zhong Y, Li C, Yan F
and He X: Hypoxia promotes non-small cell lung cancer cell
stemness, migration, and invasion via promoting glycolysis by
lactylation of SOX9. Cancer Biol Ther. 25:23041612024. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Duan W, Liu W, Xia S, Zhou Y, Tang M, Xu
M, Lin M, Li X and Wang Q: Warburg effect enhanced by AKR1B10
promotes acquired resistance to pemetrexed in lung cancer-derived
brain metastasis. J Transl Med. 21:5472023. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zheng H, Peng X, Yang S, Li X, Huang M,
Wei S, Zhang S, He G, Liu J, Fan Q, et al: Targeting
tumor-associated macrophages in hepatocellular carcinoma: Biology,
strategy, and immunotherapy. Cell Death Discov. 9:652023.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Li F, Zhang H, Huang Y, Li D, Zheng Z, Xie
K, Cao C, Wang Q, Zhao X, Huang Z, et al: Single-cell transcriptome
analysis reveals the association between histone lactylation and
cisplatin resistance in bladder cancer. Drug Resist Updat.
73:1010592024. View Article : Google Scholar
|
|
103
|
Li K, Chen MK, Li LY, Lu MH, Shao ChK, Su
ZL, He D, Pang J and Gao X: The predictive value of semaphorins 3
expression in biopsies for biochemical recurrence of patients with
low- and intermediate-risk prostate cancer. Neoplasma. 60:683–689.
2013. View Article : Google Scholar
|
|
104
|
Pan J, Zhang J, Lin J, Cai Y and Zhao Z:
Constructing lactylation-related genes prognostic model to
effectively predict the disease-free survival and treatment
responsiveness in prostate cancer based on machine learning. Front
Genet. 15:13431402024. View Article : Google Scholar
|
|
105
|
Chaudagar K, Hieromnimon HM, Khurana R,
Labadie B, Hirz T, Mei S, Hasan R, Shafran J, Kelley A, Apostolov
E, et al: Reversal of lactate and PD-1-mediated macrophage
immunosuppression controls growth of PTEN/p53-deficient prostate
cancer. Clin Cancer Res. 29:1952–1968. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Chaudagar K, Hieromnimon HM, Kelley A,
Labadie B, Shafran J, Rameshbabu S, Drovetsky C, Bynoe K, Solanki
A, Markiewicz E, et al: Suppression of tumor cell
lactate-generating signaling pathways eradicates murine
PTEN/p53-deficient aggressive-variant prostate cancer via
macrophage phagocytosis. Clin Cancer Res. 29:4930–4940. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Zhang XW, Li L, Liao M, Liu D, Rehman A,
Liu Y, Liu ZP, Tu PF and Zeng KW: Thermal proteome profiling
strategy identifies CNPY3 as a cellular target of gambogic acid for
inducing prostate cancer pyroptosis. J Med Chem. 67:10005–10011.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Jiao Y, Ji F, Hou L, Lv Y and Zhang J:
Lactylation-related gene signature for prognostic prediction and
immune infiltration analysis in breast cancer. Heliyon.
10:e247772024. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Deng J and Liao X: Lysine lactylation
(Kla) might be a novel therapeutic target for breast cancer. BMC
Med Genomics. 16:2832023. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Yu L, Jing C, Zhuang S, Ji L and Jiang L:
A novel lactylation-related gene signature for effectively
distinguishing and predicting the prognosis of ovarian cancer.
Transl Cancer Res. 13:2497–2508. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Zhu Y and Song B, Yang Z, Peng Y, Cui Z,
Chen L and Song B: Integrative lactylation and tumor
microenvironment signature as prognostic and therapeutic biomarkers
in skin cutaneous melanoma. J Cancer Res Clin Oncol.
149:17897–17919. 2023. View Article : Google Scholar
|
|
112
|
Feng H, Chen W and Zhang C: Identification
of lactylation gene CALML5 and its correlated lncRNAs in cutaneous
melanoma by machine learning. Medicine (Baltimore). 102:e359992023.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Pan L, Feng F, Wu J, Fan S, Han J, Wang S,
Yang L, Liu W, Wang C and Xu K: Demethylzeylasteral targets lactate
by inhibiting histone lactylation to suppress the tumorigenicity of
liver cancer stem cells. Pharmacol Res. 181:1062702022. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Gong H, Xu HM, Ma YH and Zhang DK:
Demethylzeylasteral targets lactate to suppress the tumorigenicity
of liver cancer stem cells: It is attributed to histone
lactylation? Pharmacol Res. 194:1068692023. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Wang C, Feng F, Pan L and Xu K: Reply to
the letter titled: Demethylzeylasteral targets lactate to suppress
the tumorigenicity of liver cancer stem cells: Is it attributed to
histone lactylation? Pharmacol Res. 194:1068682023. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Xu H, Li L, Wang S, Wang Z, Qu L, Wang C
and Xu K: Royal jelly acid suppresses hepatocellular carcinoma
tumorigenicity by inhibiting H3 histone lactylation at H3K9la and
H3K14la sites. Phytomedicine. 118:1549402023. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Guo Z, Tang Y, Wang S, Huang Y, Chi Q, Xu
K and Xue L: Natural product fargesin interferes with H3 histone
lactylation via targeting PKM2 to inhibit non-small cell lung
cancer tumorigenesis. Biofactors. 50:592–607. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Gu J, Zhou J, Chen Q, Xu X, Gao J, Li X,
Shao Q, Zhou B, Zhou H, Wei S, et al: Tumor metabolite lactate
promotes tumorigenesis by modulating MOESIN lactylation and
enhancing TGF-β signaling in regulatory T cells. Cell Rep.
39:1109862022. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Yue Q, Wang Z, Shen Y, Lan Y, Zhong X, Luo
X, Yang T, Zhang M, Zuo B, Zeng T, et al: Histone H3K9 lactylation
confers temozolomide resistance in glioblastoma via LUC7L2-Mediated
MLH1 intron retention. Adv Sci (Weinh). 11:e23092902024. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Sun T, Liu B, Li Y, Wu J, Cao Y, Yang S,
Tan H, Cai L, Zhang S, Qi X, et al: Oxamate enhances the efficacy
of CAR-T therapy against glioblastoma via suppressing
ectonucleotidases and CCR8 lactylation. J Exp Clin Cancer Res.
42:2532023. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Hu X, Wang J, Chu M, Liu Y, Wang ZW and
Zhu X: Emerging role of ubiquitination in the regulation of
PD-1/PD-L1 in cancer immunotherapy. Mol Ther. 29:908–919. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Yu Y, Huang X, Liang C and Zhang P:
Evodiamine impairs HIF1A histone lactylation to inhibit
Sema3A-mediated angiogenesis and PD-L1 by inducing ferroptosis in
prostate cancer. Eur J Pharmacol. 957:1760072023. View Article : Google Scholar
|
|
123
|
Cui Z, Li Y, Lin Y, Zheng C, Luo L, Hu D,
Chen Y, Xiao Z and Sun Y: Lactylproteome analysis indicates histone
H4K12 lactylation as a novel biomarker in triple-negative breast
cancer. Front Endocrinol (Lausanne). 15:13286792024. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Galle E, Wong CW, Ghosh A, Desgeorges T,
Melrose K, Hinte LC, Castellano-Castillo D, Engl M, de Sousa JA,
Ruiz-Ojeda FJ, et al: H3K18 lactylation marks tissue-specific
active enhancers. Genome Biol. 23:2072022. View Article : Google Scholar
|
|
125
|
Fan W, Zeng S, Wang X, Wang G, Liao D, Li
R, He S, Li W, Huang J, Li X, et al: A feedback loop driven by H3K9
lactylation and HDAC2 in endothelial cells regulates VEGF-induced
angiogenesis. Genome Biol. 25:1652024. View Article : Google Scholar
|
|
126
|
Dai W, Wu G, Liu K, Chen Q, Tao J, Liu H
and Shen M: Lactate promotes myogenesis via activating H3K9
lactylation-dependent up-regulation of Neu2 expression. J Cachexia
Sarcopenia Muscle. 14:2851–2865. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Li X, Chen M, Chen X, He X, Li X, Wei H,
Tan Y, Min J, Azam T, Xue M, et al: TRAP1 drives smooth muscle cell
senescence and promotes atherosclerosis via HDAC3-primed histone H4
lysine 12 lactylation. Eur Heart J. 45:4219–4235. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Chen H, Li Y, Li H, Chen X, Fu H, Mao D,
Chen W, Lan L, Wang C, Hu K, et al: NBS1 lactylation is required
for efficient DNA repair and chemotherapy resistance. Nature.
631:663–669. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Chen Y, Wu J, Zhai L, Zhang T, Yin H, Gao
H, Zhao F, Wang Z, Yang X, Jin M, et al: Metabolic regulation of
homologous recombination repair by MRE11 lactylation. Cell.
187:294–311.e21. 2024. View Article : Google Scholar
|
|
130
|
Zhou C, Li W, Liang Z, Wu X, Cheng S, Peng
J, Zeng K, Li W, Lan P, Yang X, et al: Mutant KRAS-activated
circATXN7 fosters tumor immunoescape by sensitizing tumor-specific
T cells to activation-induced cell death. Nat Commun. 15:4992024.
View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Deng D, Luo Y, Hong Y, Ren X, Zu X and
Feng J: Lactylation: A new direction for tumor-targeted therapy.
Biochim Biophys Acta Rev Cancer. 1880:1893992025. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Hou X, Hong Z, Zen H, Zhang C, Zhang P, Ma
D and Han Z: Lactylation in cancer biology: Unlocking new avenues
for research and therapy. Cancer Commun (Lond). Aug 19–2025.(Epub
ahead of print). View Article : Google Scholar
|
|
133
|
Jin J, Yan P, Wang D, Bai L, Liang H, Zhu
X, Zhu H, Ding C, Wei H and Wang Y: Targeting lactylation
reinforces NK cell cytotoxicity within the tumor microenvironment.
Nat Immunol. 26:1099–1112. 2025. View Article : Google Scholar
|
|
134
|
Li X, Zhang C, Mei Y, Zhong W, Fan W, Liu
L, Feng Z, Bai X, Liu C, Xiao M, et al: Irinotecan alleviates
chemoresistance to anthracyclines through the inhibition of
AARS1-mediated BLM lactylation and homologous recombination repair.
Signal Transduct Target Ther. 10:2142025. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Peng X and Du J: Histone and non-histone
lactylation: Molecular mechanisms, biological functions, diseases,
and therapeutic targets. Mol Biomed. 6:382025. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Chen X, Yuan Y, Zhou F, Li L, Pu J, Zeng Y
and Jiang X: Lactylation: From homeostasis to pathological
implications and therapeutic strategies. MedComm (2020).
6:e702262025. View Article : Google Scholar : PubMed/NCBI
|