Introduction
Lung cancer is a prevalent cause of cancer-related
mortality and poses a notable health risk (1). Lung cancer comprises several
histological subtypes, including adenocarcinoma, squamous carcinoma
and large cell carcinoma, which are collectively known as non-small
cell lung cancer (NSCLC) and small cell lung cancer (2). NSCLC accounts for 80–90% of all lung
cancer cases (3). Because of the
poor prognosis of NSCLC and limited treatment options due to issues
such as late-stage drug resistance, there is a need to identify
novel therapeutic targets and strategies (3,4). For
diseases such as lung cancer, optimization of treatment strategies
often requires multifaceted considerations.
The regulation of biological functions in lung
cancer cells involves multiple levels and molecules. A preclinical
study revealed the promise of microRNA (miRNA/miR) mimics and
modulators in lung cancer therapy, potentially enhancing
conventional treatments by reversing drug resistance and increasing
cancer cell sensitivity (1). miRNA
therapy has entered the clinical trial phase, demonstrating its
safety and potential for therapeutic applications. For instance,
the miRNA-16 mimic exhibited preliminary efficacy and tolerability
in phase I clinical trials for NSCLC and malignant mesothelioma
(5). The positive outcomes of the
aforementioned preliminary trial provide evidence for miRNA
replacement therapy. miR-199a exhibits tumor-suppressive effects in
NSCLC by inhibiting growth and metastasis in vivo (6). miR-199a can inhibit the development
of lung cancer, particularly by suppressing the proliferation,
migration and infiltration of lung cancer cells, as well as by
inhibiting tumor angiogenesis and promoting lung cancer cell
apoptosis, thereby influencing lung cancer cell drug resistance
(7). miR-199a comprises two mature
forms, miR-199a-3p and miR-199a-5p, both of which serve important
biological roles (7). A previous
study has revealed that miR-199a-3p/5p was downregulated in NSCLC
tissues, cell lines and patient samples, and its overexpression
inhibited the malignant behavior of cells (6). Overexpression of miR-199a-3p markedly
inhibits cell proliferation, migration and invasion in esophageal
squamous cell carcinoma (8).
Additionally, miR-199a-3p is substantially downregulated in
nasopharyngeal carcinoma tissues and cells, and it can inhibit the
migration and invasion of nasopharyngeal carcinoma cells by
downregulating stearoyl-CoA desaturase 1 expression (9). However, the specific mechanism of
action of miR-199a-3p in NSCLC remains to be elucidated. Therefore,
the present study focused on miR-199a-3p and explored its potential
as a therapeutic target.
Fat mass and obesity-associated protein (FTO) was
the first identified RNA N6-methyladenosine
(m6A) demethylase in eukaryotic cells, and regulates
m6A levels in mRNAs and serves a key role in tumor
progression and drug resistance, including in lung cancer (10,11).
m6A is the most abundant internal mRNA modification that
regulates processes such as alternative splicing and translation
(11). Transcriptome-wide
m6A maps, known as the epitranscriptome, reveal the
distribution and patterns of m6A in cellular RNAs. These
maps reveal specific mRNAs regulated by m6A, providing
mechanistic links between m6A and cell differentiation,
cancer progression and other processes (12). FTO promotes adipogenesis by
regulating the adipogenic pathway (13). m6A and FTO levels are
dysregulated in multiple cancer types, including lung cancer
(11). A study by Xu et al
(14) reported that FTO expression
was lower in NSCLC samples compared with healthy tissues and was
negatively associated with poor prognosis. Functional assays have
revealed that FTO inhibits NSCLC tumor cell proliferation and
metastasis in vitro and in vivo (14). However, a previous study has
reported an oncogenic role of FTO in acute myeloid leukemia
(15). This discrepancy suggests
that FTO functions are tissue- or cell-context-dependent.
Myeloid zinc finger 1 (MZF1), a SCAN-domain zinc
finger transcription factor associated with numerous types of
cancers, shows decreased expression in pancreatic cancer and NSCLC
tumors compared to normal tissues (16). High MZF1 expression is associated
with lower overall survival in patients with glioblastoma
multiforme (17). However, the
specific role of MZF1 in lung cancer remains to be fully
elucidated.
Claudin domain-containing 1 (CLDND1), also known as
claudin-25, has been identified as a novel member of the claudin
protein family (18). Claudins
serve important roles in intercellular adhesion. Elevated
expression of CLDND1 is associated with malignant progression in
estrogen receptor-negative, hormone therapy-resistant breast
cancer. Aberrant CLDND1 levels are also linked to vascular
pathologies (19). A study
investigating the regulation of CLDND1 expression identified a
strong enhancer region near its promoter that shares high homology
with the ETS domain-containing protein Elk-1 (ELK1) binding
sequence, suggesting that CLDND1 may be a potential target for
anticancer drug development (19).
However, to the best of our knowledge, no studies have investigated
the specific role of CLDND1 in the development of lung cancer.
The present study aimed to investigate a novel
signaling axis involving miR-199a-3p, FTO, MZF1, and CLDND1 in
NSCLC. Bioinformatics tools including miRwalk were employed to
predict potential downstream targets of miR-199a-3p. The SRAMP
online tool was utilized to assess the potential for m6A
modification in MZF1 mRNA, while the JASPAR database was used to
analyze the binding affinity of MZF1 to the CLDND1 promoter region.
It was hypothesized that FTO, as an m6A demethylase, may
regulate MZF1 expression via m6A modification, thereby
influencing CLDND1 transcription and contributing to NSCLC
progression. The study was designed to experimentally validate this
proposed miR-199a-3p/FTO/m6A/MZF1/CLDND1 axis, with the
goal of elucidating its functional role in NSCLC and exploring its
potential as a diagnostic or therapeutic target.
Materials and methods
Ethics statement
The present study was approved by the Ethics
Committee of Yanbian University Affiliated Hospital (approval no.
2021094; Yanji, China), and written informed consent was obtained
from all patients. The study adhered to the principles outlined in
The Declaration of Helsinki.
Clinical samples
Lung cancer tissue specimens were collected from 15
patients with NSCLC (adenocarcinoma) who underwent surgical
resection at Yanbian University Affiliated Hospital (Yanji, China)
between January 2022 and June 2023. For each patient, a paired
paracancerous tissue specimen (≥5 cm from the tumor margin and
pathologically confirmed to be free of tumor cell infiltration) was
simultaneously collected.
Inclusion criteria were: i) histopathological
confirmation of primary NSCLC (adenocarcinoma), ii) no prior
neoadjuvant chemotherapy, radiotherapy, or targeted therapy before
surgery, and iii) availability of complete clinicopathological
data. Exclusion criteria were: i) presence of other malignant
tumors or severe systemic diseases, ii) history of autoimmune or
infectious diseases, and iii) inadequate tissue quantity or RNA
quality. Immediately after resection, all tissue specimens were
snap-frozen in liquid nitrogen and stored at −80°C until further
analysis to preserve molecular integrity.
A priori power analysis was performed using G*Power
3.1 (Heinrich Heine University Düsseldorf, Germany), setting α=0.05
and an effect size=0.8, which indicated that n=15 achieves a
statistical power >85%. Patients' ages ranged from 35 to 69
years (median=52 years). All diagnoses were confirmed by the
Department of Pathology. The clinical characteristics of patients
with NSCLC are shown in Table
SI.
Cell culture and grouping
CAL12T (cat. no. MZ-8194), HCC44 (cat. no. MZ-8190),
NCIH1993 (cat. no. MZ-8234) and A549 (cat. no. B163886) lung cancer
cells, and BEAS-2B human normal bronchial epithelial cells (cat.
no. MZ-0677), were obtained from Ningbo Mingzhou Biotechnology Co.,
Ltd. All cells were cultured in DMEM; cat. no. 11965092; Gibco,
Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (FBS; cat. no. 10099141C; Gibco, Thermo Fisher Scientific,
Inc.) and 1% penicillin-streptomycin (100 U/ml penicillin, 100
µg/ml streptomycin; cat. no. 15140122; all Gibco) at 37°C in a
humidified incubator containing 5% CO2. Cells were
sub-cultured at 70–80% confluence using 0.25% trypsin-EDTA
(HyClone; Cytiva), and only cells in the logarithmic growth phase
were used for experiments. All cell lines were authenticated via
short tandem repeat analysis and regularly tested for the absence
of mycoplasma contamination (Table
SII).
The A549 cells were divided into the following
groups based on the experimental needs: i) miR-199a-3p mimic group
transfected with miR-199a-3p mimic; ii) negative control (NC) mimic
group transfected with miR-199a-3p mimic NC sequence; iii) NC mimic
+ overexpression (oe)-NC group transfected with miR-199a-3p mimic
NC sequence and FTO overexpression NC plasmid; iv) miR-199a-3p
mimic + oe-NC group transfected with miR-199a-3p mimic and FTO
overexpression NC plasmid; v) NC mimic + oe-FTO group transfected
with miR-199a-3p mimic NC sequence and FTO overexpression plasmid;
vi) miR-199a-3p mimic + oe-FTO group transfected with miR-199a-3p
mimic and FTO overexpression plasmid; vii) oe-NC group transfected
with overexpression NC plasmid; viii) oe-FTO group transfected with
FTO overexpression plasmid; ix) oe-MZF1 group transfected with MZF1
overexpression plasmid; x) oe-NC + short hairpin RNA (sh)-NC group
transfected with FTO overexpression and MZF1 silencing NC plasmid;
xi) oe-FTO + sh-NC group transfected with FTO overexpression
plasmid and MZF1 silencing NC plasmid; xii) oe-NC + sh-MZF1 group
transfected with FTO overexpression NC plasmid and MZF1 silencing
plasmid; xiii) oe-FTO + sh-MZF1 group transfected with FTO
overexpression plasmid and MZF1 silencing plasmid; xiv) oe-MZF1 +
sh-NC group transfected with MZF1 overexpression plasmid and CLDND1
silencing NC plasmid; xv) oe-NC + sh-CLDND1 group transfected with
MZF1 overexpression and CLDND1 silencing plasmid; and xvi) oe-MZF1
+ sh-CLDND1 group transfected with MZF1 overexpression plasmid and
CLDND1 silencing plasmid.
The FTO and MZF1 oe plasmids were constructed using
the pCMV6-AC-GFP vector (Hunan Fenghui Biotechnology Co., Ltd.;
cat. no. ZT877), and the MZF1 and CLDND1 silencing plasmids
(sh-MZF1, sh-CLDND1) along with their negative controls (sh-NC)
were generated using the pGPU6-GFP-Neo backbone (Hunan Fenghui
Biotechnology Co., Ltd.; cat. no. BR150). Synthetic miR-199a-3p
mimic and NC mimic were purchased from Shanghai GenePharma Co.,
Ltd., and prepared at a working concentration of 50 nM. All nucleic
acids were transfected using Lipofectamine® 2000 (cat.
no. 11668019; Invitrogen, Thermo Fisher Scientific, Inc.) following
the manufacturer's protocol. For each well of a 6-well plate, 4 µg
plasmid DNA or 50 nM miRNA mimic was mixed with 10 µl Lipofectamine
2000 in 250 µl serum-free Opti-MEM (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C with 5% CO2 for 6 h before the
medium was replaced with complete DMEM, and harvested 24–48 h
post-transfection for subsequent assays (Fig. S1) (20). Sequence information is shown in
Table SIII.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from A549 cells and human
NSCLC tumor and paired paracancerous tissue using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's instructions. cDNA was synthesized with the
PrimeScript RT reagent kit with gDNA Eraser (cat. no. RR047A;
Takara Bio, Inc.) using the following program: genomic DNA removal
at 42°C for 2 min, reverse transcription at 37°C for 15 min, and
enzyme inactivation at 85°C for 5 sec. For miRNA analysis, cDNA was
generated using the miRNA First Strand cDNA Synthesis (Tailing
Reaction) kit (cat. no. B532451-0020; Sangon Biotech Co., Ltd.)
with a tailing/RT program of 37°C for 60 min followed by 85°C for 5
min. qPCR was performed on a 7900HT Fast Real-Time PCR System
(Applied Biosystems, Thermo Fisher Scientific, Inc.) using
SYBR™ Green PCR Master Mix (Applied Biosystems). The
thermal cycling conditions were: 95°C for 10 min, then 40 cycles of
95°C for 15 sec and 60°C for 60 sec; melt-curve analysis was
conducted from 60–95°C with a gradual ramp. The relative gene
expression levels were calculated using the 2−ΔΔCq
method (21), with U6 as the
internal reference for miRNA and GAPDH as the reference for mRNA.
All primers were purchased from Sangon Biotech Co., Ltd. (Table I) (22). The experiment was repeated three
times.
 | Table I.Primer sequences for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences for reverse
transcription-quantitative PCR.
| Gene | Primer sequence,
5′à3′ |
|---|
| miR-199a-3p |
|
| (human) |
|
|
Forward |
ACAGTAGTCTGCACATTGGTTA |
|
Reverse |
GTGCAGGGTCCGAGGT |
| FTO (human) |
|
|
Forward |
GTTCACAACCTCGGTTTAGTTC |
|
Reverse |
CATCATCATTGTCCACATCGTC |
| MZF1 (human) |
|
|
Forward |
TTCCGGTGCTTCCGCTATG |
|
Reverse |
CTCCTTGGAGCGTACCTCT |
| CLDND1 (human) |
|
|
Forward |
TGGTATAGCCCACCAGAAAGGAC |
|
Reverse |
CAATCCCGCTATTGTGGTTTCCG |
| U6 (human) |
|
|
Forward |
CTCGCTTCGGCAGCACA |
|
Reverse |
AACGCTTCACGAATTTGCGT |
| GAPDH (human) |
|
|
Forward |
GGAGCGAGATCCCTCCAAAAT |
|
Reverse |
GGCTGTTGTCATACTTCTCATGG |
Western blot analysis
Total protein was extracted from human NSCLC tumor
and paired paracancerous tissue using RIPA lysis buffer (cat. no.
P0013C; Beyotime Institute of Biotechnology) containing 1 mM
phenylmethylsulfonyl fluoride. Following centrifugation at 12,000 ×
g for 10 min at 4°C, the supernatant was collected, and the protein
concentration was quantified using a BCA Protein Assay Kit (cat.
no. 23227; Thermo Fisher Scientific, Inc.). Equal amounts of
protein (30 µg per lane) were loaded and separated by 10%
SDS-polyacrylamide gel electrophoresis (PAGE), followed by transfer
onto polyvinylidene difluoride (PVDF) membranes. The membranes were
blocked with 5% non-fat dry milk (BD Difco™; BD
Biosciences) in Tris-buffered saline with 0.1% Tween-20 (TBST) at
room temperature for 1 h, then incubated overnight at 4°C with
primary antibodies as follows: Rabbit anti-FTO (1:1,000; cat. no.
ab126605; Abcam), anti-MZF1 (1:800; cat. no. ab64866; Abcam) and
anti-CLDND1 (1:1,000; cat. no. 13255; Cell Signaling Technology,
Inc.). Anti-GAPDH (1:2,500; cat. no. ab9485; Abcam) was used as an
internal reference. After washing with TBST, membranes were
incubated with HRP)-conjugated goat anti-rabbit IgG H&L
(1:2,000; cat. no. ab97051; Abcam) for 1 h at room temperature.
Equal volumes of liquids A and B from an ECL fluorescence testing
kit [cat. no. abs920; Absin Bioscience Inc.] were mixed in a dark
room and added dropwise to the membrane. Protein signals were
captured using the Bio-Rad Image Analysis System (Bio-Rad
Laboratories, Inc.) with an exposure time ranging between 10 and 30
sec, dynamically adjusted based on signal intensity to ensure
optimal image quality without saturation. Image grayscale values
were analyzed using Quantity One v4.6.2 software (Bio-Rad
Laboratories, Inc.), and relative protein content was expressed as
the grayscale value of the corresponding protein band divided by
the grayscale value of the GAPDH protein band. The relative protein
content was calculated based on the gray value ratio of the target
protein to GAPDH (23).
Dual-luciferase reporter assay
The miRwalk biological website (http://mirwalk.umm.uni-heidelberg.de/)
was utilized for analysis and predicted that the miR-199a-3p
sequence contained binding sites for FTO. To verify this
interaction, wild-type (Wt) and mutant (Mut) 3′-UTR fragments of
FTO were synthesized and cloned into the pGL3-Control vector (cat.
no. E1741; Promega Corporation) to generate pGL3-FTO-Wt and
pGL3-FTO-Mut constructs. The Mut plasmid was obtained by
site-directed mutagenesis within the miR-199a-3p seed region (for
example, A replaced with T). Synthetic miR-199a-3p mimic
(5′-ACAGUAGUCUGCACAUUGGUUA-3′) and negative control mimic (miR-NC)
(5′-UUCUCCGAACGUGUCACGUTT-3′) were purchased from Shanghai
GenePharma Co., Ltd. For luciferase reporter analysis, A549 cells
were co-transfected with 100 ng of PGLO-FTO-Wt or PGLO-FTO-Mut
vector and 50 nM miR-199a-3p mimic or miR-NC using
Lipofectamine® 2000 (cat. no. 11668019; Invitrogen;
Thermo Fisher Scientific, Inc.), the same reagent and protocol
described above (24).
The binding site of the transcription factor MZF1
was obtained from the JASPAR website (jaspar.genereg.net/). To
evaluate the regulatory effect of MZF1 on the CLDND1 promoter,
oe-NC and oe-MZF1 plasmids were co-transfected with reporter
constructs containing either the Wt or Mut CLDND1 promoter
sequences cloned upstream of the luciferase gene in the pGL3-Basic
vector (cat. no. E1751; Promega), which lacks a promoter and is
suitable for promoter-driven transcriptional analysis. A pRL-TK
Renilla luciferase plasmid (cat. no. E2241; Promega) was
co-transfected as an internal control. Cells were collected and
lysed 48 h post-transfection, and dual-luciferase activity was
quantified using the same assay kit.
Firefly luciferase activity was first determined by
adding 100 µl Luciferase Assay Reagent II to each lysate, followed
by Renilla luciferase measurement with 100 µl Stop &
Glo® Reagent using a GloMax® 20/20
Luminometer (Promega). The relative luciferase activity was
calculated as the ratio of firefly to Renilla luminescence,
expressed as relative light units (RLU). Each experiment was
independently repeated three times to ensure reproducibility
(25).
Chromatin immunoprecipitation
(ChIP)
A ChIP kit (cat. no. KT101; Guangzhou Saicheng
Biotechnology Co., Ltd.) was used to detect the enrichment of MZF1
in the promoter region of the CLDND1 gene. When A549 cells reached
70–80% confluence, they were crosslinked with 1% formaldehyde for
10 min at room temperature, and the reaction was quenched with 125
mM glycine for 5 min. Cells were collected by centrifugation at
12,000 × g for 10 min at 4°C, washed twice with cold PBS, and lysed
in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1)
containing protease inhibitor cocktail. After incubation on ice for
10 min, the lysates were subjected to sonication at 20 kHz with
30/30 sec off cycles for a total of 10 min at 4°C to shear
chromatin and obtain DNA fragments of 200–500 bp. For each
immunoprecipitation, chromatin from ~1×106 cells (≈500
µl total volume) was diluted in ChIP dilution buffer and incubated
overnight at 4°C with either 2 µg rabbit anti-MZF1 antibody (cat.
no. ab64866, Abcam, UK) or 2 µg normal rabbit IgG (cat. no.
ab172730, Abcam) as a NC. The immune complexes were captured with
25 µl Protein G magnetic beads for 2 h at 4°C with rotation. Beads
were separated on a magnetic rack, and washed three times with
low-salt buffer (20 mM Tris-HCl, pH 8.1; 150 mM NaCl; 2 mM EDTA; 1%
Triton X-100; 0.1% SDS), high-salt buffer (same as above except 500
mM NaCl), LiCl buffer (10 mM Tris-HCl, pH 8.1; 250 mM LiCl; 1%
NP-40; 1% sodium deoxycholate; 1 mM EDTA) and TE buffer (10 mM
Tris-HCl, pH 8.0; 1 mM EDTA). Chromatin complexes were eluted twice
with 1% SDS/0.1 M NaHCO3 elution buffer at 65°C for 15
min, pooled, and reverse-crosslinked overnight at 65°C. Samples
were treated sequentially with RNase A (37°C, 30 min) and
Proteinase K (55°C, 1 h), and DNA was purified using
silica-membrane spin columns (Qiagen DNA Cleanup Kit). Purified DNA
was subjected to qPCR to detect MZF1 binding to the CLDND1 promoter
as aforementioned. This amplified fragment spans nucleotides −1017
to +192 relative to the transcription start site of the CLDND1
promoter (GenBank Accession No. NM_001040181) (26). The experiment was repeated three
times.
Methylated RNA immunoprecipitation
(Me-RIP)
The SRAMP online tool (cuilab.cn/sramp; version
2023-10) was used to predict potential m6A modification
sites in MZF1 mRNA. Total RNA was isolated from A549 cells using
the TRIzol method as previously described (27), and mRNA was isolated and purified
using the PolyATtract mRNA Isolation System [cat. no. A-Z5300; Aide
Technology (Beijing) Co., Ltd.]. An aliquot of 5 µg purified mRNA
was diluted in 500 µl MeRIP lysis buffer (150 mM NaCl, 10 mM
Tris-HCl pH 7.4, 0.1% NP-40, 1 mM EDTA, 1× RNase inhibitor, and 1×
protease inhibitor cocktail) and incubated with 30 µl Protein A/G
magnetic beads (Thermo Fisher Scientific) pre-bound to antibodies
for immunoprecipitation. Anti-m6A antibody (2 µg;1:500;
cat. no. ab151230; Abcam) or anti-IgG antibody (2 µg;1:100; cat.
no. ab109489; Abcam) was incubated with Protein A/G beads in
binding buffer at 4°C for 1 h with gentle rotation to form
antibody-bead complexes. The prepared RNA-antibody-bead mixture was
incubated overnight at 4°C. The beads were magnetically separated
and washed three times with low-salt buffer (50 mM Tris-HCl, pH
7.4; 150 mM NaCl; 0.1% NP-40; 0.5 mM EDTA) and once with high-salt
buffer (same composition but 500 mM NaCl) to remove non-specific
binding. Each wash was performed for 5 min at 4°C with rotation.
Beads were suspended in 100 µl elution buffer (6.7 mM
m6A-free N6-methyladenosine solution, 50 mM
Tris-HCl, pH 7.4, 10 mM EDTA) and incubated at 55°C for 1 h to
release methylated RNA. The eluate was subjected to
phenol-chloroform extraction and RNA was precipitated with ethanol,
followed by resuspension in RNase-free water. RNA concentration and
purity were measured with a NanoDrop 2000 spectrophotometer (Thermo
Fisher Scientific). RNA was eluted with elution buffer, purified by
phenol-chloroform extraction and analyzed by RT-qPCR for MZF1 as
aforementioned. Primer sequences targeting the conserved exonic
regions of MZF1 were designed using Primer-BLAST (National Center
for Biotechnology Information, database version 2024-03) and are
listed in Table I (28,29).
Photoactivatable
ribonucleoside-enhanced crosslinking and immunoprecipitation
(PAR-CLIP)
A549 cells were incubated with 200 µM 4-thiouridine
for 16 h, then UV-irradiated at 365 nm (0.4 J/cm2) on
ice. Cells were lysed in PAR-CLIP buffer (20 mM Tris-HCl, pH 7.5,
150 mM NaCl, 1 mM MgCl2, 0.5% NP-40, 1 mM DTT, RNase and
protease inhibitors) and cleared by centrifugation (12,000 × g, 10
min, 4°C). Supernatant (500 µl) were incubated overnight at 4°C
with 2 µg anti-FTO antibody (ab126605, Abcam, UK) or 2 µg rabbit
IgG (cat. no. ab172730, Abcam) and 30 µl Protein A/G magnetic beads
(Thermo Fisher Scientific). Beads were magnetically separated,
washed three times with high-salt buffer (50 mM Tris-HCl pH 7.4,
500 mM NaCl, 1 mM EDTA, 0.1% NP-40) and once with low-salt buffer
(150 mM NaCl). The precipitated RNA was labeled with (γ-32P)-ATP
using T4 polynucleotide kinase (New England Biolabs) for 30 min at
37°C, resolved by 10% SDS-PAGE, transferred to nitrocellulose, and
visualized by autoradiography. Crosslinked RNA was recovered after
Proteinase K digestion (0.5 mg/ml, 37°C, 30 min) and
phenol-chloroform extraction. Recovered RNA was reverse-transcribed
with the PrimeScript™ RT Reagent Kit (RR047A; Takara
Bio, Japan) and analyzed by RT-qPCR as aforementioned (30). The experiment was repeated three
times.
Transwell assay
Cell migration and invasion were assessed using
Transwell 24-well cell culture inserts (6.5 mm diameter, 8.0 µm
pore; cat. no. 3422; Corning, Inc.). For the invasion assay,
Matrigel® gel (cat. no. 356234; Corning, Inc.) was added
at 37°C for 30–60 min. A549 cells in the logarithmic phase were
trypsinized, counted, and resuspended in serum-free DMEM. For each
insert, 200 µl of cell suspension containing 1×105 cells
was added to the upper chamber. The lower chamber was filled with
800 µl DMEM supplemented with 20% FBS to serve as a
chemoattractant. All plates were incubated at 37°C in a 5%
CO2 atmosphere for 24 h. For the migration assay, the
procedure was identical except that the inserts were not pre-coated
with Matrigel. After incubation, non-migrated or non-invaded cells
on the upper surface were gently removed with a cotton swab. The
membranes were fixed with 4% paraformaldehyde for 15 min at room
temperature, stained with 0.1% crystal violet solution (cat. no.
C0121; Beyotime Biotechnology) at room temperature for 20 min, and
washed twice with PBS. Cells that migrated or invaded the lower
surface of the membrane were visualized and counted under an
inverted light microscope (Eclipse TE2000; Nikon Corporation,
Tokyo, Japan). To ensure accuracy, cells from at least four
randomly selected microscopic areas were counted (31).
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was evaluated using CCK-8 (cat.
no. CA1210; Beijing Solarbio Science & Technology Co., Ltd.).
A549 cells in the logarithmic growth phase were seeded into 96-well
plates at a density of 1×104 cells/well in 100 µl
complete medium and cultured for 24 h to allow cell adherence and
recovery prior to transfection. Cells were then transfected
according to the experimental grouping, and proliferation was
assessed beginning 48 h after transfection. At 0, 24, 48, and 72 h
after the onset of proliferation monitoring (i.e., after
transfection), 10 µl CCK-8 reagent was added to each well, followed
by incubation at 37°C for 3 h. The absorbance at 450 nm was
measured using a microplate reader (Multiskan™ FC,
Thermo Fisher Scientific, USA). The absorbance was directly
proportional to the number of proliferating cells in the culture
medium, and a cell growth curve was plotted based on these values
(32). The experiment was repeated
three times.
Flow cytometry
After 48 h of transfection, the A549 cells were
digested with 0.25% trypsin (cat. no. 15050057; Gibco; Thermo
Fisher Scientific Inc.) and collected in a flow-through tube. Cell
suspensions were centrifuged at 300 × g for 5 min at 4°C, and the
supernatant was discarded. An Annexin-V-FITC apoptosis detection
kit (cat. no. ab14085; Abcam) was used to formulate the Annexin
V-FITC/PI staining solution at a ratio of 1:2:50 by mixing
Annexin-V-FITC, PI and HEPES buffer solutions according to the
manufacturer's instructions. A total of 1×106 cells were
resuspended per 100 µl of the staining solution, and after 15 min
of incubation at room temperature, 1 ml HEPES buffer solution (cat.
no. PB180325; Procell Life Science & Technology Co., Ltd.) was
added. Fluorescence was detected using a BD FACSCanto™
II flow cytometer (BD Biosciences), with FITC and PI emissions
collected at 525 nm and 620 nm, respectively, after excitation at
488 nm. Data were analyzed using FlowJo software (version 10.8.1;
BD Biosciences). Results were expressed as the percentage of
apoptotic cells (Annexin V+/PI+ and Annexin
V+/PI− fractions) to ensure quantitative
reproducibility (33). The
experiment was repeated three times.
Statistical analysis
All experiments were conducted with at least three
independent replicates (n=3), and data are presented as the mean ±
SD. For comparisons between two independent (unpaired) groups, an
unpaired two-tailed Student's t-test was used. For paired designs,
a paired two-tailed Student's t-test was applied. By contrast, for
three or more treatment groups, one-way ANOVA was employed,
supplemented with Tukey's post hoc multiple comparison test, for
comparisons involving multiple time points. For comparisons
involving both time and treatment factors, two-way ANOVA was used.
The Sidak methods were selected for post hoc multiple comparisons
at each time point. All statistical tests were two-sided, with
P<0.05 considered to indicate a statistically significant
difference, and actual P-values were retained to three decimal
places. Error bars in the graphs indicate the-SD. All statistical
analyses were performed using GraphPad Prism 6.0 (Dotmatics).
Results
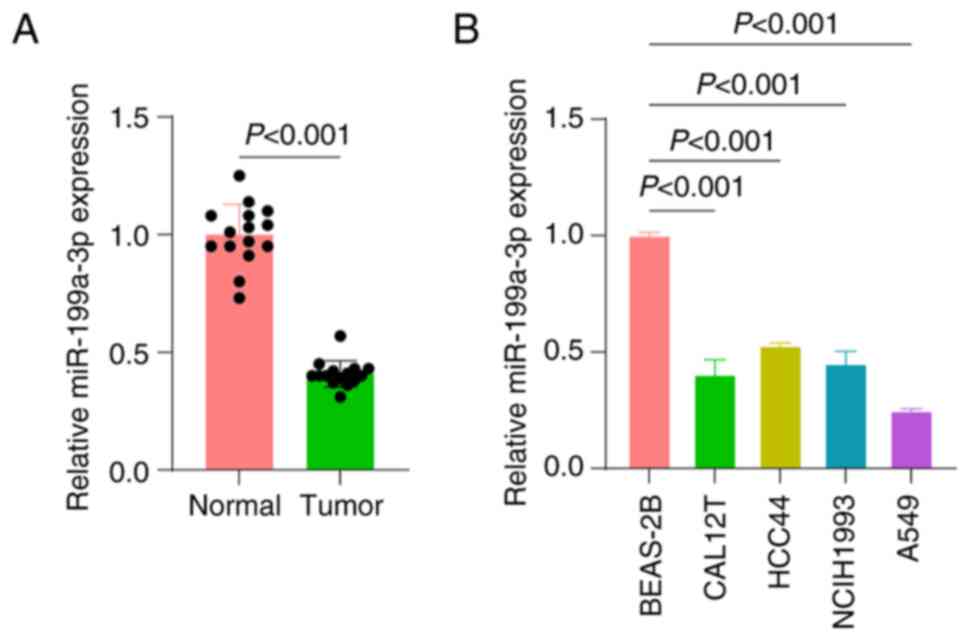
miR-199a-3p expression is
significantly downregulated in NSCLC tissues and cells
In the study of the molecular mechanisms of lung
cancer, the expression patterns of miRNAs have received notable
attention, and the aberrant expression of several miRNAs in lung
cancer has been reported to be closely related to cancer
progression (34). The RT-qPCR
results in the present study showed that the expression levels of
miR-199a-3p were significantly downregulated in NSCLC tissues
compared with normal tissues (Fig.
1A). Furthermore, miR-199a-3p expression was also assessed in
various lung cancer cell lines. The present results showed that in
the lung cancer cell lines, the expression levels of miR-199a-3p
were significantly downregulated, with the A549 cell line having
the lowest expression level (Fig.
1B). Therefore, the A549 cell line was selected for subsequent
in-depth study.
miR-199a-3p inhibits A549 cell
proliferation, migration and invasion by targeting FTO
inhibition
To elucidate the mechanism of action of miR-199a-3p
in A549 cells, bioinformatics tools were used to predict the
potential downstream target genes of miR-199a-3p, which led to the
identification of FTO as a potential direct target (Fig. 2A). A previous study reported an
oncogenic role of FTO in acute myeloid leukemia (15). To validate whether FTO exhibits
similar dysregulation in NSCLC, FTO mRNA and protein expression
levels were analyzed in paired NSCLC and adjacent normal tissue, as
well as in lung cell lines. The results of RT-qPCR and western
blotting revealed that both FTO mRNA and protein expression were
significantly upregulated in NSCLC tissues compared with the
adjacent normal tissues (Fig. 2B).
Consistently, FTO mRNA expression was markedly higher in NSCLC cell
lines (A549, CAL12T, HCC44, and NCI-H1993) than in the normal human
bronchial epithelial cell line BEAS-2B (Fig. 2C).
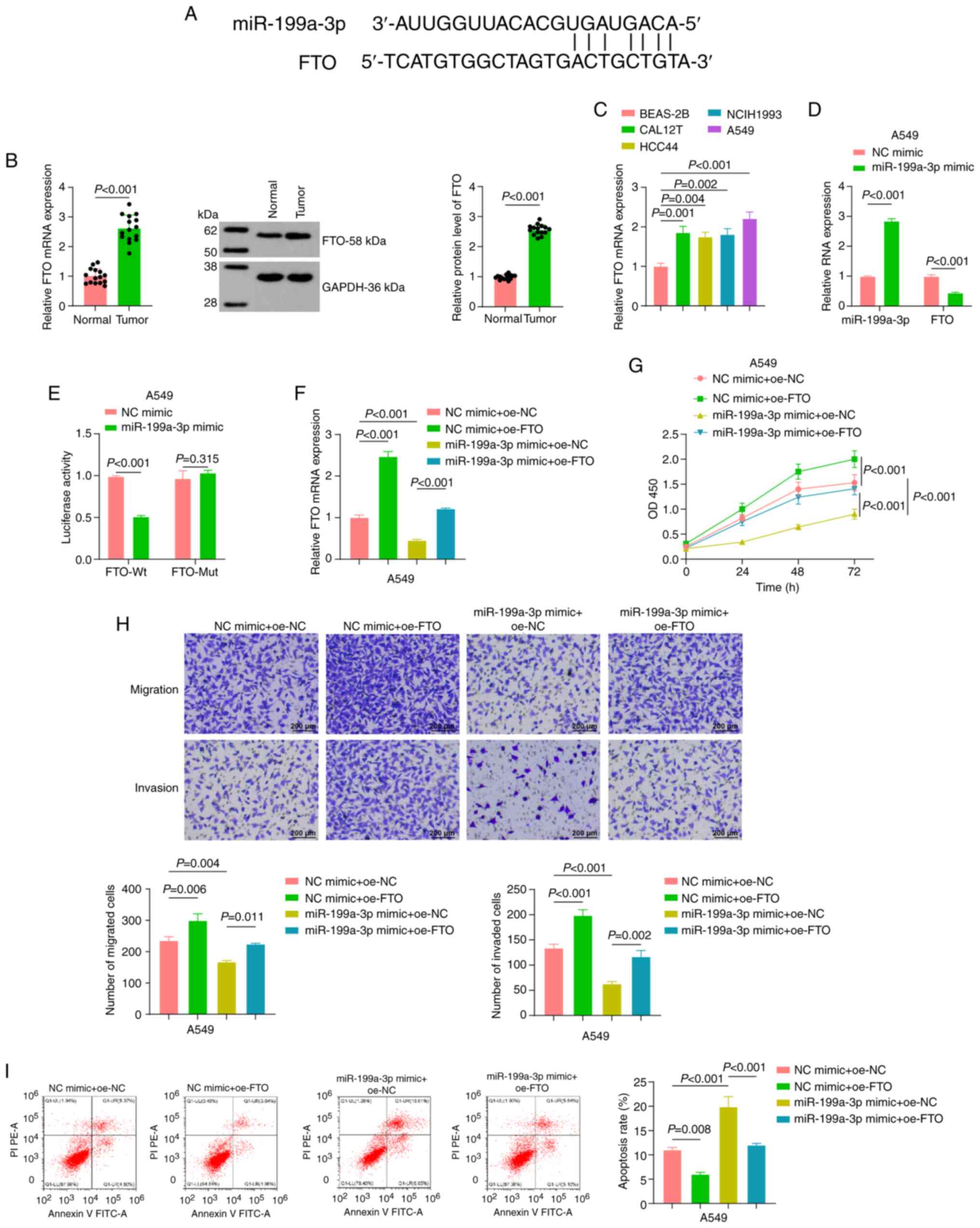 | Figure 2.Effect of miR-199a-3p on the
biological function of A549 cells by downregulating FTO. (A)
Bioinformatics tool ‘miRwalk’ predicted the possible downstream
target gene of miR-199a-3p: FTO. (B) RT-qPCR and western blot
analysis were performed to detect the expression levels of FTO in
non-small cell lung cancer tissues, with paracancerous tissues as
the control (n=15). (C) RT-qPCR was performed to detect the
expression levels of FTO in lung cancer cells. (D) After
miR-199a-3p mimic transfection, RT-qPCR was used to detect the
expression levels of miR-199a-3p and FTO. (E) A dual-luciferase
reporter assay verified the targeting relationship between
miR-199a-3p and FTO. (F) RT-qPCR was used to assess the expression
levels of FTO in A549 cells in each miR mimic + oe plasmid
combination group. (G) Cell Counting Kit-8 assay assessing the
proliferation of A549 cells in each miR mimic + oe plasmid
combination group. (H) Transwell assays indicated the migration and
invasion of A549 cells in each miR mimic + oe plasmid combination
group. Scale bar, 200 µm. (I) Flow cytometry was performed to
detect the apoptosis of A549 cells in each miR mimic + oe plasmid
combination group. Data are presented as the mean ± SEM, n=3
independent experiments. oe, overexpression; RT-qPCR, reverse
transcription-quantitative PCR; miR, microRNA; FTO, fat mass and
obesity-associated protein; NC, negative control; Wt, wild-type;
Mut, mutant; OD, optical density. |
In subsequent experiments, A549 cells were
transfected with miR-199a-3p mimic and NC mimic, and the expression
levels of miR-199a-3p and FTO in transfected cells were analyzed by
RT-qPCR. The results showed that the expression levels of
miR-199a-3p were significantly increased in the miR-199a-3p mimic
group compared with the NC mimic group. At the same time, FTO
expression was significantly decreased in the miR-199a-3p mimic
group compared with the NC mimic group, suggesting miR-199a-3p
negatively regulated FTO expression (Fig. 2D).
Dual-luciferase reporter assays were conducted to
verify the direct targeting relationship between miR-199a-3p and
FTO. The Wt and Mut sequences of the FTO 3′-UTR were co-transfected
with miR-199a-3p mimic into A549 cells. The results revealed that
in the cells co-transfected with the FTO-Wt 3′-UTR, the luciferase
activity was significantly lower in the miR-199a-3p mimic group
compared with the NC mimic group. However, in the cells
co-transfected with the FTO-Mut 3′-UTR, there was no significant
difference in luciferase activity between the two mimic groups,
indicating that miR-199a-3p specifically targeted the FTO 3′-UTR
(Fig. 2E).
RT-qPCR results revealed that, compared with those
in the NC mimic + oe-NC group, the expression levels of FTO were
significantly reduced in cells in the miR-199a-3p mimic + oe-NC
group. When FTO was overexpressed, its expression level in the
miR-199a-3p mimic + oe-FTO group was significantly higher than that
in the miR-199a-3p mimic + oe-NC group (Fig. 2F). Cell proliferation, migration,
and invasion were significantly reduced, whereas apoptosis was
markedly increased, in the miR-199a-3p mimic + oe-NC group compared
with the NC mimic + oe-NC group. By contrast, cells transfected
with NC mimic + oe-FTO exhibited increased proliferation,
migration, and invasion and decreased apoptosis relative to the
corresponding controls, indicating a tumor-promoting effect of FTO
overexpression. Furthermore, co-transfection of miR-199a-3p mimic
with oe-FTO partially restored the proliferative, migratory, and
invasive capacities that had been suppressed by miR-199a-3p alone
(Fig. 2G-I).
In summary, the present study demonstrated that
miR-199a-3p inhibited the proliferation, migration and invasion of
A549 cells by targeting FTO. This revealed the important role of
the miR-199a-3p/FTO axis in regulating the behavior of A549 cells
and provided potential novel targets for molecularly targeted
therapy of NSCLC.
m6A demethylase FTO
promotes MZF1 expression by reducing the levels of m6A
modification of MZF1
A previous study reported that MZF1 promotes
tumorigenesis and metastasis (35), and an online website analysis
showed that MZF1 mRNA can undergo m6A modification
(Fig. 3A). Therefore, we
hypothesized that FTO, an m6A demethylase, affects tumor
progression by regulating the m6A modification of
MZF1.
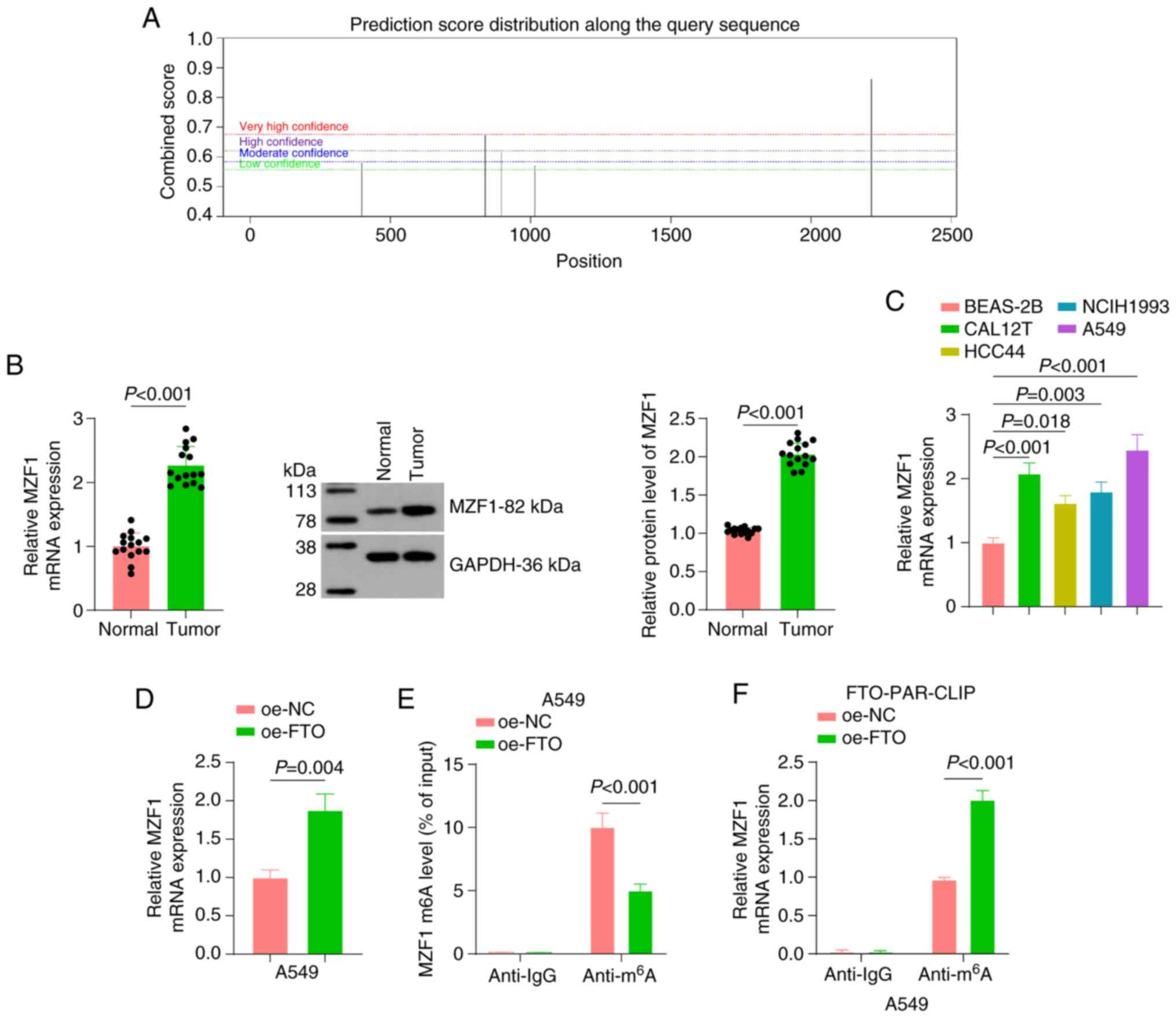 | Figure 3.Mechanism of m6A
modification regulation of MZF1 by FTO. (A) The SRAMP online tool
(http://www.cuilab.cn/sramp) predicted
potential m6A modification sites on MZF1 mRNA. (B)
RT-qPCR and western blot analysis were conducted to assess the
expression levels of MZF1 in NSCLC tissues, with paracancerous
tissues as the control (n=15). (C) RT-qPCR analysis of MZF1 mRNA
expression in normal bronchial epithelial (BEAS-2B) and NSCLC cell
lines (CAL12T, HCC44, NCIH1993 and A549). (D) RT-qPCR was used to
determine MZF1 expression after overexpression of FTO. (E)
Methylated RNA immunoprecipitation assay detecting the
m6A modification level of MZF1 after overexpression of
FTO. (F) PAR-CLIP assay detecting the direct interaction between
FTO and MZF1 mRNA. All cell experiments were repeated three times.
oe, overexpression; m6A, N6-methyladenosine;
MZF1, myeloid zinc finger 1; FTO, fat mass and obesity-associated
protein; RT-qPCR, reverse transcription-quantitative PCR; NSCLC,
non-small cell lung cancer; NC, negative control; PAR-CLIP,
photoactivatable ribonucleoside-enhanced crosslinking and
immunoprecipitation. |
Expression levels of MZF1 were examined in NSCLC
tissues and cell lines by RT-qPCR and western blot analyses. As
shown in Fig. 3B, both the mRNA
and protein levels of MZF1 were significantly higher in NSCLC tumor
tissues than in their paired adjacent normal tissues. Consistently,
MZF1 mRNA expression was markedly upregulated in NSCLC cell lines
(CAL12T, HCC44, A549, and NCI-H1993) compared with BEAS-2B, with
the highest expression observed in A549 cells (Fig. 3C). Subsequently, FTO was
overexpressed in the cells; overexpression of FTO significantly
increased the mRNA expression levels of MZF1 (Fig. 3D).
To determine how FTO regulated MZF1, the
m6A modification level of MZF1 after FTO overexpression
was examined using a Me-RIP assay. FTO overexpression significantly
reduced the m6A modification of MZF1 mRNA compared with
the oe-NC group (Fig. 3E).
Additionally, a PAR-CLIP assay was employed to investigate the
direct interaction between FTO and MZF1 mRNA. The
anti-m6A antibody pulled down significantly more MZF1
mRNA than the anti-IgG control, and the enrichment was markedly
higher in the oe-FTO group than in the oe-NC group (Fig. 3F).
Taken together, the present data demonstrated that
FTO could promote MZF1 expression by reducing the m6A
modification level of MZF1 mRNA.
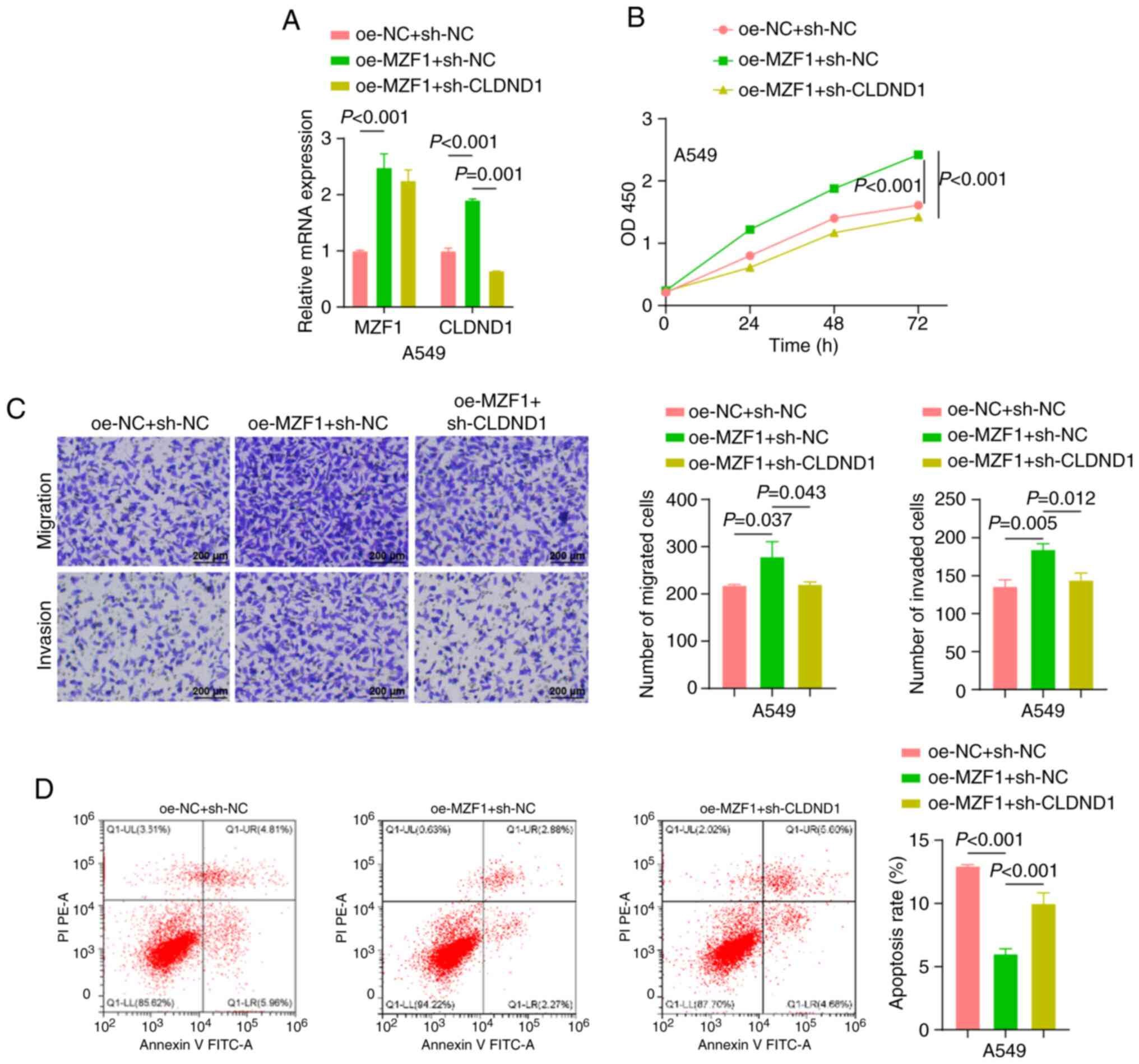
Silencing of MZF1 reverses the
promoting effect of FTO on A549 cell proliferation, migration and
invasion
To further ascertain the effect of the FTO/MZF1 axis
on the biological function of A549 cells, three groups were
established: oe-NC + sh-NC, oe-FTO + sh-NC, and oe-FTO + sh-MZF1.
RT-qPCR demonstrated that the expression levels of both FTO and
MZF1 were significantly elevated in the cells in the oe-FTO + sh-NC
group compared with the oe-NC + sh-NC group, and the expression
levels of MZF1 were significantly lower in the cells in the oe-FTO
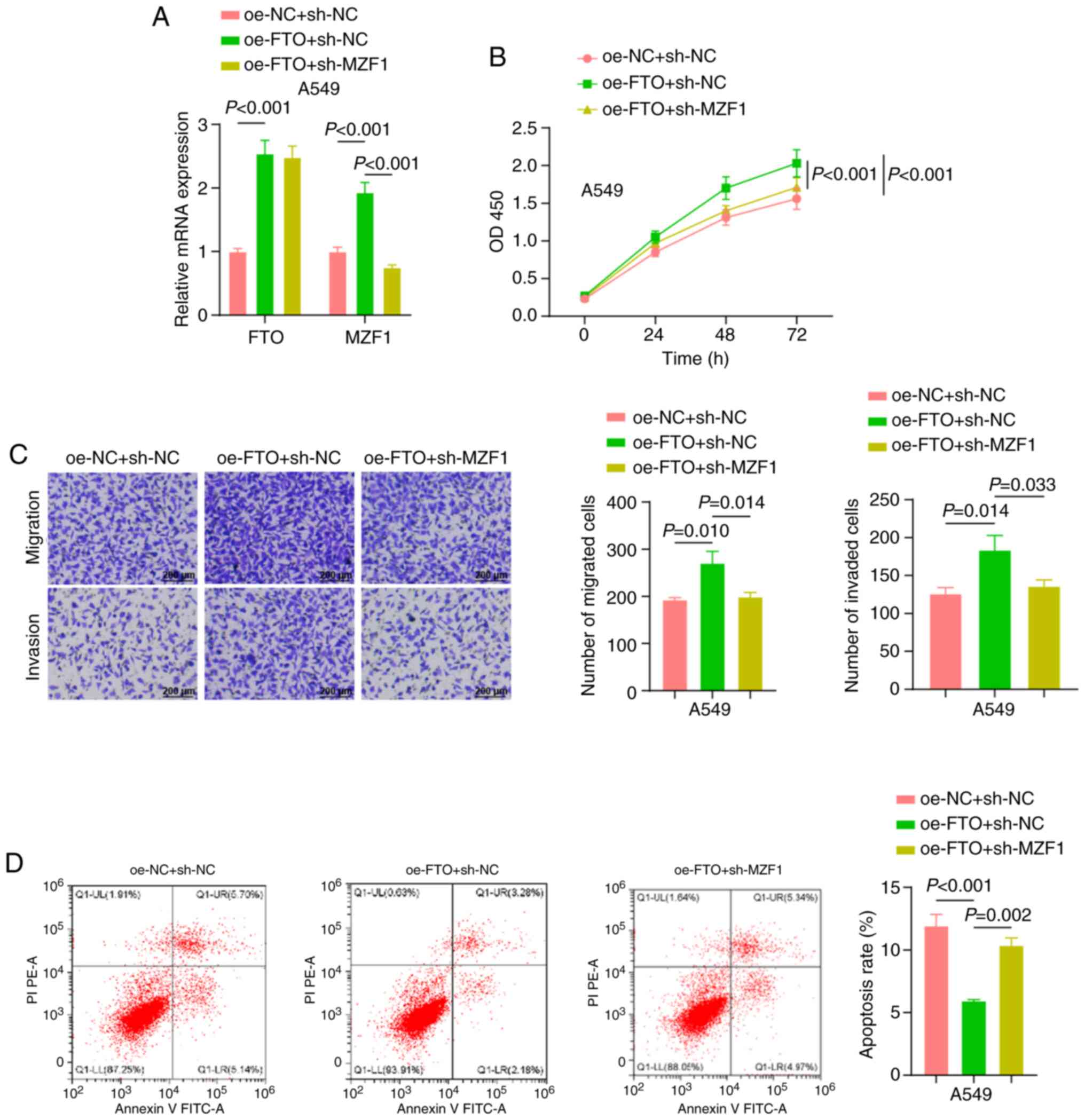
+ sh-MZF1 group compared with the oe-FTO + sh-NC group (Fig. 4A).
The biological functions of A549 cells were
evaluated using CCK-8, Transwell and flow cytometry assay. The
results revealed that, compared with the oe-NC + sh-NC group, the
oe-FTO + sh-NC group exhibited significantly higher cell
proliferation, migration and invasion, as well as a significantly
lower apoptosis. When MZF1 was silenced, the effect of FTO was
partially reversed, as demonstrated by the significant decrease in
proliferation, migration and invasion in the oe-FTO + sh-MZF1 group
compared with the oe-FTO + sh-NC group, as well as a significant
increase in apoptosis (Fig.
4B-D).
Taken together, the present study demonstrated that
silencing of MZF1 reversed the promoting effect of FTO on A549 cell
proliferation, migration and invasion.
MZF1 promotes A549 cell proliferation,
migration and invasion by facilitating CLDND1 transcription
MZF1 is a transcription factor that can influence
tumor progression by promoting downstream gene expression (36). The binding site of the
transcription factor MZF1 was obtained from the JASPAR website
(https://jaspar.genereg.net/) (Fig. 5A), and MZF1 showed a high binding
affinity (GAAATCCCCTATT) for the CLDND1 promoter region. CLDND1
expression was significantly higher in NSCLC tumor tissues than in
adjacent normal tissue (Fig. 5B).
Consistently, CLDND1 mRNA levels were markedly elevated in A549 and
other NSCLC cell lines (NCI-H1993, CAL12T, HCC44) compared with
BEAS-2B cells (Fig. 5C).
![Mechanism of the transcriptional
regulation of CLDND1 by MZF1. (A) The JASPAR website (https://jaspar.genereg.net/) was used to retrieve the
binding site of the transcription factor MZF1. (B) RT-qPCR and
western blot analysis were performed to detect the expression
levels of CLDND1 in non-small cell lung cancer tissues, using
paracancerous tissues as the control (n=15). (C) RT-qPCR analysis
of CLDND1 mRNA expression in normal bronchial epithelial (BEAS-2B)
and NSCLC cell lines (CAL12T, HCC44, NCIH1993 and A549). (D) After
overexpression of MZF1, RT-qPCR was performed to measure MZF1 and
CLDND1 expression. (E) Dual-luciferase reporter assay examining the
effect of MZF1 on CLDND1 promoter activity. (F) Chromatin
immunoprecipitation assays determining the enrichment of MZF1 in
the CLDND1 promoter region [Chr3:167, 450, 703–167, 450, 714
(hg38)]. All cell experiments were repeated three times. oe,
overexpression; CLDND1, claudin domain-containing 1; MZF1, myeloid
zinc finger 1; RT-qPCR, reverse transcription-quantitative PCR; NC,
negative control; Wt, wild-type; Mut, mutant.](/article_images/mmr/33/1/mmr-33-01-13736-g04.jpg) | Figure 5.Mechanism of the transcriptional
regulation of CLDND1 by MZF1. (A) The JASPAR website (https://jaspar.genereg.net/) was used to retrieve the
binding site of the transcription factor MZF1. (B) RT-qPCR and
western blot analysis were performed to detect the expression
levels of CLDND1 in non-small cell lung cancer tissues, using
paracancerous tissues as the control (n=15). (C) RT-qPCR analysis
of CLDND1 mRNA expression in normal bronchial epithelial (BEAS-2B)
and NSCLC cell lines (CAL12T, HCC44, NCIH1993 and A549). (D) After
overexpression of MZF1, RT-qPCR was performed to measure MZF1 and
CLDND1 expression. (E) Dual-luciferase reporter assay examining the
effect of MZF1 on CLDND1 promoter activity. (F) Chromatin
immunoprecipitation assays determining the enrichment of MZF1 in
the CLDND1 promoter region [Chr3:167, 450, 703–167, 450, 714
(hg38)]. All cell experiments were repeated three times. oe,
overexpression; CLDND1, claudin domain-containing 1; MZF1, myeloid
zinc finger 1; RT-qPCR, reverse transcription-quantitative PCR; NC,
negative control; Wt, wild-type; Mut, mutant. |
To explore the regulatory effect of MZF1 on CLDND1,
MZF1 was overexpressed in A549 cells. RT-qPCR results revealed that
MZF1 overexpression significantly elevated the mRNA levels of
CLDND1 (Fig. 5D). In addition, a
dual-luciferase reporter assay revealed that overexpression of MZF1
significantly increased the luciferase activity of the wild-type
(CLDND1-Wt) promoter reporter compared with the oe-NC group,
whereas no significant change was observed in the mutant
(CLDND1-Mut) reporter lacking the predicted MZF1 binding motif
(Fig. 5E).
ChIP experiments indicated that MZF1 directly
regulated the CLDND1 promoter, showing a significant increase in
MZF1 enrichment in the CLDND1 promoter region (Fig. 5F).
In the cell function experiments, the effects of
different transfection combinations on the proliferation, migration
and invasion of A549 cells were evaluated. RT-qPCR results revealed
that the expression levels of both MZF1 and CLDND1 were
significantly elevated in the cells of the oe-MZF1 + sh-NC group
compared with those in the oe-NC + sh-NC group. Furthermore, the
expression levels of CLDND1 were lower in the oe-MZF1 + sh-CLDND1
group than in the oe-MZF1 + sh-NC group (Fig. 6A). Functional experiments
demonstrated that cell proliferation, migration and invasion were
significantly increased, and apoptosis was significantly decreased
in the oe-MZF1 + sh-NC group compared with the oe-NC + sh-NC group.
When CLDND1 was silenced, the effect of MZF1 on A549 cell
proliferation, migration, invasion and apoptosis was partially
reversed (Fig. 6B-D).
In summary, these data indicated that MZF1 enhanced
the proliferation, migration and invasion of A549 cells by
promoting the transcription of CLDND1, and that the silencing of
CLDND1 effectively inhibited these effects.
Discussion
Despite advances in research and treatment, lung
cancer remains a notable threat to human health, with a low 5-year
survival rate (37). Therefore,
there is a need to develop reliable preclinical models and
effective treatments. The findings of the present study contributed
to the understanding of the molecular mechanisms underlying lung
cancer and may provide novel targets for the development of
molecularly targeted therapies (38).
Aberrant miR-199a-3p expression has been observed in
various cancer types and exhibits tumor suppressor effects. In one
study, miR-199a-3p was shown to inhibit tumor growth in vivo
and suppress the invasion and migration of nasopharyngeal carcinoma
cells by downregulating the expression of stearoyl-CoA desaturase
1, thus providing a potential target for the treatment of
nasopharyngeal carcinoma (9).
Furthermore, miR-199a-3p is downregulated in various human
malignancies, including hepatocellular carcinoma (HCC), leading to
poor prognosis in patients with HCC (39). In HCC, knockdown of zinc finger and
homeoboxes-1 (ZHX1) inhibits the upregulation of p53-upregulated
modulator of apoptosis (PUMA) following miR-199a-3p transfection.
Furthermore, silencing of either ZHX1 or PUMA reverses the effects
of miR-199a-3p (40). One study
has predicted that miR-199a-5p and miR-199a-3p can serve as early
diagnostic markers or therapeutic targets in NSCLC (6). However, the specific mechanism by
which miR-199a-3p acts in lung cancer remains to be fully
elucidated. Evidence supports the anticancer role of the
miR-199a/Rheb/mTOR axis in NSCLC. Rheb, a common target of
miR-199a-5p and miR-199a-3p, is involved in regulating the mTOR
signaling pathway. Knockdown of Rheb recapitulated the
tumor-suppressive effects of miR-199a overexpression, including
inhibited proliferation and migration. Furthermore, miR-199a-5p/3p
enhances the sensitivity of EGFR-T790M mutant NSCLC cells to
(6). Taken together, these
findings demonstrate that miR-199a-5p/3p can serve as an oncogene
that inhibits the mTOR signaling pathway by targeting Rheb, thus
inhibiting the regulatory process of NSCLC (6).
The present study explored a novel regulatory
mechanism: miR-199a-3p targeted FTO to regulate the m6A
modification of MZF1, thus inhibiting CLDND1 expression, which
provided a novel perspective to understand the function of
miR-199a-3p in NSCLC. Observations revealed that miR-199a-3p was
downregulated in NSCLC tissues and cells, and it had a direct
targeting relationship with FTO. By targeting and inhibiting FTO,
miR-199a-3p suppressed the migration, proliferation and invasion of
A549 cells. FTO promoted MZF1 expression by decreasing the level of
m6A modification of MZF1. In addition, silencing of MZF1
reversed the promoting effect of FTO on lung cancer cell migration,
proliferation and invasion. Furthermore, MZF1 promoted A549 cell
migration, proliferation and invasion by promoting CLDND1
transcription.
Mechanistically, estrogen receptor α (ESR1) serves
as the target for FTO, where heightened FTO expression decreases
the m6A levels in ESR1 mRNA (14). A study by Xu et al (14) reported two distinct m6A
modification sites, 5409A and 5247A, located in the 3′-UTR of ESR1,
with FTO capable of reducing their methylation. In addition, the
m6A readers insulin-like growth factor 2 mRNA-binding
protein (IGF2BP)3 and YTH domain-containing family protein 1
(YTHDF1) identify and bind to these m6A sites, thereby
enhancing ESR1 mRNA stability and promoting tumor growth. ESR1 was
also found to exhibit diagnostic potential for NSCLC. In summary,
the study by Xu et al (14)
elucidated an important mechanism by which the
FTO/YTHDF1/IGF2BP3/ESR1 axis regulates the epithelial transcriptome
and highlighted the potential of m6A-dependent NSCLC
therapeutic strategies.
The data in the present study revealed that FTO
stabilized MZF1 by removing its m6A modification,
thereby linking RNA methylation changes to the CLDND1-driven
processes of cell migration and invasion. Notably, the study by Xu
et al (14) proposed that
FTO exerted an oncogenic effect by stabilizing ESR1, while another
study has suggested that FTO has a tumor-suppressive effect in
adenocarcinoma models (41). These
discrepancies may stem from cell-type specificity, for example,
adenocarcinoma vs. squamous cell carcinoma or different
microenvironments, as well as variations in downstream targets. The
findings of the present study supported the oncogenic role of FTO
through the stabilization of MZF1 and underscored the need for
future validation of the translational potential of FTO in models
based on EGFR mutation types or immune therapy resistance
backgrounds. In summary, FTO may possess dual regulatory functions
in lung cancer, with specific modes of action potentially depending
on cancer subtype, target gene context and microenvironmental
differences, which are factors that should be further clarified in
future research.
Regulation of MZF1 signaling has been associated
with the progression of numerous human malignancies, such as
triple-negative breast and cervical cancer (42). Silencing of MZF1 inhibits cell
proliferation, whereas its aberrant expression contributes to
cancer development (43). Limited
research exists on the specific mechanisms underlying the
m6A modification of MZF1 in lung cancer, which supports
the novelty of the present study in exploring novel therapeutic
regulatory mechanisms in this context. Furthermore, CLDND1, as a
tight junction-associated protein, interacts with other tight
junction proteins, such as occludin and claudins, to collectively
construct and maintain the intact structure of intercellular tight
junctions, forming a selective barrier that strictly regulates the
transport of substances between cells and the conduction of
intercellular signals, which is important for maintaining the
stability of the tissue internal environment (18,44).
The role of CLDND1 during cancer progression has
also attracted attention. Changes in CLDND1 expression are closely
related to tumor development in certain cancer types, such as
breast cancer (19). The
inhibitory effect of gefitinib, an EGFR tyrosine kinase inhibitor,
on CLDND1 expression has been evaluated using a luciferase reporter
and ELK1 overexpression in ChIP assays. ELK1, identified as an
enhancer region activator, transiently elevated CLDND1 expression
at both the mRNA and protein levels. EGF-induced phosphorylation of
ELK1 further increased CLDND1 expression, which was inhibited by
gefitinib (19). Thus,
EGF-dependent activation of ELK1 leads to the induction of CLDND1
expression. These findings pave the way for the development of
novel anticancer drugs targeting CLDND1 (19). The present study revealed a
significant increase in the enrichment of MZF1 in the promoter
region of CLDND1 using ChIP experiments, supporting its direct
action on the CLDND1 promoter. The present study also identified a
novel role for CLDND1 in the malignant progression of tumors. A
previous study has focused on the effects of CLDND1 on endothelial
cell adhesion and cell junctions (45). However, the present study
demonstrated that once transcriptionally activated by MZF1, CLDND1
also promoted malignant tumor phenotypes.
Based on the aforementioned analysis, miR-199a-3p
was lowly expressed in NSCLC cells, which led to a weakening of its
inhibitory effect on FTO, which in turn increased the expression
levels of FTO. FTO, as an m6A demethylase, removed
m6A modifications on the MZF1 mRNA, improved the mRNA
stability and translational efficiency of MZF1 and increased the
protein levels of MZF1. Subsequently, MZF1, as a transcription
factor, upregulated the expression levels of CLDND1. The high
expression of CLDND1 promoted malignant biological behaviors such
as migration, proliferation and invasion of NSCLC cells. In
summary, miR-199a-3p regulated the m6A modification of
MZF1 by targeting FTO, resulting in the inhibition of CLDND1
expression, which ultimately affected the biological functions of
NSCLC cells.
Although the present study primarily focused on the
axis of miR-199a-3p/FTO-mediated m6A modification of
MZF1 and its transcriptional regulation of CLDND1, the present
study did not systematically exclude the involvement of other key
factors. The stability and translational efficiency of MZF1 may
have also been regulated by m6A reader proteins,
including the IGF2BP family and YTHDF1/2/3 (46). m6A methyltransferases
and demethylases such as methyltransferase-like protein 3/14 and
alkylated DNA repair protein alkB homolog 5 may have also
indirectly intervened by regulating the expression of MZF1
(47); and iii) in addition to
MZF1, there may have been other cis-acting elements and binding
proteins on the CLDND1 promoter (19,48).
As the present study did not conduct m6A site
prediction, enrichment analysis or RNA immunoprecipitation
validation of the reader proteins, the relevant regulatory
mechanisms require further exploration.
The present study also had certain limitations. The
small sample size in the present study may undermine the
universality of the molecular associations among miR-199a-3p, FTO,
MZF1, and CLDND1, potentially limiting the reliability of clinical
relevance analysis. Furthermore, the present study relied on in
vitro cell experiments and lacked data from in vivo
animal models, making it difficult to comprehensively evaluate the
role of this pathway in living tumors. Additionally, the signaling
pathways in NSCLC cells are complex, and the factors in the present
study may have participated in multiple pathways, necessitating
further research with expanded samples to explore their
interrelationships and mechanisms.
In conclusion, miR-199a-3p targeted and inhibited
FTO expression, thereby blocking the m6A demethylation
modification of MZF1 and suppressing MZF1 expression and the
transcriptional activation of downstream CLDND1 (Fig. S2). This ultimately inhibited the
migration, proliferation and invasion of NSCLC cells. The
aforementioned mechanism suggested that the
miR-199a-3p/FTO/MZF1/CLDN1 signaling pathway serves an important
role in the development and progression of NSCLC. Unlike the
findings of a study by Xu et al (14) indicating that FTO participates in
NSCLC progression by regulating ESR1, the present study is, to the
best of our knowledge, the first to propose that miR-199a-3p
targets and inhibits FTO, relieving its m6A
demethylation effect on MZF1 and indirectly inhibiting the
MZF1-mediated transcriptional activation of CLDND1. To the best of
our knowledge, this multi-level regulatory axis of miRNA-epigenetic
modification/transcription factor/target gene has not been reported
before. The present study explored a novel upstream regulator,
miR-199a-3p, and downstream oncogenic mechanism, the MZF1/CLDND1
axis, of FTO in NSCLC, providing novel insights for
m6A-targeted therapy.
Future studies could validate the clinical
significance of this signaling pathway in larger sample cohorts,
study the association between the expression levels of miR-199a-3p,
FTO, MZF1 and CLDND1, and clinical parameters such as TNM staging
to enhance its clinical applicability, establish lung cancer
xenograft models to simulate the human tumor microenvironment and
validate the biological functions and therapeutic potential of this
pathway, and test this signaling axis in models with EGFR mutations
or immune therapy resistance to explore its roles in different lung
cancer subtypes and treatment resistance scenarios, providing novel
targets and strategies for precision therapy. m6A reader
protein in RNA pull-down experiments will be supplemented in the
future to further refine the analysis of the regulatory pathway
network.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Application Foundation
Project of Yanbian University in 2024 (grant no. ydkj202458).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YC conceived and designed the study. XL performed
the experiments. HZ, WY and EZ conducted the data analysis and
interpretation. WY and EZ edited the manuscript. YC and HZ confirm
the authenticity of all the raw data. All authors read and approved
the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Yanbian University Affiliated Hospital (approval no.
2021094; Yanji, China) and written informed consent was obtained
from the patients. The study followed The Declaration of
Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yang H, Liu Y, Chen L, Zhao J, Guo M, Zhao
X, Wen Z, He Z, Chen C and Xu L: MiRNA-based therapies for lung
cancer: Opportunities and challenges? Biomolecules. 13:8772023.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ruiz-Cordero R and Devine WP: Targeted
therapy and checkpoint immunotherapy in lung cancer. Surg Pathol
Clin. 13:17–33. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang K, Mei Z, Zheng M, Liu X, Li D and
Wang H: FTO-mediated autophagy inhibition promotes non-small cell
lung cancer progression by reducing the stability of SESN2 mRNA.
Heliyon. 10:e275712024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zahmatyar M, Kharaz L, Abiri Jahromi N,
Jahanian A, Shokri P and Nejadghaderi SA: The safety and efficacy
of binimetinib for lung cancer: A systematic review. BMC Pulm Med.
24:3792024. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
van Zandwijk N, Pavlakis N, Kao SC, Linton
A, Boyer MJ, Clarke S, Huynh Y, Chrzanowska A, Fulham MJ, Bailey
DL, et al: Safety and activity of microRNA-loaded minicells in
patients with recurrent malignant pleural mesothelioma: a
first-in-man, phase 1, open-label, dose-escalation study. Lancet
Oncol. 18:1386–1396. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu X, Wang X, Chai B, Wu Z, Gu Z, Zou H,
Zhang H, Li Y, Sun Q, Fang W and Ma Z: miR-199a-3p/5p regulate
tumorgenesis via targeting Rheb in non-small cell lung cancer. Int
J Biol Sci. 18:4187–4202. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Meng W, Li Y, Chai B, Liu X and Ma Z:
miR-199a: A tumor suppressor with noncoding RNA network and
therapeutic candidate in lung cancer. Int J Mol Sci. 23:85182022.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hou G, Wang Y, Zhang M, Hu Y, Zhao Y, Jia
A, Wang P, Zhao W, Zhao W and Lu Z: miR-199a-3p suppresses
progression of esophageal squamous cell carcinoma through
inhibiting mTOR/p70S6K pathway. Anticancer Drugs. 32:157–167. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hu W, Wang Y, Zhang Q, Luo Q, Huang N,
Chen R, Tang X, Li X and Luo H: MicroRNA-199a-3p suppresses the
invasion and metastasis of nasopharyngeal carcinoma through
SCD1/PTEN/AKT signaling pathway. Cell Signal. 110:1108332023.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wei H, Li Z, Liu F, Wang Y, Ding S, Chen Y
and Liu J: The role of FTO in tumors and its research progress.
Curr Med Chem. 29:924–933. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Azzam SK, Alsafar H and Sajini AA: FTO m6A
demethylase in obesity and cancer: Implications and underlying
molecular mechanisms. Int J Mol Sci. 23:38002022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zaccara S, Ries RJ and Jaffrey SR:
Publisher correction: Reading, writing and erasing mRNA
methylation. Nat Rev Mol Cell Biol. 24:7702023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang Z, Yu GL, Zhu X, Peng TH and Lv YC:
Critical roles of FTO-mediated mRNA m6A demethylation in regulating
adipogenesis and lipid metabolism: Implications in lipid metabolic
disorders. Genes Dis. 9:51–61. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu X, Qiu S, Zeng B, Huang Y, Wang X, Li
F, Yang Y, Cao L, Zhang X, Wang J and Ma L:
N6-methyladenosine demethyltransferase FTO mediated
m6A modification of estrogen receptor alpha in non-small
cell lung cancer tumorigenesis. Oncogene. 43:1288–1302. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang ZW, Zhao XS, Guo H and Huang XJ: The
role of m6A demethylase FTO in chemotherapy resistance
mediating acute myeloid leukemia relapse. Cell Death Discov.
9:2252023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu D, Tan H, Su W, Cheng D, Wang G, Wang
J, Ma DA, Dong GM and Sun P: MZF1 mediates oncogene-induced
senescence by promoting the transcription of p16(INK4A). Oncogene.
41:414–426. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang CH, Wu HC, Hsu CW, Chang YW, Ko CY,
Hsu TI, Chuang JY, Tseng TH and Wang SM: Inhibition of MZF1/c-MYC
Axis by cantharidin impairs cell proliferation in glioblastoma. Int
J Mol Sci. 23:147272022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ohnishi M, Ochiai H, Matsuoka K, Akagi M,
Nakayama Y, Shima A, Uda A, Matsuoka H, Kamishikiryo J, Michihara A
and Inoue A: Claudin domain containing 1 contributing to
endothelial cell adhesion decreases in presence of cerebellar
hemorrhage. J Neurosci Res. 95:2051–2058. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Matsuoka H, Yamaoka A, Hamashima T, Shima
A, Kosako M, Tahara Y, Kamishikiryo J and Michihara A:
EGF-dependent activation of ELK1 contributes to the induction of
CLDND1 expression involved in tight junction formation.
Biomedicines. 10:17922022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yin H, Chen L, Piao S, Wang Y, Li Z, Lin
Y, Tang X, Zhang H, Zhang H and Wang X: M6A RNA
methylation-mediated RMRP stability renders proliferation and
progression of non-small cell lung cancer through regulating
TGFBR1/SMAD2/SMAD3 pathway. Cell Death Differ. 30:605–617. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gong Y, Li X and Xie L: Circ_0001897
regulates high glucose-induced angiogenesis and inflammation in
retinal microvascular endothelial cells through
miR-29c-3p/transforming growth factor beta 2 axis. Bioengineered.
13:11694–11705. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Robinson R, Srinivasan M, Shanmugam A,
Ward A, Ganapathy V, Bloom J, Sharma A and Sharma S: Interleukin-6
trans-signaling inhibition prevents oxidative stress in a mouse
model of early diabetic retinopathy. Redox Biol. 34:1015742020.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu P, Peng QH, Tong P and Li WJ:
Astragalus polysaccharides suppresses high glucose-induced
metabolic memory in retinal pigment epithelial cells through
inhibiting mitochondrial dysfunction-induced apoptosis by
regulating miR-195. Mol Med. 25:212019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao G, Liang J, Shan G, Gu J, Xu F, Lu C,
Ma T, Bi G, Zhan C and Ge D: KLF11 regulates lung adenocarcinoma
ferroptosis and chemosensitivity by suppressing GPX4. Commun Biol.
6:5702023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang W, Ruan X, Li Y, Zhi J, Hu L, Hou X,
Shi X, Wang X, Wang J, Ma W, et al: KDM1A promotes thyroid cancer
progression and maintains stemness through the Wnt/β-catenin
signaling pathway. Theranostics. 12:1500–1517. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lin S, Xu H, Qin L, Pang M, Wang Z, Gu M,
Zhang L, Zhao C, Hao X, Zhang Z, et al: UHRF1/DNMT1-MZF1
axis-modulated intragenic site-specific CpGI methylation confers
divergent expression and opposing functions of PRSS3 isoforms in
lung cancer. Acta Pharm Sin B. 13:2086–2106. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang F, Hou W, Li X, Lu L, Huang T, Zhu M
and Miao C: SETD8 cooperates with MZF1 to participate in
hyperglycemia-induced endothelial inflammation via elevation of
WNT5A levels in diabetic nephropathy. Cell Mol Biol Lett.
27:302022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xue X, Wang M, Zhang X, Ma L and Wang J:
PAR-CLIP assay in ferroptosis. Methods Mol Biol. 2712:29–43. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hou Y, Zhang Q, Pang W, Hou L, Liang Y,
Han X, Luo X, Wang P, Zhang X, Li L and Meng X: YTHDC1-mediated
augmentation of miR-30d in repressing pancreatic tumorigenesis via
attenuation of RUNX1-induced transcriptional activation of Warburg
effect. Cell Death Differ. 28:3105–3124. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu L and Xie D, Xie H, Huang W, Zhang J,
Jin W, Jiang W and Xie D: ARHGAP10 inhibits the proliferation and
metastasis of CRC cells via blocking the activity of RhoA/AKT
signaling pathway. Onco Targets Ther. 12:11507–11516. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang G, Wang L, Zhao L, Yang F, Lu C, Yan
J, Zhang S, Wang H and Li Y: Silibinin induces both apoptosis and
necroptosis with potential anti-tumor efficacy in lung cancer.
Anticancer Agents Med Chem. 24:1327–1338. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Iqbal MA, Arora S, Prakasam G, Calin GA
and Syed MA: MicroRNA in lung cancer: Role, mechanisms, pathways
and therapeutic relevance. Mol Aspects Med. 70:3–20. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kan A, Liu S, He M, Wen D, Deng H, Huang
L, Lai Z, Huang Y and Shi M: MZF1 promotes tumour progression and
resistance to anti-PD-L1 antibody treatment in hepatocellular
carcinoma. JHEP Rep. 6:1009392023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Brix DM, Bundgaard Clemmensen KK and
Kallunki T: Zinc finger transcription factor MZF1-A specific
regulator of cancer invasion. Cells. 9:2232020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Meng FT, Jhuang JR, Peng YT, Chiang CJ,
Yang YW, Huang CY, Huang KP and Lee WC: Predicting lung cancer
survival to the future: Population-based cancer survival modeling
study. JMIR Public Health Surveill. 10:e467372024. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li H, Chen Z, Chen N, Fan Y, Xu Y and Xu
X: Applications of lung cancer organoids in precision medicine:
From bench to bedside. Cell Commun Signal. 21:3502023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen G, Zhang Z, Li Y, Wang L and Liu Y:
LncRNA PTPRG-AS1 promotes the metastasis of hepatocellular
carcinoma by enhancing YWHAG. J Oncol. 2021:36243062021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guan J, Liu Z, Xiao M, Hao F, Wang C, Chen
Y, Lu Y and Liang J: MicroRNA-199a-3p inhibits tumorigenesis of
hepatocellular carcinoma cells by targeting ZHX1/PUMA signal. Am J
Transl Res. 9:2457–2465. 2017.PubMed/NCBI
|
|
41
|
Wu G, Yan Y, Cai Y, Peng B, Li J, Huang J,
Xu Z and Zhou J: ALKBH1-8 and FTO: Potential therapeutic targets
and prognostic biomarkers in lung adenocarcinoma pathogenesis.
Front Cell Dev Biol. 9:6339272021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yue CH, Liu JY, Chi CS, Hu CW, Tan KT,
Huang FM, Pan YR, Lin KI and Lee CJ: Myeloid zinc finger 1 (MZF1)
maintains the mesenchymal phenotype by down-regulating IGF1R/p38
MAPK/ERα signaling pathway in high-level MZF1-expressing TNBC
cells. Anticancer Res. 39:4149–4164. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mudduluru G, Vajkoczy P and Allgayer H:
Myeloid zinc finger 1 induces migration, invasion, and in vivo
metastasis through Axl gene expression in solid cancer. Mol Cancer
Res. 8:159–169. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Matsuoka H, Shima A, Uda A, Ezaki H and
Michihara A: The retinoic acid receptor-related orphan receptor α
positively regulates tight junction protein claudin
domain-containing 1 mRNA expression in human brain endothelial
cells. J Biochem. 161:441–450. 2017.PubMed/NCBI
|
|
45
|
Shima A, Matsuoka H, Yamaoka A and
Michihara A: Transcription of CLDND1 in human brain endothelial
cells is regulated by the myeloid zinc finger 1. Clin Exp Pharmacol
Physiol. 48:260–269. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Huang H, Weng H, Sun W, Qin X, Shi H, Wu
H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al: Recognition of RNA
N6-methyladenosine by IGF2BP proteins enhances mRNA
stability and translation. Nat Cell Biol. 20:285–295. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu J, Ren D, Du Z, Wang H, Zhang H and
Jin Y: m6A demethylase FTO facilitates tumor progression
in lung squamous cell carcinoma by regulating MZF1 expression.
Biochem Biophys Res Commun. 502:456–464. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sun J, Liu P and Wang X: MiR-595 and
Cldnd1: Potential related factors for bone loss in postmenopausal
women with hip osteoporotic fracture. PLoS One. 19:e03131062024.
View Article : Google Scholar : PubMed/NCBI
|

















![Mechanism of the transcriptional
regulation of CLDND1 by MZF1. (A) The JASPAR website (https://jaspar.genereg.net/) was used to retrieve the
binding site of the transcription factor MZF1. (B) RT-qPCR and
western blot analysis were performed to detect the expression
levels of CLDND1 in non-small cell lung cancer tissues, using
paracancerous tissues as the control (n=15). (C) RT-qPCR analysis
of CLDND1 mRNA expression in normal bronchial epithelial (BEAS-2B)
and NSCLC cell lines (CAL12T, HCC44, NCIH1993 and A549). (D) After
overexpression of MZF1, RT-qPCR was performed to measure MZF1 and
CLDND1 expression. (E) Dual-luciferase reporter assay examining the
effect of MZF1 on CLDND1 promoter activity. (F) Chromatin
immunoprecipitation assays determining the enrichment of MZF1 in
the CLDND1 promoter region [Chr3:167, 450, 703–167, 450, 714
(hg38)]. All cell experiments were repeated three times. oe,
overexpression; CLDND1, claudin domain-containing 1; MZF1, myeloid
zinc finger 1; RT-qPCR, reverse transcription-quantitative PCR; NC,
negative control; Wt, wild-type; Mut, mutant.](/article_images/mmr/33/1/mmr-33-01-13736-g04.jpg)