Introduction
Lung cancer is the leading cause of cancer morbidity
and mortality. According to the results of a study published in
2024 based on global cancer statistics, there were ~2.5 million new
cases of lung cancer and >1.8 million associated deaths in 2022,
accounting for ~1/8 (12.4%) of all cancer diagnoses and 1/5 (18.7%)
of all cancer-related deaths worldwide (1). The incidence and mortality rates of
lung cancer vary among different countries and regions, with higher
rates observed among men and in countries with a higher human
development index. With the growth in human development index, it
is expected that lung cancer incidence and mortality will markedly
increase in the coming decades (2).
Recurrence and metastasis are the leading causes of
poor prognosis and death in lung cancer. Although a number of
studies have explored cancer metastasis, the underlying mechanism
of action has not yet been fully elucidated (3). Homeobox (HOX) genes are a superfamily
of transcription factors containing homeodomains that control cell
proliferation, differentiation and morphological changes during the
early stages of embryonic development (4). The dysregulation of HOX genes
enhances cell survival and proliferation, and inhibits cell
differentiation. Previous studies have revealed that numerous HOX
genes are abnormally expressed in various types of human tumor,
such as bladder, breast, lung, liver, colorectal, prostate and
ovarian cancer (5–8).
As a recently identified HOX transcription factor,
the intestine-specific HOX (ISX) gene is specifically expressed in
the adult and fetal intestine (9).
ISX is a proto-oncogene that is upregulated in hepatocellular
carcinoma (HCC) through the induction of proinflammatory cellular
factors. Notably, it can bind directly to the cyclin D1 promoter in
the nucleus, thereby regulating the proliferation of tumor cells,
and it is also an important activator of proliferation and
tumorigenesis in vivo (10). In addition, ISX is highly expressed
in lung cancer cells and patient tissue; its upregulation can
accelerate the migration and invasion of lung cancer cells and
higher levels of ISX expression in lung cancer tissues indicate
more obvious lymph node metastasis (11). Previous studies have reported that
ISX expression can induce an epithelial-mesenchymal transition
(EMT) response, promoting tumor cell migration and invasiveness
(10,11); however, the underlying mechanisms
are not yet fully understood. Therefore, it is important to explore
the potential mechanism underlying the regulation of metastasis for
the prevention and treatment of lung cancer.
The present study aimed to establish a cell model
with enhanced migration and invasion properties by
lentivirus-mediated overexpression of the ISX oncogene, which was
assessed using cellular and molecular biology technologies. The
current study examined how ISX induces and participates in the
development of the tumor microenvironment, thereby promoting tumor
cell migration and invasion and progression. Transcriptomics
analysis was employed to further investigate potential ISX-promoted
lung cancer migration biomarkers and molecular mechanisms.
Furthermore, the therapeutic efficacy of currently available lung
cancer drugs was verified in a cell model with stable
overexpression of ISX. The study aimed to identify novel options to
treat lung cancer, and to provide a scientific basis for its
prevention and treatment, which may reduce the cost to patients,
their families and society.
Materials and methods
Cell culture
A549 human non-small cell lung cancer (NSCLC) cells
(cat. no. ZQ003) were purchased from Shanghai Zhongqiao Xinzhou
Biotechnology Co., Ltd. The cells were cultured in Dulbecco's
Modified Eagle Medium supplemented with 10% fetal bovine serum
(FBS) and 1% penicillin/streptomycin (100 U/ml) (all from Gibco;
Thermo Fisher Scientific, Inc.) at 37°C and 5% CO2. The
medium was changed every 3 days. When the cells reached the
logarithmic growth phase, they were passaged and used for
subsequent experiments.
Lentiviral infection
GFP-labeled lentiviral vectors overexpressing ISX
[Lv-ISX (Ubc-NM_001303508.2-3FLAG-CBh-gcGFP-IRES-puromycin; Gene:
ISX Human, gene ID: 91464; transcribed transcript: NM_001303508.2)]
and a negative control lentiviral vector
(Ubc-MCS-3FLAG-CBh-gcGFP-IRES-puromycin) were obtained from
Shanghai GeneChem Co., Ltd. A549 cells in the logarithmic growth
phase were harvested, trypsinized and counted. The cell density was
adjusted to 2×105 cells/well for inoculation into 6-well
plates, 2 ml complete medium was added, and the plates were
incubated at 37°C and 5% CO2 overnight to allow the
cells to attach to the wall and reach 30–50% confluence. First, a
preliminary experiment involving viruses with different
multiplicity of infection (MOI) gradients was conducted to
determine the minimum effective infection dose (MOI, 10). Based on
the pre-experiment result, 1 ml medium containing the virus at an
MOI of 10 was added to each well. After 24 h of incubation, the
medium was removed and fresh medium was added. The transfection
efficiency was observed under a fluorescence microscope after 48 h.
If the proportion of GFP-labeled cells reaches >80%, the cells
were harvested for further validation by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
western blotting. The validated cells were either screened in a
cell culture medium supplemented with 1 µg/ml puromycin for
continued culture or cryopreserved in liquid nitrogen for use in
subsequent experiments.
Western blot analysis
Total protein was extracted from the cells using
RIPA buffer (cat. no. G2002; Wuhan Servicebio Technology Co., Ltd.)
according to the manufacturer's instructions, and the protein
concentration was measured using a BCA assay. Protein samples were
subjected to SDS-PAGE on 12% gels at a loading volume of 10 µl/well
(total protein, 30 µg) and then transferred to PVDF membranes. The
membranes were incubated for 1 h at room temperature with 5%
non-fat milk/TBS-0.1% Tween-20 (TBST) and were then washed five
times with TBST. Subsequently, the membranes were incubated
overnight at 4°C with anti-ISX (1:100; cat. No. sc-398934; Santa
Cruz Biotechnology, Inc.) and anti-β-actin (1:500; cat. No.
sc-58673; Santa Cruz Biotechnology, Inc.) antibodies, followed by
incubation with a horseradish peroxidase-coupled secondary antibody
(1:2,000; cat. No. 7076; Cell Signaling Technology, Inc.) for 1 h
at room temperature. Finally, the membranes were incubated with ECL
solution (cat. no. G2014; Wuhan Servicebio Technology Co., Ltd.),
and the images were exposed and developed. Protein expression was
analyzed semi-quantitatively using ImageJ2 (Fiji; National
Institutes of Health).
RT-qPCR analysis
RT-qPCR detection was performed using the RNA
extracted from A549 cells with or without ISX overexpression, as
well as from A549 cells overexpressing ISX treated with 25 µg/ml
gefitinib (Iressa®; cat. no. HY-50895; MedChemExpress)
for 48 h in a cell incubator at 37°C. Total RNA was extracted from
these cells using the EZ-10 DNA-free RNA Mini-Preps Kit (Sangon
Biotech Co., Ltd.) according to the manufacturer's instructions. A
total of 1 µg RNA was then reverse transcribed into cDNA using a
PrimeScript™ RT Kit with gDNA Eraser (Takara Bio, Inc.)
according to the manufacturer's protocol. Briefly, total RNA, 5X
PrimeScript RT Master Mix (Perfect Real Time) and RNase Free
dH2O were used to form a reaction system, which was
incubated at 42°C for 15 min, followed by incubation at 85°C for 5
sec. Subsequently, qPCR was performed using TB Green Premix Ex
Taq™ II (Takara Bio, Inc.) according to the
manufacturer's protocol. The reaction system included 2 µl cDNA, 1
µl primer, 12.5 µl 2X TB Green Premix Ex Taq II (Tli RNaseH Plus)
and 8.5 µl ddH2O, and the thermocycling conditions used
were as follows: 95°C for 30 sec, followed by 40 cycles at 95°C for
5 sec and 60°C for 30 sec. Melt curve analysis was performed at the
end of amplification by increasing the temperature from 65 to 95°C
in 0.5°C increments every 5 sec to confirm the specificity of the
amplification products. The primer sequences used were as follows:
GAPDH, forward (F) 5′-TGCACCACCAACTGCTTAGC-3′, reverse (R)
5′-GGCATGGACTGTGGTCATGAG-3′; ISX, F 5′-CCTGCTCTCTGCAGGGGT-3′, R
5′-CTGTCCATATCACTCCTCCTGGC-3′; TWIST, F 5′-GGAGTCCGCAGTCTTACGAG-3′,
R 5′-TCTGGAGGACCTGGTAGAGG-3′; Slug, F
5′-ACACATTACCTTGTGTTTGCAAGATCT-3′, R
5′-TGTCTGCAAATGCTCTGTTGCAGTG-3′; vimentin (VIM), F
5′-GAGAACTTTGCCGTTGAAGC-3′, R 5′-GCTTCCTGTAGGTGGCAATC-3′; zinc
finger E-box binding HOX 1 (ZEB1), F
5′-GAAAATGAGCAAAACCATGATCCTA-3′, R 5′-CAGGTGCCTCAGGAAAAATGA-3′; and
E-cadherin, F 5′-AGAACGCATTGCCACATACACTC-3′, R
5′-CATTCTGATCGGTTACCGTGATC-3′. The relative expression levels of
the genes were examined using the 2−ΔΔCq method with
GAPDH as the reference gene (12).
Transcriptome sequencing
Three replicates of A549 cells with lentivirus
infection-mediated ISX overexpression and three replicates of A549
cells infected with the negative control lentiviral vector were
used for transcriptome sequencing. These six samples were sent to
Beijing Novogene Technology Co., Ltd. for transcriptome sequencing
and data analysis. Briefly, RNA was extracted using the TRNzol
universal reagent; cat. no. 4992730; Tiangen Biotech Co., Ltd.)
method, and the total amounts and integrity of extracted RNA were
assessed using the RNA Nano 6000 Assay Kit on the Bioanalyzer 2100
system (Agilent Technologies, Inc.). Upon successful detection,
library construction was performed using the NEB Next
Ultra™ RNA Library Prep Kit (Illumina, Inc.). After the
construction of the library, the library was quantified using the
Qubit2.0 Fluorometer (Thermo Fisher Scientific, Inc.), then diluted
to 1.5 ng/µl, and the insert size of the library was detected using
the Agilent 2100 bioanalyzer. RT-qPCR (Touch q-PCR system CFX96;
Bio-Rad Laboratories, Inc.) was used to accurately quantify the
effective concentration of the library (the loading concentration
of the final library was >2 nM) to ensure the quality of the
library. RT-qPCR was performed using TB Green Premix Ex
Taq™ II (Takara Bio, Inc.), and the thermocycling
conditions used were as follows: 95°C for 30 sec, followed by 40
cycles at 95°C for 5 sec and 60°C for 30 sec. Subsequently, the
libraries were pooled according to the required effective
concentration and target downstream data volume for Illumina
sequencing [Illumina NovaSeq X plus; Illumina MiSeq Reagent Kit v3
(cat. no. MS-102-3003); both from Illumina, Inc.], and 150 bp
paired-end reads were generated. The image data of the sequenced
fragments measured by the high-throughput sequencer were converted
into sequence data (reads) by CASAVA base recognition (version 1.6;
Illumina, Inc.). The raw data were then filtered by removing reads
containing adapters or ‘N’ (which indicates that base information
could not be determined), as well as low-quality reads (reads with
Qphred ≤20 bases accounting for >50% of the entire read length).
Subsequently, the Q20 (≥90%) and Q30 (≥85%) values, and the GC
content (45–55%) were calculated for the clean and filtered data,
and it was ensured that all subsequent analyses were of a high
quality and based on clean data. The HISAT2 software (v2.0.5;
http://daehwankimlab.github.io/hisat2/) was then used
to rapidly and accurately align the clean reads with the reference
genome to obtain the localization information of the reads on the
reference genome. Intergroup differences and intra-group sample
replication were further evaluated through Pearson's correlation
analysis and principal component analysis.
The Venn diagram was implemented using the
VennDiagram R package (version 1.7.3; http://cran.r-project.org/web/packages/VennDiagram)
and the gene expression heatmaps were generated using the ggplot2
(version 3.5.1; http://cran.r-project.org/web/packages/ggplot2/index.html)
and pheatmap R packages (version 1.0.2; http://cran.r-project.org/package=pheatmap).
After quantifying the gene expression levels, differential
expression between the two comparison groups was analyzed using
DESeq2 software (version 1.20.0; http://packages.renjin.org/package/org.renjin.bioconductor/DESeq2).
The Benjamini-Hochberg method was then used to adjust the obtained
P-value (Padj) to control the false discovery rate. The threshold
for significant differential expression was set to Padj ≤0.05 and
|log2 (fold change)|≥1. The names of each differentially expressed
gene (DEG) and the word ‘cancer’ were used as key words when
searching Google Scholar (https://scholar.google.com/) to explore the
association between these genes and cancer. The retrieved
literature, including research articles and review papers, was
further screened based on titles and abstracts, and finally
confirmed through the content of the documents. The clusterProfiler
(version 3.8.1; http://github.com/YuLab-SMU/clusterProfiler) software
was then used to perform Gene Ontology (GO) and Kyoto Encyclopedia
of Genomes (KEGG) pathway enrichment analyses on the DEGs. A Padj
value of <0.05 was considered significant. The STRING database
(v11.5; http://string-db.org/) was further used
to construct the protein-protein interaction (PPI) network, and the
top five central genes were identified using the Degree algorithm
in the CytoHubba plugin of Cytoscape software (version 0.1;
http://apps.cytoscape.org/).
Cell Counting Kit 8 (CCK8) assay
The active ingredients of the following established
lung cancer drugs were purchased from MedChemExpress: Bevacizumab
(Avastin®), erlotinib (Tarceva®), cetuximab
(Erbitux®) and gefitinib. Initially, their effects on
the viability of uninfected A549 cells were assessed. Specifically,
A549 cells with or without ISX overexpression in the logarithmic
growth phase were inoculated into 96-well plates at a density of
3,000 cells/well and were cultured for 24 h. Cells not
overexpressing ISX were then treated with bevacizumab, erlotinib,
cetuximab and gefitinib (100 µl) at concentrations of 0, 5, 25, 30,
35, 40, 45 and 50 µg/ml, and were cultured in the incubator for 48
h. In addition, cells overexpressing ISX were treated with
gefitinib under the same conditions. After 48 h, 10 µl CCK8 reagent
(cat. no. CK04; Dojindo Laboratories, Inc.) was added to each well.
Several wells with only the culture medium added and no cells were
used as blank controls. After incubation at 37°C for 2–4 h, the
optical density (OD) at 450 nm was measured using an ultraviolet
spectrophotometer. The cell viability was calculated using the
following formula: Cell viability (%)=[(experimental group OD-blank
group OD)/(control group OD-blank group OD)] ×100.
Wound healing assay
Cells overexpressing ISX in the logarithmic growth
phase were collected, digested with trypsin into a single-cell
suspension and inoculated at a density of 1×105
cells/well into a 6-well culture plate to ensure that the fusion
rate of the inoculated cells reached >90% overnight.
Subsequently, a sterile pipette tip was used to make a straight
scratch on the cell monolayer, followed by a wash with
phosphate-buffered saline to remove the exfoliated cells. An
initial image of the scratch was captured under a fluorescence
microscope and recorded as 0 h. Subsequently, the medium was
replaced with low-serum (1% FBS) medium with or without gefitinib.
After culturing for a further 48 h, images were captured from the
same position as at 0 h to record the closure of the scratches.
Images of the scratch area were analyzed using ImageJ software and
the scar healing analysis plug-in (MRI_Wound_Healing_Tool;
http://www.mri.cnrs.fr/en/data-analysis/software-and-tools/271-mri-tools/409-wound-healing-tool.html)
was used to measure cell migration. Migration rate (%)=(48 h
scratch area-0 h scratch area)/0 h scratch area ×100.
Cell invasion assay
A Transwell invasion assay was conducted using
Transwell cell chambers (24-well plates; pore size, 8 µm) that were
coated with Matrigel. Briefly, Matrigel was diluted in PBS at a 1:8
ratio under 4°C conditions (on ice). Subsequently, 50 µl was added
evenly to the upper chamber surface of the Transwell chamber and
incubated at 37°C for 4 h to allow the Matrigel to polymerize into
a gel film. A549 cells overexpressing ISX in the logarithmic growth
phase were assessed. Briefly, after digestion, cells were
resuspended in serum-free medium, and 250 µl cells
(8×104/well) were transferred to the upper chamber of
the Transwell. Different concentrations of gefitinib diluted in
medium containing 10% FBS (0, 25 and 50 µg/ml) were added to the
lower chamber. The cells were incubated for 48 h to allow the cells
to penetrate the Matrigel and to migrate into the lower chamber.
Subsequently, the non-invasive cells were removed from the upper
chamber with a cotton swab, and the lower chamber membrane was
fixed with 4% paraformaldehyde for 30 min followed by staining with
0.2% crystal violet for 10 min at room temperature. Observations
were made and images were captured under a light microscope.
Finally, the crystal violet was completely eluted by thoroughly
shaking the solution in 3% acetic acid at room temperature for 5
min. The eluate was then used to measure the OD value at 570 nm
using a microplate reader.
Statistical analysis
All data are presented as the mean ± SEM (n≥3) of
three independent experiments. The results were statistically
analyzed using GraphPad Prism 9.0 software (Dotmatics). Statistical
differences between the two groups were analyzed using an unpaired
Student's t-test. Statistical differences among multiple groups
were analyzed using ANOVA followed by Tukey's multiple comparisons
post hoc test to identify the groups with significant differences.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Overexpression of the carcinogenic
factor ISX promotes the expression of EMT-related genes
To explore the potential role of the oncogene ISX in
regulating downstream genes associated with EMT in lung cancer
metastasis and its molecular mechanisms, ISX overexpression was
induced in A549 lung cancer cells using lentiviral infection and
the expression of EMT markers was verified. The MOI and optimal
conditions for lentiviral infection of cells were first determined
through preliminary experimentation, the results of which are shown
in Fig. 1A. Using an MOI of 10
resulted in healthy cells with high fluorescence expression and an
infection efficiency of >80%. Subsequently, the mRNA and protein
expression levels of ISX were verified by RT-qPCR and western
blotting. The results showed that both the mRNA and protein
expression levels of ISX were significantly increased after
lentiviral infection (Fig. 1B and
C). Subsequent experiments were conducted using A549 cells
overexpressing ISX. The effects of ISX overexpression on the
expression of EMT markers were further validated (Fig. 1D). Consistent with the findings of
previous reports (10,11), ISX overexpression significantly
increased the expression levels of the EMT markers TWIST, Slug, VIM
and ZEB1, while downregulating the expression of the epithelial
cell marker E-cadherin.
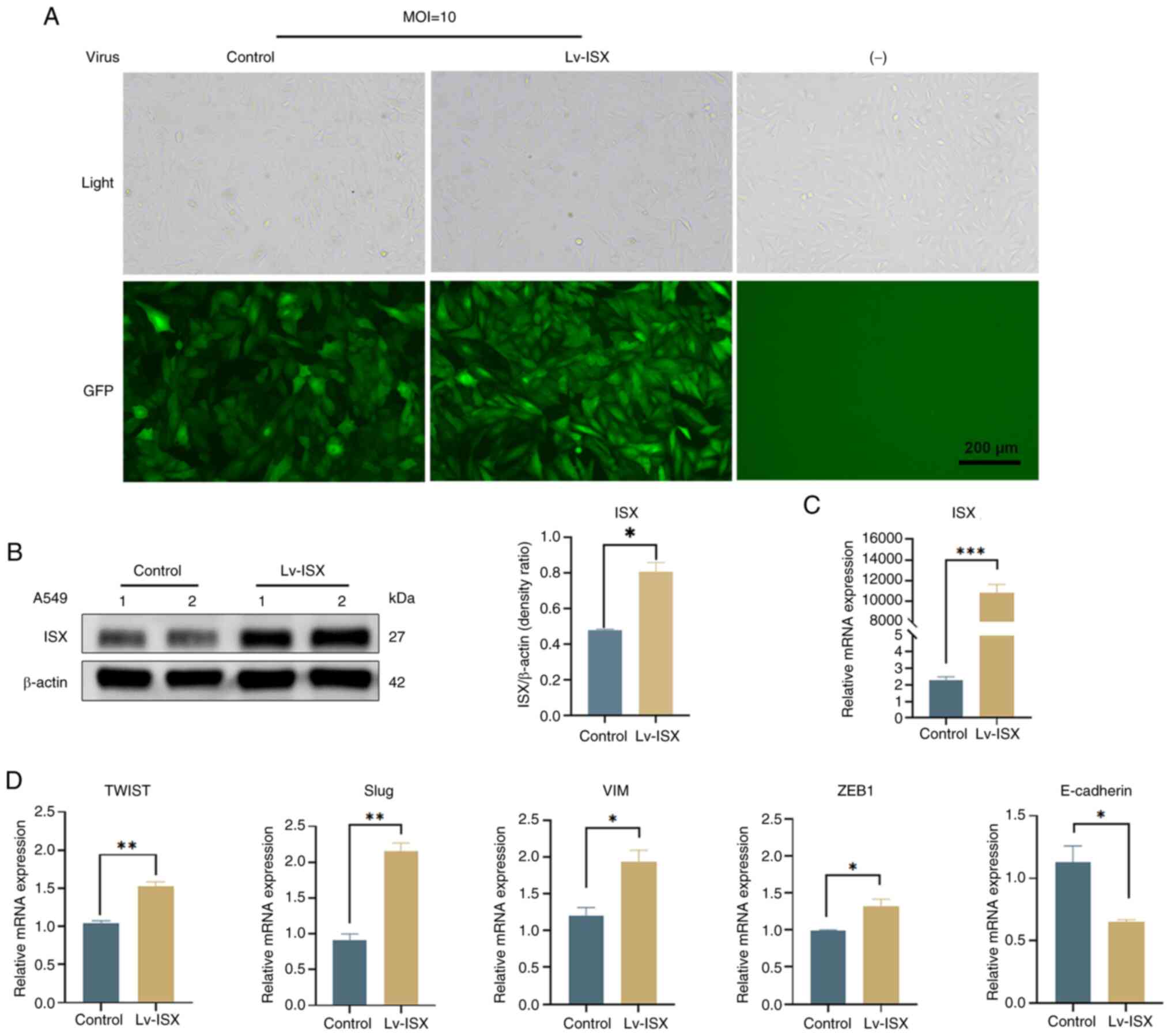 | Figure 1.Overexpression of the oncogenic
factor ISX promotes the expression of EMT-related genes. (A) ISX
overexpression in A549 cells was mediated by Lv infection and
observed by GFP fluorescence intensity. Scale bar, 200 µm. (B) ISX
protein expression levels were verified by western blot analysis.
(C) ISX mRNA expression levels were verified by RT-qPCR detection.
(D) Genes related to EMT (TWIST, Slug, VIM and ZEB1) and epithelial
cells (E-cadherin) were detected by RT-qPCR. *P<0.05;
**P<0.01, ***P<0.001. EMT, epithelial-mesenchymal transition;
ISX, intestine-specific homeobox; Lv, lentivirus; MOI, multiplicity
of infection; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; VIM, vimentin; ZEB1, zinc finger E-box
binding homeobox 1. |
ISX overexpression significantly
alters the gene expression profile of human lung cancer cells
The present study aimed to investigate the
mechanisms related to the promotion of tumor cell metastasis and
progression by the oncogenic factor ISX, and to discover their
possible underlying mechanisms and biomarkers. Briefly,
transcriptome sequencing was performed on A549 cells overexpressing
ISX via lentiviral infection and on A549 cells infected with the
control lentivirus vector (n=3 samples/group). The sequencing data
were uploaded to the Sequence Read Archive of the National Center
for Biotechnology Information under the accession number
PRJNA1282884.
Table SI
summarizes the sequencing metrics and quality check results for the
raw RNA sequencing reads. After excluding reads containing
aptamers, one N base or low quality, 46,016,622, 39,526,880, and
44,502,284 clean reads were obtained in the control groups (Con1,
Con2, and Con3), whereas 45,903,940, 44,323,396, and 43,279,598
clean reads were obtained in the ISX overexpression groups
(Lv_ISX1, Lv_ISX2, and Lv_ISX3). The sequencing error rate for each
library was 0.01%, and >95% of the clean read data had
Phred-like quality scores at the Q30 level (error probability of
0.001), indicating high sequencing quality. The total mapping rate
of the reference genome ranged between 94.64 and 95.72%, while a
smaller percentage (<5%) of the reference genome's multiple loci
were mapped (Table SII, Multi
map), further verifying the reliability of the sequencing quality.
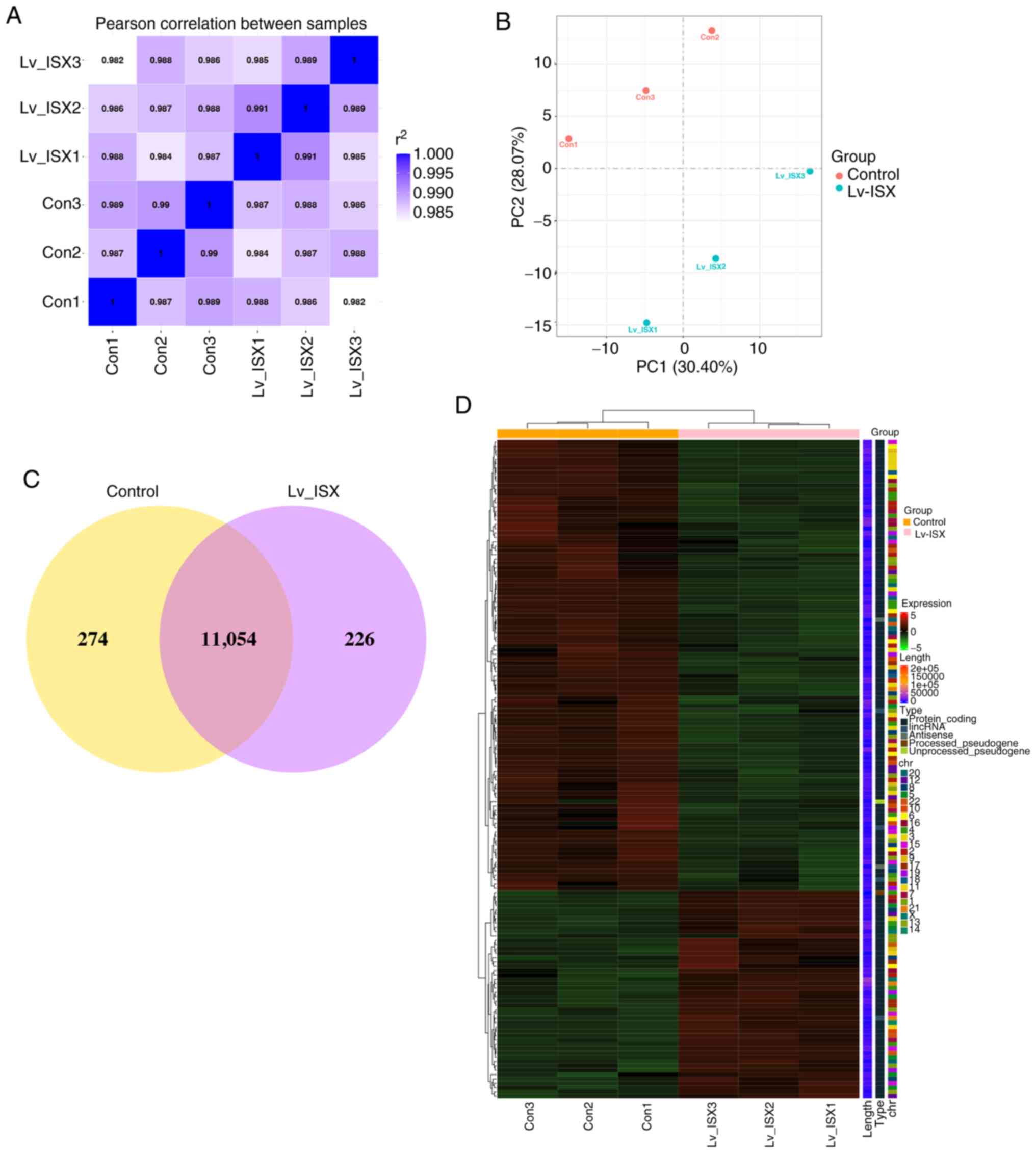
Pearson's correlation analysis and principal component analysis
were performed on the two groups of samples to investigate their
similarities and differences. The results showed that samples
within the same group were highly similar, whereas samples in
different groups were dissimilar (Fig.
2A and B). The Venn diagram (Fig.
2C) showed a total of 11,054 genes in both groups. The
clustering heatmap further confirmed the reproducibility of the
samples, and the global gene expression patterns were revealed to
be significantly different between the treatment and control groups
(Fig. 2D).
Gene expression analysis was performed on the
transcripts to further characterize the DEGs in the ISX
overexpression group compared with in the control group. A total of
152 DEGs were identified, 48 of which were upregulated and 104 of
which were downregulated (Fig.
3A). The heatmap in Fig. 3B
and Table SIII show the
upregulated DEGs in the ISX overexpression group. The results of
the literature review indicated that most of the DEGs were
associated with tumor metastasis. Subsequently, the biological
functions and signaling pathways of these genes were
comprehensively analyzed by GO functional annotation and KEGG
pathway analyses.
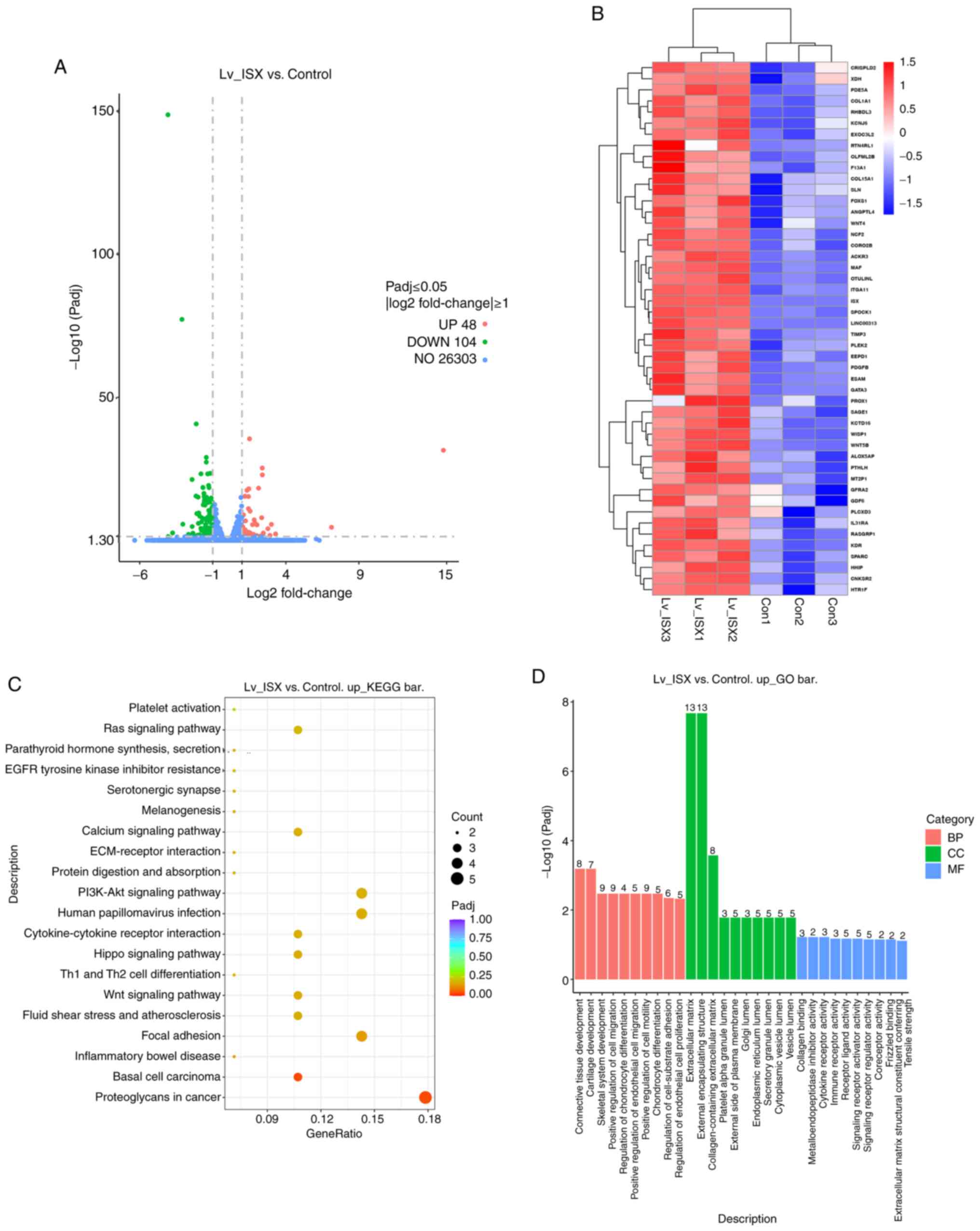 | Figure 3.Screening of DEGS in cells with ISX
overexpression, and enrichment analyses of biological functions and
signaling pathways. (A) Volcano plot showing the number of genes
with significant differences in expression levels between the ISX
overexpression group and the control group, as well as the specific
distribution of DEGs. (B) Heatmap showing the expression of DEGs
that were upregulated in the ISX overexpression groups. (C) KEGG
analysis showing the top 20 enriched pathways of the DEGs that were
upregulated in the ISX overexpression group compared with the
control group. Bubble size represents the number of genes, color
shade represents Padj, and the enrichment ratio on the y-axis
represents the number of genes/total number of genes. (D) GO term
enrichment analysis of the DEGs that were upregulated in the ISX
overexpression group compared with the control group. The numbers
on the columns indicate the number of enriched genes in that
category. BP, biological process; CC, cellular component; Con,
control; DEG, differentially expressed gene; GO, Gene Ontology;
ISX, intestine-specific homeobox; KEGG, Kyoto Encyclopedia of Genes
and Genomes; Lv, lentivirus; MF, molecular function. |
KEGG pathway analysis revealed the primary pathways
enriched in the upregulated DEGs, including ‘proteoglycans in
cancer’, ‘basal cell carcinoma’, ‘focal adhesion’, ‘Wnt signaling
pathway’, ‘cytokine-cytokine receptor interaction’, ‘PI3K-Akt
signaling pathway’, ‘ECM-receptor interaction’, ‘calcium signaling
pathway’ and ‘Ras signaling pathway’ (Fig. 3C). Further functional enrichment
analyses revealed that these interacting genes were primarily
involved in the following biological processes: ‘Connective tissue
development’, ‘cartilage development’, ‘positive regulation of cell
migration’, ‘regulation of chondrocyte differentiation’, ‘positive
regulation of endothelial cell migration’ and ‘positive regulation
of cell motility’. Enriched cellular components included
‘extracellular matrix’, ‘endoplasmic reticulum lumen’ and
‘collagen-containing extracellular matrix’. At the molecular
function level, these genes were associated with ‘collagen
binding’, ‘metalloendopeptidase inhibitor activity’, ‘cytokine
receptor activity’, ‘immune receptor activity’, ‘receptor ligand
activity’ and ‘extracellular matrix structural constituent
conferring tensile strength’ (Fig.
3D). These results suggested that the overexpression of ISX may
contribute to the processes of invasion, migration and cartilage
formation in lung cancer cells.
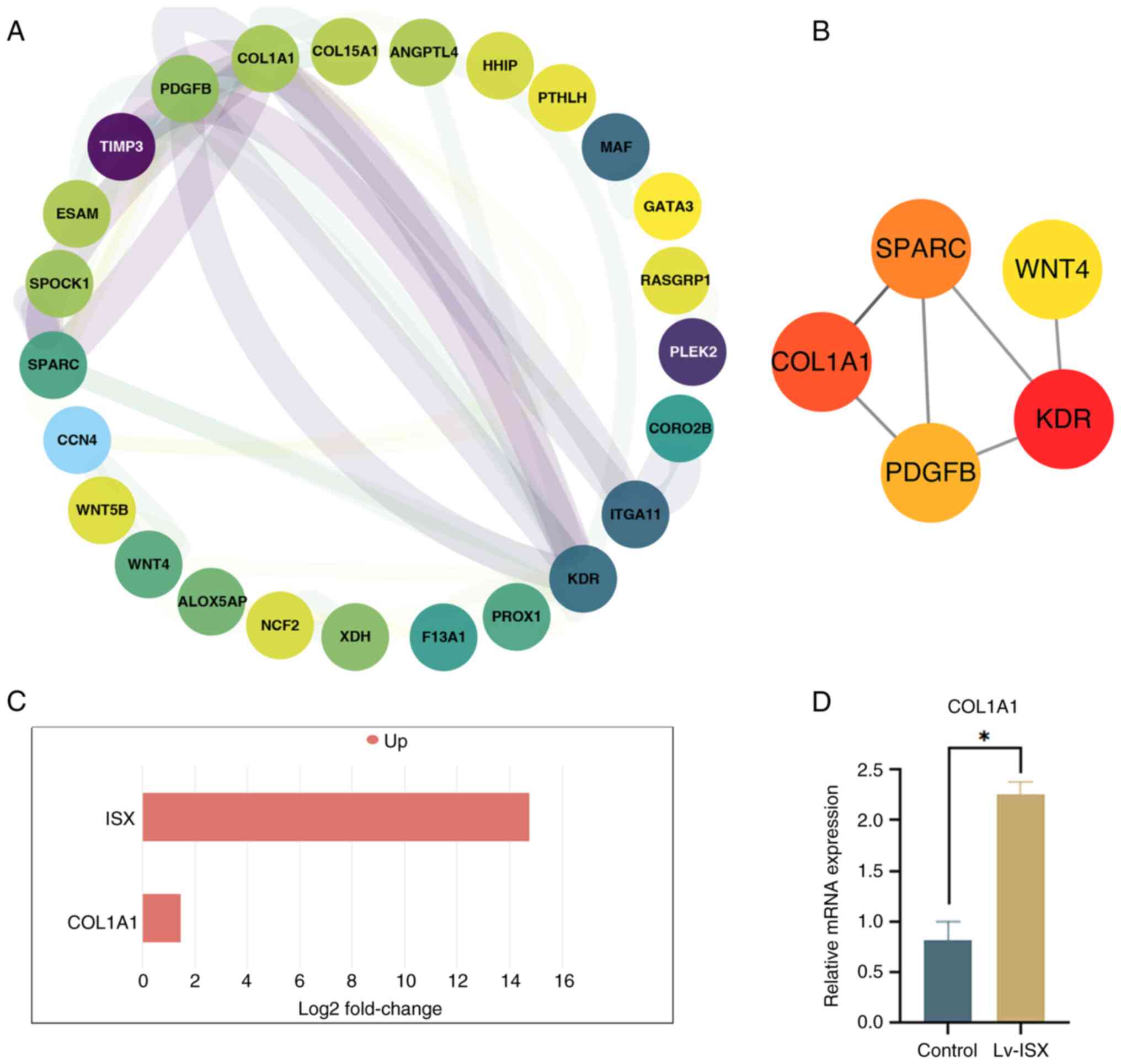
ISX may promote lung cancer migration
and invasion by upregulating type I collagen α1 chain (COL1A1)
To further investigate the possible mechanism by
which ISX overexpression promotes tumor cell migration and
invasion, a PPI network was constructed for the upregulated DEGs by
using the software program Cytoscape (Fig. 4A). Based on the degree-based
topological algorithm, the top five core genes (KDR, COL1A1, SPARC,
PDGFB and WNT4), which showed the most significant interactions,
were identified using the Cytoscape CytoHubba application (Fig. 4B). Notably, COL1A1 was recognized
as one of the hub genes; COL1A1 is a key protein encoding fibrous
collagen, the main component of type I collagen, and it server a
crucial role in maintaining cell morphology, intercellular
connections, tissue structure stability and extracellular matrix
(ECM) homeostasis (13). Notably,
COL1A1 dysfunction has been shown to be associated with various
diseases, including fibrosis, osteogenesis imperfecta and
osteoporosis, which are bone diseases caused by COL1A1 deficiency
(14). In recent years, multiple
reports have shown that COL1A1 is upregulated in various tumor
tissues and cells, prompting researchers to pay increasing
attention to its role in cancer (15–18).
The transcriptome sequencing results showed that ISX
overexpression led to the upregulated expression of COL1A1
(Fig. 4C), which was further
verified by RT-qPCR (Fig. 4D).
Studies have shown that the abnormal upregulation of COL1A1 in
cancer is closely related to regulating tumor cell proliferation,
differentiation and migration (15). These findings provide important
insights into the specific mechanisms by which COL1A1 may be
involved in ISX-induced regulation of tumor migration and invasion.
Its potential mechanism awaits further systematic analysis in
future research.
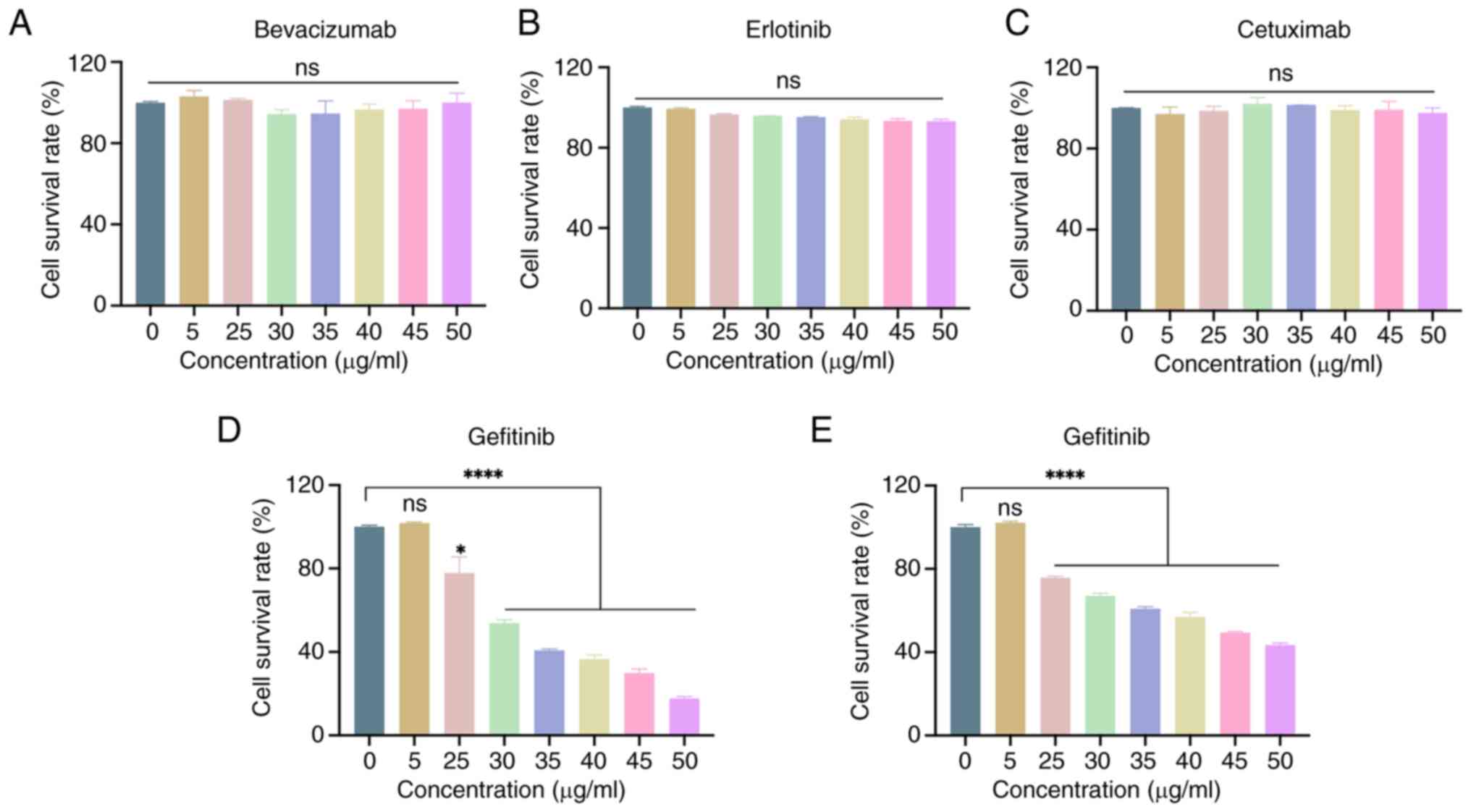
Screening of mature tumor-targeting
drugs that may be used to treat ISX-induced lung cancer
Metastasis of tumor cells is the primary cause of a
poor prognosis for tumors. Notably, the upregulation of ISX may
accelerate the migration and invasion of lung cancer cells;
therefore, four mature tumor-targeting drugs were used to treat the
ISX-induced lung cancer cell model, with the aim of identifying
better therapeutic drugs that can resist tumor metastasis. The
drugs used in the current study included Iressa (gefitinib),
Tarceva (erlotinib) and Avastin (bevacizumab), which are already
used to treat lung cancer. Although Erbitux (cetuximab) is approved
for treating colorectal cancer and head and neck cancer, as an
anti-EGFR monoclonal antibody, it has been extensively studied in
combination with chemotherapy for the treatment of advanced NSCLC
(19,20). The current study first evaluated
the effects of these treatments on the viability of un-infected
A549 cells. As shown in Fig. 5A-D,
only gefitinib affected A549 cell viability within the tested
concentration range (0–50 µg/ml). The concentration range
significantly affected A549 cell viability in a dose-dependent
manner, starting at 25 µg/ml. Therefore, gefitinib was subsequently
used for the treatment of cells stably overexpressing ISX. The
results showed that gefitinib was also effective in reducing the
viability of A549 cells stably overexpressing ISX at concentrations
of ≥25 µg/ml (Fig. 5E), although
the reduction in cell viability was not as high as that in
non-infected A549 cells.
The effects of gefitinib (the active ingredient of
Iressa®) on the migration and invasion characteristics
of lung cancer cells with stable overexpression of ISX were then
evaluated through wound healing and Transwell assays. The results,
as shown in Fig. 6A and B,
indicated that gefitinib can significantly inhibit the migration
and invasion of ISX-overexpressing A549 lung cancer cells in a
concentration-dependent manner. Furthermore, the effects of
gefitinib on the expression of ISX, COL1A1 and EMT-related genes
were detected by RT-qPCR to preliminarily explore the mechanism of
gefitinib in inhibiting the migration and invasion of lung cancer
cells stably overexpressing ISX. Although gefitinib had no
inhibitory effect on the expression of ISX and EMT-related genes
(data not shown), the results showed that it could significantly
reduce the upregulation of COL1A1 gene expression caused by ISX
(Fig. 6C). These data initially
suggested that the mature tumor-targeting drug Iressa®
may be a promising therapeutic drug that can resist tumor
metastasis, particularly in patients with tumor metastasis caused
by high ISX expression. Moreover, the downregulation of COL1A1 gene
expression may be considered an important molecular mechanism for
its effective resistance to tumor metastasis, which is worthy of
in-depth exploration in future studies.
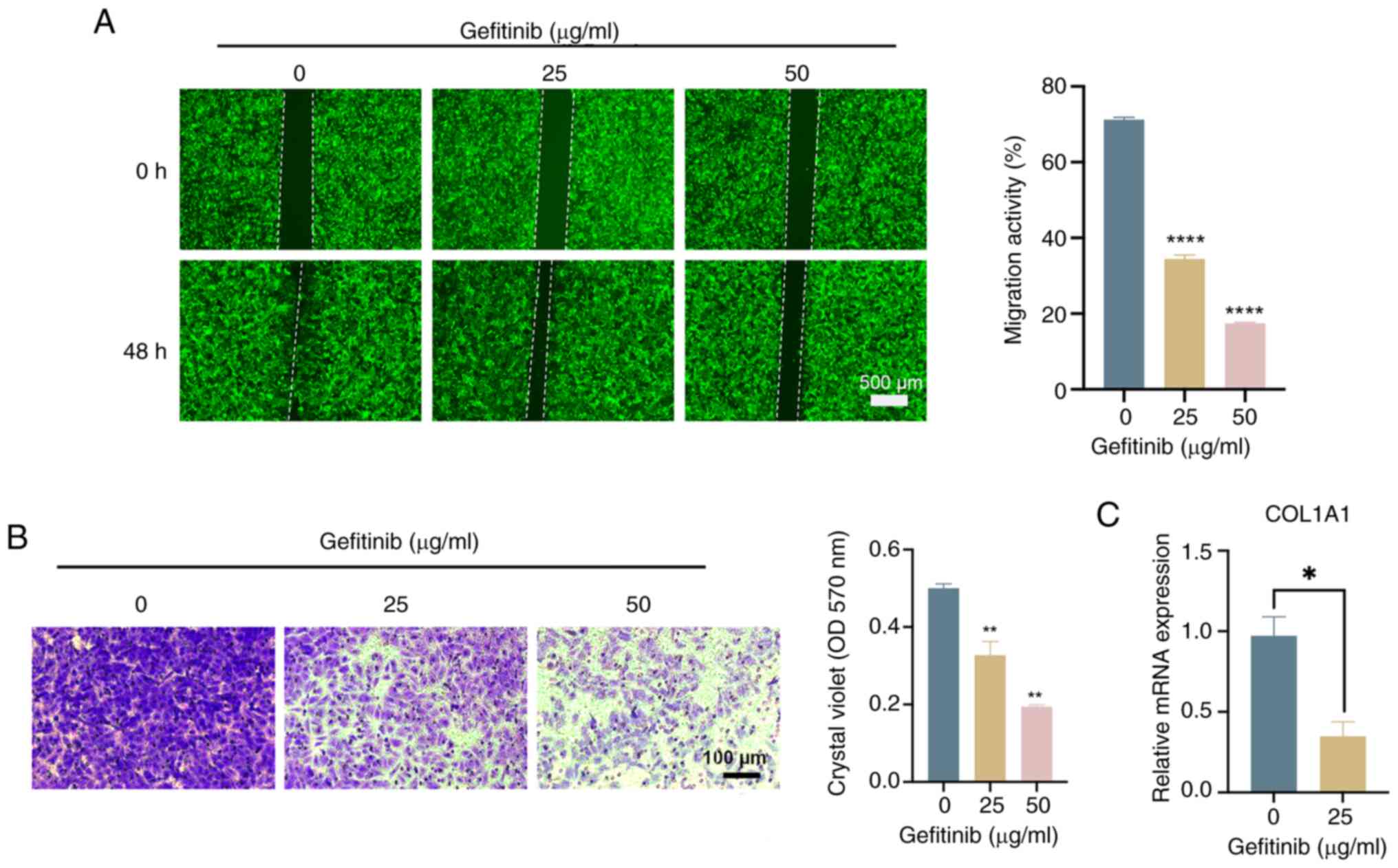 | Figure 6.Mature tumor-targeting drug
Iressa® (gefitinib) may inhibit the migration and
invasion of ISX-overexpressing human lung cancer cells by
regulating COL1A1. A549 cells infected with a GFP-labeled ISX
overexpression vector were treated with different concentrations of
gefitinib (0, 25, 50 µg/ml) for 48 h, and (A) cell migration (wound
healing; scale bar, 500 µm, and (B) invasion (Transwell; scale bar,
100 µm) assays were performed. (C) Reverse
transcription-quantitative polymerase chain reaction was used to
detect the effect of gefitinib (25 µg/ml, 48 h) on the mRNA
expression levels of COL1A1 in ISX-overexpressing A549 cells.
*P<0.05; **P<0.01; ****P<0.0001. COL1A1, type I collagen
α1 chain; ISX, intestine-specific homeobox; OD, optical
density. |
Discussion
As a gut-specific HOX transcription factor, ISX is a
proto-oncogene induced by the inflammatory factor IL-6, and
previous researchers have identified the effect of ISX on cancer
development. In HCC cells, the kynurenine (KYN) pathway promotes
oncogenic activity by binding to aryl hydrocarbon receptor (AHR) in
an ISX-dependent positive feedback loop. In addition, it has been
shown that the ISX-KYN axis upregulates immunosuppressive activity
by targeting CD86 and programmed death ligand 1 (PD-L1) expression,
thereby suppressing CD8+ T-cell proliferative responses.
Thus, ISX-KYN-AHR signaling acts together with CD86 and PD-L1 to
promote the carcinogenesis and immunosuppression of HCC cells
(10). In lung cancer, ISX is
highly expressed in lung cancer cells and patient tissues, and its
upregulation is positively associated with lung cancer cell
migration and invasion. A mechanistic study has shown that the
P300/CBP-associated factor (PCAF)-ISX-bromodomain-containing
protein 4 (BRD4) axis is an important regulator of lung cancer
metastasis and cell plasticity (11). PCAF-mediated acetylation of the
transcription factor ISX promotes ISX-BRD4 translocation to the
nucleus, which activates EMT genes and induces metastasis (11).
Cancer cell migration is a complex process in which
the EMT of cancer cells is the initial step. Although numerous
signaling molecules and pathways (such as PI3K, Snail,
hypoxia-inducible factor 1α and Smad interacting protein 1) and
transcription factors (TWIST1/2, SNAIL1/2, ZEB1/2 and FOXC2) have
been proposed to regulate and induce EMT (21), the detailed regulatory mechanisms
remain largely unknown. In the present study, lung cancer cells
stably overexpressing ISX were constructed by lentivirus infection,
and EMT-related gene expression was detected by RT-qPCR. It was
verified that ISX expression induced EMT response, and promoted
tumor cell migration and invasion. To further investigate the
molecular regulatory mechanisms by which the oncogene ISX enhances
tumor cell invasion and migration, transcriptome sequencing and
analysis was performed using A549 cells overexpressing ISX. The
results revealed that COL1A1 was upregulated in ISX-overexpressing
lung cancer cells, which was verified by RT-qPCR. The Cytoscape
CytoHubba analysis further identified COL1A1 as a hub gene (one of
the top five core genes) in the PPI pathway, indicating that COL1A1
may be involved in the molecular mechanism of ISX-regulated tumor
metastasis.
COL1A1 is one of the main components of the ECM, and
the dynamic balance of ECM serves a central role in maintaining the
tumor microenvironment. Numerous studies have shown that COL1A1 is
upregulated in numerous types of cancers and that it can affect
various signaling pathways related to those types of cancer,
including gastric cancer, lung adenocarcinoma, ovarian cancer,
colorectal cancer, breast cancer and thyroid cancer (15,16,22–26).
In HCC, knocking down COL1A1 markedly inhibits cell invasion and
migration, and downregulates VIM, a protein that promotes the EMT
phenotype, thus suggesting that COL1A1 may promote tumor cell
migration and invasion by promoting EMT (27). A multi-omics analysis previously
identified COL1A1 as a key gene for lung adenocarcinoma development
and progression (28).
Subsequently, the clinical importance of COL1A1 expression in lung
cancer samples and its association with clinical prognosis were
determined by detecting the expression levels of COL1A1 in lung
cancer samples. The results demonstrated that the mRNA levels and
gene amplification of COL1A1 in lung cancer tissues are higher than
those in normal lung tissues. Furthermore, COL1A1 was shown to be
highly expressed in lung cancer tissues and serum, and high
expression of COL1A1 was revealed to be associated with poor
progression-free survival and chemotherapy resistance in patients
with lung cancer. Therefore, COL1A1 may be used as a novel
diagnostic, prognostic and chemoresistance biomarker for human lung
cancer (29). Taken together,
these findings suggested that ISX may induce EMT by upregulating
COL1A1 and could alter the tumor microenvironment, thereby
promoting tumor migration and more aggressive lung cancer behavior.
This represents a novel mechanism discovered through transcriptome
sequencing in the current study. However, only A549 cells were used
in subsequent experiments. Further validation through clinical
trials is required to determine the specific type of lung cancer
associated with this mechanism; this will be a key area assessed in
our future research.
Notably, the PI3K/AKT signaling pathway was enriched
in the upregulated DEGs of the ISX-overexpressing lung cancer
cells. It has been shown that knockdown of COL1A1 can inhibit the
progression of gastric cancer by regulating the PI3K/AKT signaling
pathway (22). In addition, an
association between COL1A1 and the PI3K/Akt signaling pathway has
been detected in ovarian cancer. This suggests that COL1A1 may
regulate the proliferation, anti-apoptotic ability and migratory
potential of ovarian cancer cells via the PI3K/Akt signaling
pathway (16). Therefore, the
PI3K/AKT signaling pathway may also be involved in the mechanism by
which ISX regulates COL1A1 expression in lung cancer. Although
studies have shown that the PI3K/AKT pathway serves an important
role in tumors (30), to the best
of our knowledge, the specific relationship between ISX and COL1A1
and this pathway in lung cancer has not yet been reported. Future
studies should further explore the molecular mechanism of ISX
regulating lung cancer progression through COL1A1 and PI3K/AKT
signaling pathways.
In conclusion, the present study demonstrated that
the expression levels of COL1A1 were increased in lung cancer cells
overexpressing ISX, and indicated that the PI3K/AKT signaling
pathway may be involved in ISX-regulated COL1A1 expression,
promoting tumor migration and invasion. In addition, four currently
established drugs for lung cancer treatment were screened. Among
them, Iressa (gefitinib) significantly inhibited the viability,
migration and invasion of lung cancer cells stably overexpressing
ISX. The expression of COL1A1 was also significantly downregulated
after gefitinib treatment, indicating that gefitinib may be a
promising cancer drug for the treatment of tumor metastasis by
inhibiting the upregulation of COL1A1 expression caused by ISX
overexpression. The present findings revealed a novel molecular
mechanism by which ISX may promote the progression and metastasis
of lung cancer. This discovery provides new theoretical insights
into the etiology and progression of cancer, offering novel
strategies for its prevention and treatment. The aim of these
approaches is to alleviate the suffering of patients with lung
cancer and to reduce the societal burden associated with the
disease.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the National
Natural Science Foundation (grant nos. 82341060, 82204883 and
82073950), the Science and Technology Planning Project of Guangdong
Province (grant nos. 2023A1515012245 and 2022B1515120055), the
Science and Technology Program of Shenzhen (grant nos.
JCYJ20250604191406009, JCYJ20210324134209026,
JCYJ20220531102217038, JCYJ20210324112414038 and
JCYJ20220818102005011), the Shenzhen Medical Research Fund (grant
no. A2403062), the Special Fund for Economic and Technological
Development of Longgang District, Shenzhen Medical Health
Technology Project (grant no. LGWJ2021-046), the Science and
Technology Planning Project of Nanshan District (grant no.
NS2024026) and the State Key Laboratory of Respiratory Disease
(grant no. SKLRD-Z-202216).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author. The sequencing data
generated in the present study may be found in the National Center
for Biotechnology Information Sequence Read Archive database under
accession number PRJNA1282884 or at the following URL: https://www.ncbi.nlm.nih.gov/sra/PRJNA1282884.
Authors' contributions
YM, XX, JL and QH conceived and designed the
experiments. YM, YC and YL performed the experiments. YH, MG and LT
analyzed and interpreted the data. YM and XX wrote the manuscript.
XX, and QH revised the manuscript. YM, YL and XX confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
2
|
Cao W, Qin K, Li F and Chen W:
Socioeconomic inequalities in cancer incidence and mortality: An
analysis of GLOBOCAN 2022. Chin Med J (Engl). 137:1407–1413. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fares J, Fares MY, Khachfe HH, Salhab HA
and Fares Y: Molecular principles of metastasis: A hallmark of
cancer revisited. Signal Transduct Target Ther. 5:282020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Steens J and Klein D: HOX genes in stem
cells: Maintaining cellular identity and regulation of
differentiation. Front Cell Dev Biol. 10:10029092022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chin FW, Chan SC and Veerakumarasivam A:
Homeobox gene expression dysregulation as potential diagnostic and
prognostic biomarkers in bladder cancer. Diagnostics (Basel).
13:26412023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Feng Y, Zhang T, Wang Y, Xie M, Ji X, Luo
X, Huang W and Xia L: Homeobox genes in cancers: From
carcinogenesis to recent therapeutic intervention. Front Oncol.
11:7704282021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yadav C, Yadav R, Nanda S, Ranga S, Ahuja
P and Tanwar M: Role of HOX genes in cancer progression and their
therapeutical aspects. Gene. 919:1485012024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jasim SA, Farhan SH, Ahmad I, Hjazi A,
Kumar A, Jawad MA, Pramanik A, Altalbawy FMA, Alsaadi SB and
Abosaoda MK: Role of homeobox genes in cancer: Immune system
interactions, long non-coding RNAs, and tumor progression. Mol Biol
Rep. 51:9642024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gill HK, Yin S, Nerurkar NL, Lawlor JC,
Lee C, Huycke TR, Mahadevan L and Tabin CJ: Hox gene activity
directs physical forces to differentially shape chick small and
large intestinal epithelia. Dev Cell. 59:2834–2849.e9. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang LT, Chiou SS, Chai CY, His E,
Yokoyama KK, Wang SN, Huang SK and Hsu SH: Intestine-specific
homeobox gene ISX integrates IL6 signaling, tryptophan catabolism,
and immune suppression. Cancer Res. 77:4065–4077. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang LT, Liu KY, Jeng WY, Chiang CM, Chai
CY, Chiou SS, Huang MS, Yokoyama KK, Wang SN, Huang SK and Hsu SH:
PCAF-mediated acetylation of ISX recruits BRD4 to promote
epithelial-mesenchymal transition. EMBO Rep. 21:e487952020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Devos H, Zoidakis J, Roubelakis MG,
Latosinska A and Vlahou A: Reviewing the regulators of COL1A1. Int
J Mol Sci. 24:100042023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Selvaraj V, Sekaran S, Dhanasekaran A and
Warrier S: Type 1 collagen: Synthesis, structure and key functions
in bone mineralization. Differentiation. 136:1007572024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Sun X, Kan C, Chen B, Qu N, Hou N,
Liu Y and Han F: COL1A1: A novel oncogenic gene and therapeutic
target in malignancies. Pathol Res Pract. 236:1540132022.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiao X, Long F, Yu S, Wu W, Nie D, Ren X,
Li W, Wang X, Yu L, Wang P and Wang G: Col1A1 as a new decoder of
clinical features and immune microenvironment in ovarian cancer.
Front Immunol. 15:14960902025. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Y, Xue J, Zhong M, Wang Z, Li J and
Zhu Y: Prognostic prediction, immune microenvironment, and drug
resistance value of collagen type I alpha 1 chain: From
gastrointestinal cancers to pan-cancer analysis. Front Mol Biosci.
8:6921202021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang C, Liu S, Wang X and Liu H, Zhou X
and Liu H: COL1A1 Is a potential prognostic biomarker and
correlated with immune infiltration in mesothelioma. Biomed Res
Int. 2021:53209412021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ribeiro Gomes J and Cruz MRS: Combination
of afatinib with cetuximab in patients with EGFR-mutant
non-small-cell lung cancer resistant to EGFR inhibitors. Onco
Targets Ther. 8:1137–1142. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Della Corte CM, Fasano M, Ciaramella V,
Cimmino F, Cardnell R, Gay CM, Ramkumar K, Diao L, Di Liello R,
Viscardi G, et al: Anti-tumor activity of cetuximab plus avelumab
in non-small cell lung cancer patients involves innate immunity
activation: Findings from the CAVE-Lung trial. J Exp Clin Cancer
Res. 41:1092022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chanvorachote P, Petsri K and Thongsom S:
Epithelial to mesenchymal transition in lung cancer: Potential
EMT-targeting natural product-derived compounds. Anticancer Res.
42:4237–4246. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ding Y, Zhang M, Hu S, Zhang C, Zhou Y,
Han M, Li J, Li F, Ni H, Fang S and Chen Q: MiRNA-766-3p inhibits
gastric cancer via targeting COL1A1 and regulating PI3K/AKT
signaling pathway. J Cancer. 15:990–998. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y, Mei X, Song W, Wang C and Qiu X:
LncRNA LINC00511 promotes COL1A1-mediated proliferation and
metastasis by sponging miR-126-5p/miR-218-5p in lung
adenocarcinoma. BMC Pulm Med. 22:2722022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li Y, Sun R, Zhao X and Sun B: RUNX2
promotes malignant progression in gastric cancer by regulating
COL1A1. Cancer Biomark. 31:227–238. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Z, Wang Y, Zhang J, Zhong J and Yang
R: COL1A1 promotes metastasis in colorectal cancer by regulating
the WNT/PCP pathway. Mol Med Rep. 17:5037–5042. 2018.PubMed/NCBI
|
|
26
|
Huang C, Yang X, Han L, Fan Z, Liu B,
Zhang C and Lu T: The prognostic potential of alpha-1 type I
collagen expression in papillary thyroid cancer. Biochem Biophys
Res Commun. 515:125–132. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma HP, Chang HL, Bamodu OA, Yadav VK,
Huang TY, Wu ATH, Yeh CT, Tsai SH and Lee WH: Collagen 1A1 (COL1A1)
is a reliable biomarker and putative therapeutic target for
hepatocellular carcinogenesis and metastasis. Cancers (Basel).
11:7862019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang Z, Liu B, Lin T, Zhang Y, Zhang L and
Wang M: Multiomics analysis on DNA methylation and the expression
of both messenger RNA and microRNA in lung adenocarcinoma. J Cell
Physiol. 234:7579–7586. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hou L, Lin T, Wang Y, Liu B and Wang M:
Collagen type 1 alpha 1 chain is a novel predictive biomarker of
poor progression-free survival and chemoresistance in metastatic
lung cancer. J Cancer. 12:5723–5731. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Noorolyai S, Shajari N, Baghbani E,
Sadreddini S and Baradaran B: The relation between PI3K/AKT
signalling pathway and cancer. Gene. 698:120–128. 2019. View Article : Google Scholar : PubMed/NCBI
|