Introduction
Nasopharyngeal carcinoma (NPC) is a non-lymphomatous
squamous cell carcinoma occurring in the epithelial lining of the
nasopharynx. NPC occurs frequently in the populations of Southern
China, Northern Africa and Alaska (1). The combination of radiotherapy and
chemotherapy is currently the standard treatment strategy. However,
the optimal regimens of chemotherapy and radiotherapy have yet to
be determined (2). The poor
survival rate and high recurrence risk require that novel
therapeutic approaches be developed. The therapeutic strategy
targeting specific molecules and immunotherapy may improve the of
outcome of NPC (3). However, the
development of novel strategies is limited due to insufficient
understanding of the genes related to NPC development and the lack
of conventional cell and animal models for monitoring genes
affecting tumor growth and metastasis.
NPC cell line S18, a subclone that was isolated from
the human NPC undifferentiated cells (CNE-2 cells) (4), exhibits easy migration and invasion
in vitro when compared to CNE-2 cells. Moreover, the S18
xenografted cancer model has a significant metastatic potential and
maintains a similar growth rate to the in situ cancer.
Regulation of the expression of target genes in the
S18 cell line, is considered to be of great significance for the
investigation of target genes in NPC. The Tet-Off Advanced System
is a well-developed gene regulation tool in eucaryon (5,6). The
Tet-Off Advanced system comprises two key components: the
doxycycline-dependent regulator, such as pTet-Off Advanced, and the
response element containing the target gene, such as pTRE-Tight-X.
The Tet-Off Advanced system aims to create a double-stable cell
line that contains integrated copies of the regulator and response
plasmids. In this cell line, transcription of the gene of interest
is maintained in the ‘off’ state by the presence of doxycycline in
the culture medium, while transcription induction may occur
following the removal of doxycycline. Although this system appears
to be beneficial, more studies are required to create an inducible
system. Moreover, the efficacy of doxycycline-controlled gene
induction is known to be affected by cell types (7–9). To
the best of our knowledge, no study has reported whether this
system can confirm a high efficacy of transcriptional induction of
target genes in NPC cell lines. In the present study, a NPC S18
Tet-Off cell line was developed, which effectively expressed a
doxycycline-dependent regulator, and had potent transcriptional
activity of genes of interest with a low background.
Materials and methods
Cell culture
Human NPC S18 cells (kindly provided by Dr Chaonan
Qian) were grown in high glucose (Gibco) Dulbecco’s modified
Eagle’s minimal essential medium (DMEM) containing 10% Tet
system-approved fetal bovine serum (FBS; Clontech). S18 Tet-Off
cells were grown in complete DMEM medium containing 100 μg/ml G418
(Clontech). The S18 Tet-Off-Luc and Tet-Off-FTH1 clones were grown
in complete DMEM containing 100 μg/ml G418 and 100 μg/ml hygromycin
(Alexis) with or without 10 ng/ml doxycycline (Alexis). For
inducible experiments, cells maintained in the ‘off’ state of gene
expression by 10 ng/ml doxycycline were passaged. Briefly, the
cells were washed twice with phosphate-buffered solution (PBS)
prior to trypsinization. Following trypsinization and harvesting,
the cells were washed twice with PBS, and grown in fresh medium
with or without various concentrations of doxycycline.
Transfection protocol
Cells were transfected using Lipofectamine 2000
(Invitrogen) according to modified manufacturer’s instructions. For
transient transfection, cells (2×104) were plated in 100
μl of complete cell growth medium on a 96-well plate and incubated
for 24 h until a cell confluence of 50–60% was achieved.
DNA-Lipofectamine 2000 was prepared by combining the diluted
plasmid DNA [0.22 mg in 25 ml of Opti-MEM (Invitrogen)] with
Lipofectamine 2000 (0.15 ml in 25 ml of Opti-MEM) followed by
incubation at room temperature for 20 min. The DNA-Lipofectamine
2000 reagent complex was then directly added to each well, followed
by mixing and subsequent incubation at 37°C. After 6 h, the medium
was replaced with fresh complete cell growth medium with or without
10 ng/ml doxycycline followed by incubation for 48 h. Firefly
luciferase activity was then determined.
For stable transfection, 5×105 cells in
500 μl of complete cell growth medium were seeded on a 24-well
plate. After 16–24 h of culture, the cells were transfected with
DNA-Lipofectamine 2000 reagent complex (0.8 or 0.84 μg of plasmid
in 50 μl of Opti-MEM mixed with 0.5 μl of Lipofectamine 2000 in 50
μl of Opti-MEM) when cell confluence of approximately 90% was
achieved. All the plasmids were digested with restriction enzyme
ScaI. One day post-transfection, the cells were washed,
trypsinized and divided equally onto two 10-cm plates. Following 24
h of incubation, either 500 μg/ml G418 or G418 plus 250 μg/ml
hygromycin was added to the medium. After 16 days,
antibiotic-resistance clones were selected, using cloning rings,
and analyzed. Under these conditions, the transfection efficacy was
>80%, with low cytotoxicity detected 48 h after transfection
with pEGFP-C1 under identical transfection conditions.
Firefly luciferase activity assay
Cells were washed twice with PBS, lysed and then
assayed for firefly luciferase activity with the dual-luciferase
reporter assay (for the transient transfection that requires
Renilla luciferase to normalize the transfection efficacy) or the
luciferase assay system, according to the manufacturer’s
instructions (Promega, Madison, WI, USA). The firefly luciferase
activity in each cell lysate was measured with the GloMax 96
Microplate Luminometer (Promega) according to the standard
protocol, and the protein concentration was determined using the
BCA method (Pierce, Rockford, IL, USA). Luminescence data were
expressed as relative light units (measured in 10 s) per milligram
of protein.
Construction of plasmids and reverse
transcription-PCR (RT-PCR)
The human ferritin heavy chain gene (FTH1) with
Kozak sequence was amplified by RT-PCR from the total RNA of S18
cells. FTH1 gene was then subcloned into the SmaI site of
pUC119 yielding pUC119-FTH1. Following sequencing, the fragment
from pUC119-FTH1 after digestion by EcoRI and KpnI
was cloned into the same site of pTRE-Tight yielding
pTRE-Tight-FTH1. For RT-PCR, the first strand cDNA was reverse
transcribed with oligo (dT) primer. The full-length FTH1 with Kozak
sequence was amplified using PrimeSTAR HS DNA Polymerase (Takara).
The primers were forward, 5′-GAATTCGCCACCATGACGACCGCGTCCACCTC-3′
and reverse, 5′-AGATCTGGTACCTTTAGCTTTCATTATCA CTGTC-3′.
Western blot analysis
Cells were lysed in RIPA buffer [50 mM Tris pH 8.0,
150 mM sodium chloride, 1.0% Triton X-100 (v/v), 0.5% sodium
deoxycholate and 0.1% SDS (w/v)], agitated for 30 min at 4°C and
centrifuged at 12,000 × g for 15 min. The concentration of total
proteins was determined using BCA. Total proteins (25 μg) in equal
volume of 2X Laemmli buffer were then denatured and subjected to
12% SDS-PAGE. The proteins were transferred onto acetyl cellulose
membranes which were subsequently blocked in 5% non-fat milk in
TBST (20 mM Tris pH 7.6, 137 mM NaCL, 0.1% Tween-20). The membranes
were incubated with primary antibodies overnight [rabbit anti-FTH1
1:1,000 (Santa Cruz Biotechnologies, Santa Cruz, CA, USA) and
rabbit anti-GAPDH 1:2,000 (GenScript, Piscataway, NJ, USA)].
Following washing, the membranes were treated with secondary
antibody [HRP-conjugated goat anti-rabbit 1:5,000 (Invitrogen)] and
visualized by enhanced chemiluminescence.
MTT assay
Cell proliferation was measured by
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
Sigma) assay. Cells were plated onto 96-well plates and maintained
in 100 μl of DMEM (supplemented with different concentrations of
doxycycline) for 3 days. The experiment was repeated three times
for each group. After 24, 48 and 72 h of incubation, 20 μl MTT was
added to each well and the incubation was maintained for an
additional 4 h at 37°C. At the end of the incubation period, 150 μl
DMSO was added to each well and throroughly mixed for 10 min.
Absorbance at 570 nm was then determined.
Results
Generation of S18 Tet-Off clones
NPC S18 cells were transfected with the pTet-Off
Advanced plasmid, the G418-resistant clones were isolated and the
inducible firefly luciferase activity was detected following
transient transfection of pTRE-Tight-Luc into each clone. Among the
52 clones identified, the firefly luciferase expression in 10
clones during induction was 20-fold higher than that at the
non-induced state (Table I),
indicating the 10 clones that expressed the functional regulator
protein. Of the 10 clones, the cell clone (A9) with a 42-fold
increase in the firefly luciferase expression was selected as the
host cell line for the development of double-stable cells (S18
Tet-Off cell line).
 | Table IAnalysis of firefly luciferase
activity induction in the S18 Tet-Off clones. |
Table I
Analysis of firefly luciferase
activity induction in the S18 Tet-Off clones.
| Cell clone | Firefly luciferase
activity (RLU/μg protein) | Regulation
factor |
|---|
|
| |
|---|
| Dox+ | Dox− | |
|---|
| A5 | 175±13 | 48,062±4,052 | 274 |
| B18 | 397±18 | 93,619±8,466 | 236 |
| B7 | 193±16 | 36,835±1,613 | 190 |
| B4 | 120±11 | 17,623±1,489 | 146 |
| A12 | 75±10 | 9,836±801 | 131 |
| B15 | 228±12 | 29,571±2,763 | 130 |
| A21 | 201±12 | 23,523±1,045 | 117 |
| A9 | 74±5 | 3,115±188 | 42 |
| B23 | 96±9 | 2,653±267 | 28 |
| B19 | 77±6 | 1,766±189 | 23 |
Generation of clones with inducible
firefly luciferase activity
To investigate whether the S18 Tet-Off cell line
maintains a high induction activity of firefly luciferase and a low
basal expression, clones were prepared by stable transfection with
pTRE-Tight-Luc into the cell line. Of the 24 clones, 4 exhibited a
>1,000-fold increase in induction (Table II). These results suggest that it
is relatively simple to develop stable clones that show at least
1,000-fold inducible transcription activity based on the host cell
line. The D7 clone with the lowest basal expression and the highest
induction activity of firefly luciferase was termed the S18
Tet-Off-Luc cell line and subcloned for further experiments.
 | Table IIAnalysis of firefly luciferase
activity induction in the S18 Tet-Off-Luc clones. |
Table II
Analysis of firefly luciferase
activity induction in the S18 Tet-Off-Luc clones.
| Cell clone | Firefly luciferase
activity (RLU/μg protein) | Regulation
factor |
|---|
|
| |
|---|
| Dox+ | Dox− | |
|---|
| D7 | 0.89±0.17 | 3,281±352 | 3,671 |
| D25 | 2.47±0.33 | 5,280±188 | 2,138 |
| D18 | 29.3±2.60 | 44,491±1,189 | 1,534 |
| D3 | 0.96±0.09 | 1,177±167 | 1,225 |
Time kinetics of inducible firefly
luciferase expression
The time course of firefly luciferase activity of
the S18 Tet-Off-Luc cell line was analyzed in the absence or
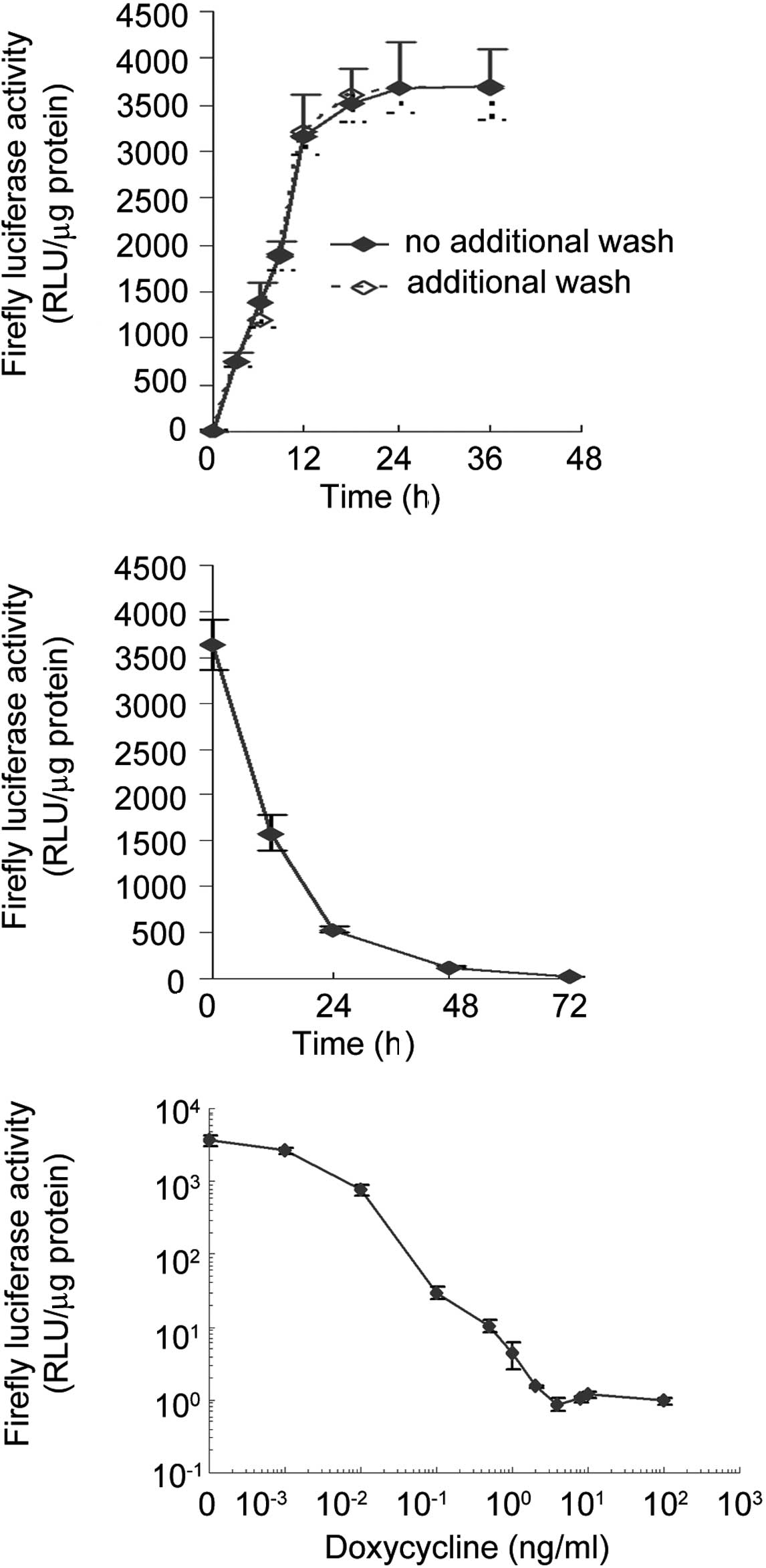
presence of doxycycline. As shown in Fig. 1A, the depletion of doxycycline led
to a rapid induction of firefly luciferase activity. Luciferase
activity increased by 775-fold within 3 h and by 3,321-fold in 12
h; the peak level was reached within 24 h. To investigate whether
residual doxycycline in culture medium inhibits the full gene
expression (maximum transcription activity), the cells were washed
once with PBS 3 h after initial removal of doxycycline, and the
luciferase activity was analyzed. As shown in Fig. 1A, the two time courses almost
overlapped. As shown in Fig. 1B, a
similar rapid reduction of firefly luciferase activity was observed
in the presence of doxycycline. The luciferase activity dropped to
43.7% of its initial level within 12 h and to <15% in 24 h.
Dose-dependent expression of firefly
luciferase by doxycycline
S18 Tet-Off-Luc cells were grown in medium
containing 10 ng/ml doxycycline for 5 days and then in medium
containing various concentrations of doxycycline for 48 h. The cell
extracts were then analyzed for firefly luciferase activity. As
shown in Fig. 1C, the firefly
luciferase activity was completely inhibited at 4 ng/ml
doxycycline, an event that was also observed at 100 ng/ml. A
stepwise reduction in the concentration of doxycycline gradually
increases the activity of firefly luciferase from complete
inhibition to an increase of 3,600-fold. This result showed that
induction of the luciferase expression by doxycycline in S18
Tet-Off-Luc cells occurred in a dose-dependent manner. The
doxycycline concentration ranged between 0 and 4 ng/ml, within
which the expression of firefly luciferase was the most sensitive
to doxycycline induction.
Expression of human ferritin heavy chain
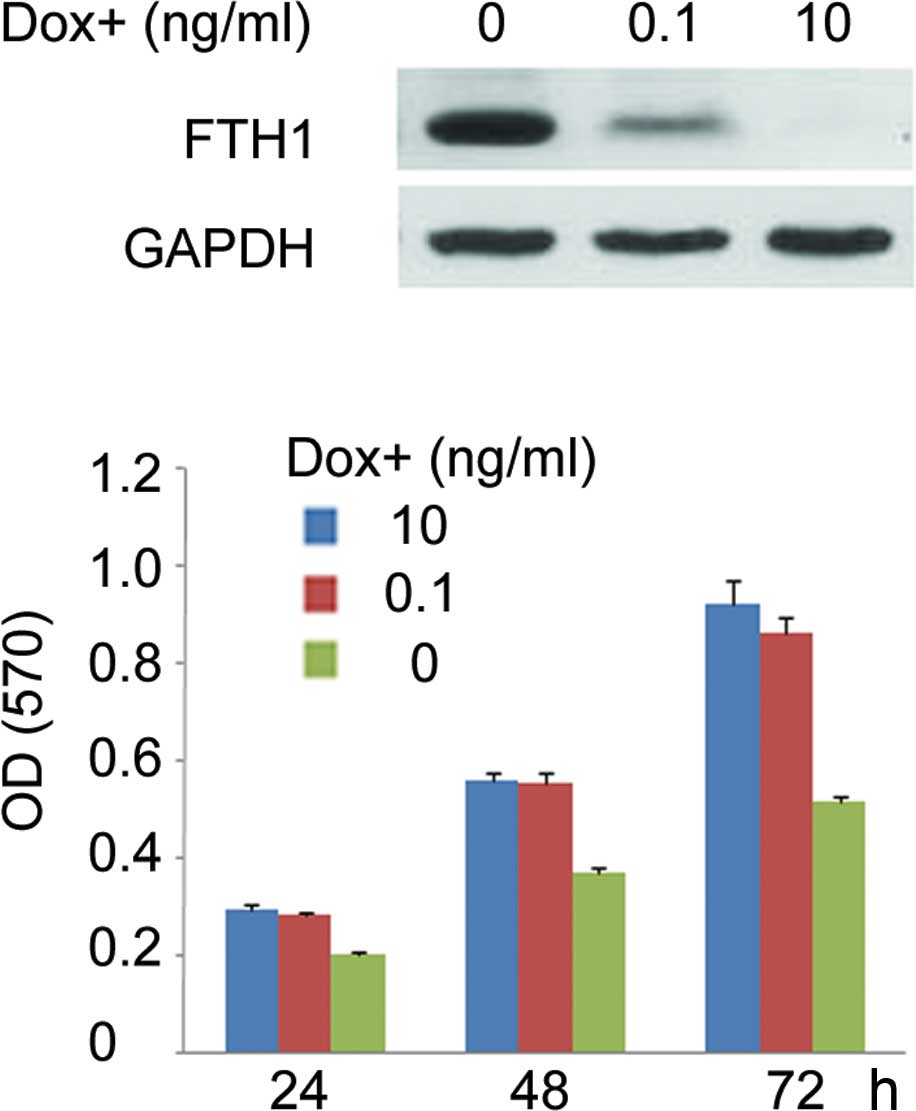
controlled by doxycycline
For further applications of the S18 Tet-Off cell
line, the clone inducibly expressing FTH1 was developed. The
construct of pTRE-Tight-FTH1 was stably transfected into S18
Tet-Off cells. FTH1 levels in clones were determined by Western
blotting. One of these clones expressing the highest levels of FTH1
in the absence of doxycycline with a low background level in the
presence of 10 ng/ml doxycycline was selected and termed S18
Tet-Off-FTH1 clone. By adjusting the concentrations of doxycycline,
the semi-quantitative expression of FTH1 was achieved in the S18
Tet-Off-FTH1 clone. Fig. 2A shows
that in the S18-Off-FTH1 cell line, FTH1 expression was maximally
inhibited at a concentration of 10 ng/ml doxycycline and increased
to a moderate level at 0.1 ng/ml, and then completely depressed
when doxycycline was removed.
Effect of human ferritin heavy chain
overexpression on NPC S18 cell proliferation
The S18-Off-FTH1 clone is maintained in culture in
the absence of doxycycline for up to 2 months without evident signs
of toxicity. However, it was observed that the clone achieved
confluence more rapidly in the presence of doxycycline than in the
absence thereof. Doxycycline at 10 ng/ml had no evident effects on
cell growth. To assess the impact of FTH1 on cell proliferation, a
MTT assay that measures mitochondrial activity was used (Fig. 2B). The results showed a correlation
between the effect on cell viability and the FTH1 levels. When
cells were grown in the absence of doxycycline, a significantly
high overexpression of FTH1 caused at least a 30% decrease in cell
growth rate at 48 h. When cells were grown at 0.1 ng/ml
doxycycline, the moderate overexpression of FTH1 did not affect
cell growth.
Discussion
Among head and neck cancers, NPC is highly
susceptible to lymph node metastasis. Undifferentiated NPCs have a
higher rate of local control after treatment, but a higher
incidence of distant metastasis than differentiated NPCs (10,11).
The undifferentiated NPC S18 cell line, not only showed
characteristics of advanced cancers, but also had a high potential
of lymph node metastasis. Therefore, S18 cells serve as an option
for the development of a cell model in which the S18 target gene is
conditionally overexpressed.
In this study, following the stable transfection of
pTet-Off Advanced plasmid into S18 cells, G418-resistant clones
were selected and analyzed using the transient luciferase
expression assay. In the investigation of 10 clones with an
increase of luciferase activity of more than 20-fold, the
expression levels in the absence of doxycycline varied by 53-fold,
whereas the expression levels at 10 ng/ml doxycycline varied by
less than 6-fold.
This clonal variation is most likely contributed to
the variation in the concentration of regulator proteins in each
clone (12). According to the
manufacturer’s instructions of the Tet-Off Advanced system
(Clontech), only clones exhibiting 20- to 50-fold induction were
selected to develop double-stable cell lines. We selected the clone
with a 42-fold induction as the host cell line. Following stable
transfection with pTRE-Tight-Luc into this clone, clones with more
than 1,000-fold induction of transcriptional activity were
developed. The basal expression level (RLU=0.89, n=4, SD=0.17) of
the double-stable cell line (S18 Tet-Off-Luc) in the presence of 4
ng/ml doxycycline was comparable to that of the background level.
This extremely low background level is similar to that of
well-established double-stable Tet controlled cell lines, such as
HeLa (clone X1) and CHO AA8 (clone 19) (5,12). As
shown in the majority of studies, if a low level of background
expression is achieved, a regulation factor of between 50- and
approximately 1,000-fold is sufficient (13). Therefore, the 42-fold clone was
termed S18 Tet-Off cell line.
Rennel and Gerwins previously reported that residual
doxycycline binding to cells or extracellular matrix prevents full
gene induction in the Tet-Off system. Moreover, a robust and rapid
transgene expression is induced in certain cell lines if
doxycycline is removed by washing 3 h after the initial removal of
doxycycline (14). We found that
the additional wash was not required for the double-stable cell
lines based on the S18 Tet-Off cell line, making the operation more
convenient.
As shown in Fig. 1C,
the transcription activity was quantitatively regulated by
doxycycline. The doxycycline at between 0 and 4 ng/ml allowed
adjustment of promoter activity within a range of three orders of
magnitude. This may allow assessment of quantitative parameters of
gene function.
To further test the application of the S18 Tet-Off
cell line, another clone that inducibly expressed candidate MRI
reporter gene FTH1 (15,16) was successfully created. As an MRI
candidate reporter, evidence of its safety warrants further
investigation of its practical applications. Thus, in the present
study, the impact of FTH1 overexpression on NPC S18 cell growth was
addressed. The highest expression of FTH1 (grown in the absence of
doxycycline) reduced cell growth. However, a moderate
overexpression of FTH1 (grown in 0.1 ng/ml doxycycline) in NPC S18
cells did not reduce cell growth. These results were in accordance
with those of Cozzi et al (18). Findings of that study showed that
the overexpression of FTH1 in HeLa cells induced an iron-deficient
phenotype with significantly reduced cell growth. In a previous
study, overexpression of FTH1 in C6 glioma cell lines (19), A549 cells (16) and stem cells (20) did not reduce cell growth. However,
Cheng et al (21) found that
a significant reduction in growth rate was observed in C6 glioma
cell lines under markedly high FTH1 transgene expression (>500%
of the level in mouse embryonic stem cells described in their
study). On the other hand, in other S18 double-stable clones, those
with low FTH1 levels did not reduce cell growth (data not shown).
It was hypothesized that a significantly high expression of FTH1
decreases cell growth in certain types of cells, whereas a moderate
level of FTH1 expression may be safe.
To investigate the role of genes relative to NPC, a
NPC cell line stably expressing the genes is a powerful tool.
Routinely, target genes under the general and non-controlled
promoter, including CMV promoter, are transfected into host cells,
and then clones with high expression levels are selected to
investigate the involvement of these clones in NPC. However, this
process leads to certain issues: i) To develop stable clones with a
high expression of level of target genes is impossible if the
target genes are toxic to cells. ii) If the gene of interest has no
evident cell cytotoxity, a high expression of target genes
continues for the whole clone-selected process. Chronic alterations
may exist, however, and these changes are often not easy to detect.
For example, a high expression of FTH1 reduces NPC S18 cell growth,
and does not form clones in the early clone-selected days. Clones
with low expression levels are usually selected. If this selection
occurs, it is difficult to judge the impact of FTH1 on cell growth.
iii) The expression levels vary largely as a result of different
vector integration sites. In order to study the biological
properties on different levels of the target gene, more cell clones
should be selected and investigated. However, this process is
time-consuming and not cost-effective. A gene-controllable system
would be beneficial. Therefore, the NPC S18 Tet-Off cell line is an
effective tool that provides a proven genetic background for gene
regulation. Using the NPC S18 Tet-Off cell line, only one-step
transfection is required to produce a cell model in which the
target genes can be specifically and tightly regulated. The
characteristically low level of basal activity is beneficial for
studies on novel genes that may disrupt the cell cycle, induce
apoptosis or exhibit cytotoxicity, in the case that experiments
depend on the absolute control of the background of the gene
expression level.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (No. 80171207 and 39970237), the
Fundamental Research Funds for the Central Universities (No.
10ykjcll), and the Open Funds of State Key Laboratory of Oncology
in Southern China.
References
|
1
|
Wei WI and Sham JS: Nasopharyngeal
carcinoma. Lancet. 365:2041–2054. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lu H, Peng LX, Yuan XB, et al: Concurrent
chemoradiotherapy in locally advanced nasopharyngeal carcinoma: a
treatment paradigm also applicable to patients in Southeast Asia.
Cancer Treat Rev. 35:345–353. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Agulnik M and Epstein JB: Nasopharyngeal
carcinoma: current management, future directions and dental
implications. Oral Oncol. 44:617–627. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qian CN, Berghuis B, Tsarfaty G, et al:
Preparing the ‘soil’: the primary tumor induces vasculature
reorganization in the sentinel lymph node before the arrival of
metastatic cancer cells. Cancer Res. 66:10365–10376. 2006.
|
|
5
|
Gossen M and Bujard H: Tight control of
gene expression in mammalian cells by tetracycline-responsive
promoters. Proc Natl Acad Sci USA. 89:5547–5551. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Urlinger S, Baron U, Thellmann M, Hasan
MT, Bujard H and Hillen W: Exploring the sequence space for
tetracycline-dependent transcriptional activators: novel mutations
yield expanded range and sensitivity. Proc Natl Acad Sci USA.
97:7963–7968. 2000. View Article : Google Scholar
|
|
7
|
Gossen M and Bujard H: Efficacy of
tetracycline-controlled gene expression is influenced by cell type:
commentary. Biotechniques. 19:213–217. 1995.PubMed/NCBI
|
|
8
|
Ackland-Berglund CE and Leib DA: Efficacy
of tetracycline- controlled gene expression is influenced by cell
type. Biotechniques. 18:196–200. 1995.PubMed/NCBI
|
|
9
|
Howe JR, Skryabin BV, Belcher SM, Zerillo
CA and Schmauss C: The responsiveness of a tetracycline-sensitive
expression system differs in different cell lines. J Biol Chem.
270:14168–14174. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Marks JE, Phillips JL and Menck HR: The
National Cancer Data Base report on the relationship of race and
national origin to the histology of nasopharyngeal carcinoma.
Cancer. 83:582–588. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Reddy SP, Raslan WF, Gooneratne S,
Kathuria S and Marks JE: Prognostic significance of keratinization
in nasopharyngeal carcinoma. Am J Otolaryngol. 16:103–108. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang XS, Hu CS, Ying HM, Zhou ZR, Ding JH
and Feng Y: Patterns of retropharyngeal node metastasis in
nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys. 73:194–201.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Freundlieb S, Schirra-Muller C and Bujard
H: A tetracycline controlled activation/repression system with
increased potential for gene transfer into mammalian cells. J Gene
Med. 1:4–12. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rennel E and Gerwins P: How to make
tetracycline-regulated transgene expression go on and off. Anal
Biochem. 309:79–84. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cohen B, Ziv K, Plaks V, et al: MRI
detection of transcriptional regulation of gene expression in
transgenic mice. Nat Med. 13:498–503. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Genove G, DeMarco U, Xu H, Goins WF and
Ahrens ET: A new transgene reporter for in vivo magnetic resonance
imaging. Nat Med. 11:450–454. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zurkiya O, Chan AWS and Hu XP: MagA is
sufficient for producing magnetic nanoparticles in mammalian cells,
making it an MRI reporter. Magn Reson Med. 59:1225–1231. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cozzi A, Corsi B, Levi S, Santambrogio P,
Albertini A and Arosio P: Overexpression of wild type and mutated
human ferritin H-chain in HeLa cells: in vivo role of ferritin
ferroxidase activity. J Biol Chem. 275:25122–25129. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cohen B, Dafni H, Meir G, Harmelin A and
Neeman M: Ferritin as an endogenous MRI reporter for noninvasive
imaging of gene expression in C6 glioma tumors. Neoplasia.
7:109–117. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Naumova AV, Reinecke H, Yarnykh V, Deem J,
Yuan C and Murry CE: Ferritin overexpression for noninvasive
magnetic resonance imaging-based tracking of stem cells
transplanted into the heart. Mol Imaging. 9:201–210.
2010.PubMed/NCBI
|
|
21
|
Liu J, Cheng ECH, Long RC, et al:
Noninvasive monitoring of embryonic stem cells in vivo with MRI
transgene reporter. Tissue Engineering Part C Methods. 15:739–747.
2009. View Article : Google Scholar : PubMed/NCBI
|