Introduction
Mature cystic teratomas of the ovary are one of the
most common benign ovarian neoplasms, accounting for approximately
10–20% of all ovarian tumors (1).
Teratoma may occur at any age in women, but predominantly occurs in
younger patients (20–40 years old) (2,3).
Common complications for mature cystic teratomas include torsion,
rupture, malignant transformation, infection and autoimmune
hemolytic anemia (4,5), the prevalence of which is
approximately 20%. The exact molecular mechanism underlying the
formation of mature cystic teratomas, however, remains to be
determined.
microRNAs (miRNAs) are a class of approximately
20–22 nucleotide small non-coding RNAs that regulate gene
expression by binding to the 3′ untranslated regions (UTRs) of
their target mRNAs and inhibiting the translation and/or promoting
the degradation of mRNAs (6,7).
miRNAs not only play significant roles in various biological
processes but are also involved in pathological processes (8,9).
Mounting evidence has shown that an abnormal expression of miRNAs
is involved in human cancer, including ovarian tumors. In their
study, Corney et al revealed that a reduced expression of
miR-34b*/c may be particularly significant for progression to the
most advanced stages and that the miR-34 family plays a crucial
role in epithelial ovarian cancer pathogenesis (10). miRNA-15a and miRNA-16 target the
Bmi-1 3′ UTR and significantly correlate with Bmi-1 protein levels
in ovarian cancer and cell lines (11). However, the role of miRNAs in mature
ovarian teratomas has yet to be elucidated.
The aim of the present study was to analyze miRNA
differential expression profiles for mature ovarian teratoma
tissues compared with normal tissues. Our data demonstrate that
certain miRNAs may be relevant to the development of mature ovarian
teratomas.
Materials and methods
Patients and methods
Three patients with mature cystic teratomas were
recruited at the Department of Gynecology, Nanjing Maternal and
Child Health Hospital of Nanjing Medical University (China) between
March and August 2010. This study was approved by the institute’s
ethics committee and informed consent was provided by each patient.
The patients were pathologically confirmed as having mature cystic
teratoma. Teratoma samples were obtained from the growth nidus of
the cyst or from the innermost cell wall, but excluded any material
from the ovarian capsule. The samples were washed with a sufficient
amount of cold saline to reduce blood contamination (12). Tissues were immediately removed and
preserved in liquid nitrogen and stored at −80°C until they were
processed.
Total RNA preparation
Total RNA was extracted using TRIzol (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer’s instructions.
Total RNA quality and quantity was measured using a nanodrop
spectrophotometer (ND-1000, Nanodrop Technologies; Wilmington, DE,
USA) and RNA integrity was determined by gel analysis.
miRNA microarray
Microarrays were performed by utilizing the miRCURY
Locked Nucleic Acid (LNA) microarray platform (Exiqon, Vedbaek,
Denmark). All procedures were carried out according to the
manufacturer’s instructions. Briefly, purified RNA was labeled with
a miRCURY™ Hy3™/Hy5™ Power labeling kit (Exiqon). After
discontinuing the labeling procedure, the Hy3™-labeled samples and
Hy5™-labeled reference pool RNA samples were then mixed pair-wise
and hybridized on the miRCURY™ LNA array (version 14.0) (Exiqon).
The hybridization and slide washing were performed according to the
miRCURY LNA array manual. The results were subjected to
unsupervised hierarchical clustering (cluster 3.0) and TreeView
analysis (Stanford University, Standford, CA, USA). Data were
normalized using the locally weighted scatter plot smoothing
(lowess) regression algorithm (MIDAS, TIGR Microarray Data Analysis
System). Following scale normalization, replicated miRNAs were
averaged. Differentially-expressed miRNAs, with statistical
significance, were identified through volcano plot filtering. The
results of hierarchical clustering were performed by MEV software
(v4.6, TIGR).
Quantitative RT-PCR of miRNA
The total RNA from tissue was extracted with TRIzol
reagent (Invitrogen) following the manufacturer’s instructions. RNA
samples were quantified spectrophotometrically at 260 nm, and RNA
integrity was verified by agarose-formaldehyde gel electrophoresis.
In the reverse transcription step, complementary DNA (cDNA) was
reversely transcribed from total RNA samples with a miRNA-specific
stem-loop primer using the TaqMan microRNA reverse transcription
kit (ABI, Forest City, CA, USA). Reverse transcription was carried
out in reaction mixtures containing 1 μg total RNA, 0.5 μl miRNA
reverse primer (1 μM), 0.3 μl RNase inhibitor (40 U/μl), 2 μl 10X
buffer, 2 μl RNasin (10 U/μl) and an appropriate amount of
RNAse-free H2O to a total volume of 20 μl. The reaction
was performed at 16°C for 30 min and at 42°C for 40 min, followed
by heat inactivation at 85°C for 5 min. Subsequently, quantitative
real-time PCR was performed using ABI 7300 real-time PCR system
(Applied Biosystems, CA, USA). The 25 μl PCR reaction included 2.5
μl RT product, 0.5 μl forward and reverse primer and 12.5 μl
SYBR-Green real-time PCR master. The reactions were incubated in a
96-well optical plate at 95°C for 10 min, followed by 40 cycles of
95°C for 15 min and 60°C for 1 min. Reactions were run at least in
triplicate. miRNA probe sets were specifically selected from
reported miRNA studies in human blastocysts (13) or human tumor-associated tissue
(14,15). The U6 expression level was used as
an internal control for miRNA expression levels.
miRNA target predictions
miRNA target genes were predicted by miRGen Targets
(http://www.diana.pcbi.upenn.edu/cgi-bin/miRGen/v3/Targets.cgi).
Statistical analysis
Data were analyzed by the Student’s t-test using the
SPSS 15.0 statistical package (SPSS Inc., Chicago, IL, USA).
P<0.05 was considered to be statistically significant.
Results
miRNA expression profiles in mature
ovarian teratoma relative to normal tissues
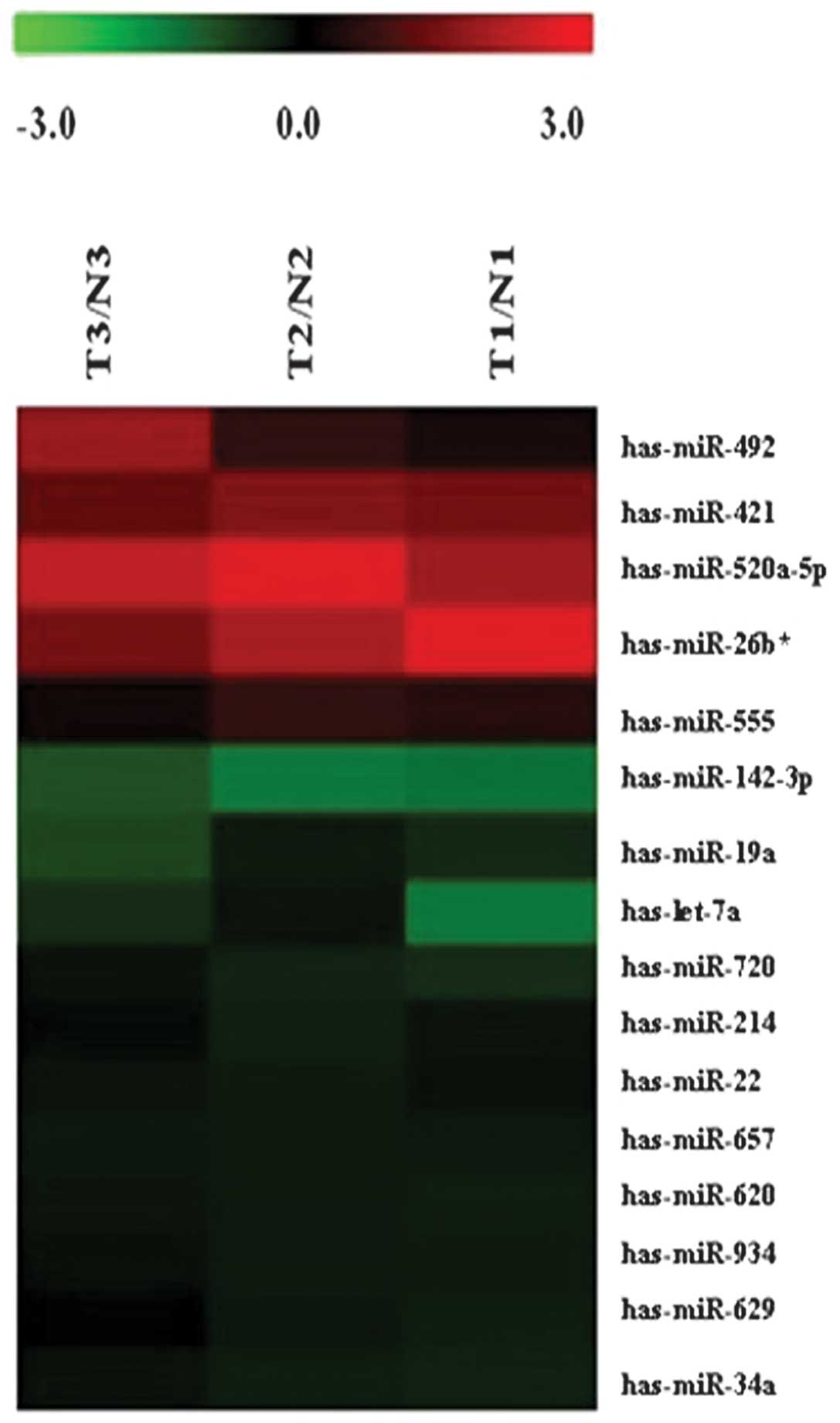
We detected 1,223 human mature miRNAs (miRbase
v16.0) using miRNA microarrays and profiled the expression of
miRNAs in mature ovarian teratoma. A total of 16 miRNA levels were
significantly different between mature cystic teratomas and normal
tissues (Fig. 1, Table I; P≤0.05). A total of 5 miRNAs were
found to be over-expressed in mature ovarian teratoma tissues, with
a 1.30- to 2.63-fold change; whereas 11 miRNAs were down-regulated,
with a decreased level of 55–87% compared with the non-tumorous
tissues.
 | Table IRelative expression of 16 miRNAs in
mature ovarian teratomas and matched normal tissues. |
Table I
Relative expression of 16 miRNAs in
mature ovarian teratomas and matched normal tissues.
| Gene name | Regulation | Fold-change | P-value |
|---|
|
has-miRNA-520a-5p | Up | 2.6342413 | 0.002060 |
| has-miRNA-26b* | Up | 1.4913426 | 0.043586 |
| has-miRNA-421 | Up | 1.3811204 | 5.03E-06 |
| has-miRNA-492 | Up | 1.3156408 | 0.008240 |
| has-miRNA-555 | Up | 1.3002984 | 0.015700 |
| has-miRNA-142-3p | Down | 0.4544302 | 0.035932 |
| has-let-7a | Down | 0.4107124 | 0.007090 |
| has-miRNA-19a | Down | 0.3691387 | 0.049331 |
| has-miRNA-34a | Down | 0.3149833 | 6.25E-0.6 |
| has-miRNA-620 | Down | 0.2925045 | 0.036400 |
| has-miRNA-934 | Down | 0.2910072 | 0.041420 |
| has-miRNA-657 | Down | 0.2809156 | 0.005450 |
| has-miRNA-720 | Down | 0.2026972 | 0.033304 |
| has-miRNA-22 | Down | 0.1849261 | 0.002740 |
| has-miRNA-629 | Down | 0.1529428 | 0.014600 |
| has-miRNA-214 | Down | 0.1347023 | 0.007200 |
Quantitative RT-PCR analysis
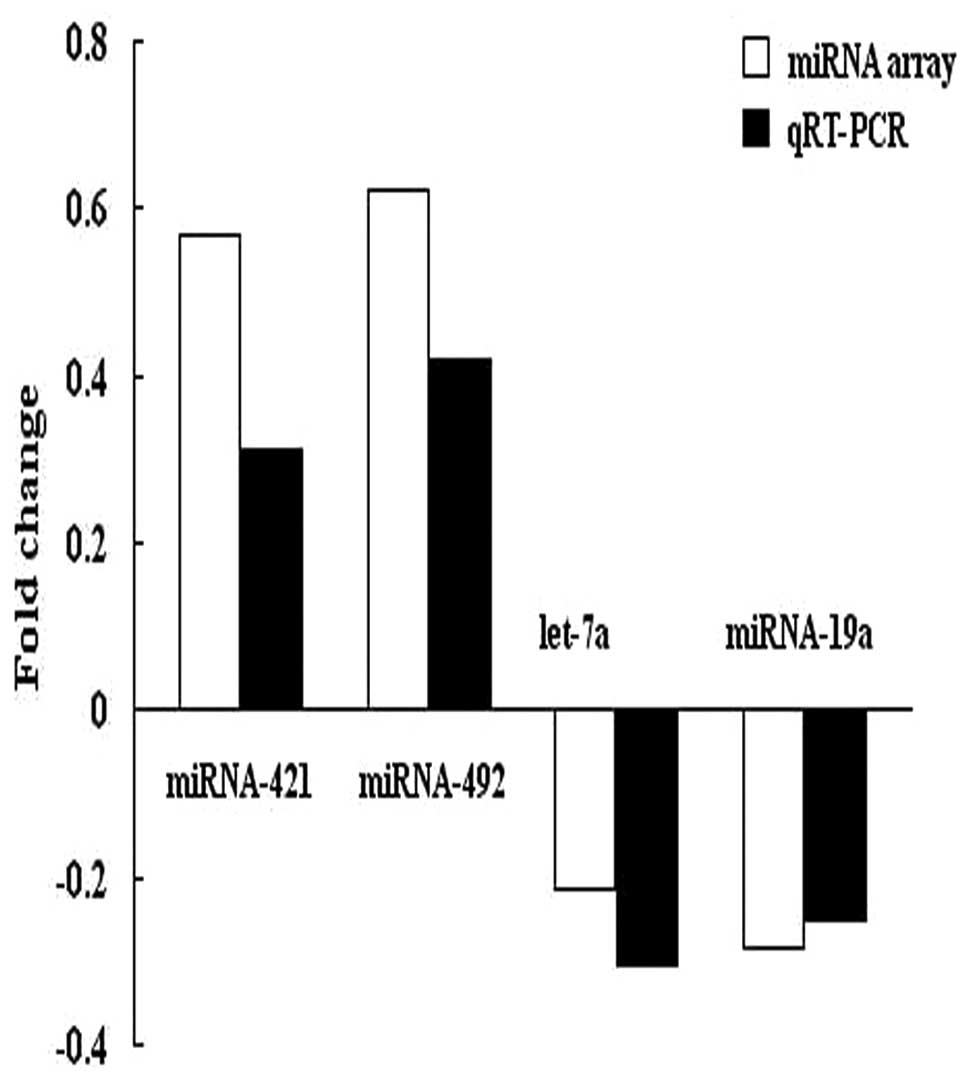
To confirm the results of microarray analysis, we
performed quantitative RT-PCR analysis on the same number of
samples using probes corresponding to miRNA-421, miRNA-492, let-7a
and miRNA-19a. The expression data obtained by qRT-PCR analysis
were comparable to the results observed in the microarray analysis
(Fig. 2).
miRNA target prediction
To explore potential target genes of miRNAs, we used
miRGen Targets (http://www.diana.pcbi.upenn.edu/cgi-bin/miRGen/v3/Targets.cgi).
Among the miRNA whose expression was significantly different
between tumor and non-tumorous samples, we found the marker gene
for benign teratomas, HDAC1, which is the target gene of
miRNA-142-3p and miRNA-34a.
Discussion
miRNAs play a crucial role in ovarian function
(including folliculogenesis, oocyte maturation, ovulation,
fertilization, cleavage, implantation and maintenance of
pregnancy). Aberrant patterns of miRNA expression have also been
reported in ovarian tumors (16–18).
In the present study, we systematically analyzed the miRNA profile
and identified significantly altered miRNAs in mature ovarian
teratoma samples compared with normal tissues. A total of 5
up-regulated miRNAs and 11 down-regulated miRNAs were found in
teratoma versus non-tumorous tissues. The microarray results were
further confirmed via qRT-PCR for four selected miRNAs (miRNA-421,
miRNA-492, let-7a and miRNA-19a).
DNA damage is a very common event in the life-cycle
of a cell. DNA damage is also the underlying cause of specific
mutations in cells leading to cancer, including ovarian cancer
(19,20). Ataxia-telangiectasia mutated (ATM)
is a serine/threonine kinase that plays a key role in the DNA
damage response process (21). In
their study, Hu et al found that human miR-421 suppresses
ATM expression by targeting the 3′ UTR of ATM transcripts (15). In our tumor samples an up-regulation
of miR-421 was observed, indicating that up-regulated miR-421 may
affect DNA damage repair by suppressing ATM expression in ovarian
teratoma tissues.
miR-421 has previously been described as
up-regulated in late-stage ovarian carcinomas compared to
early-stage cancer (22). In the
present study, we found that miR-421 was also up-regulated in
mature ovarian teratomas versus non-tumorous tissues. The results
indicate that miR-421 expression is both up-regulated in benign and
malignant tumors. Our study results further confirmed that miR-421
plays a key role during the initiation and development of ovarian
carcinomas.
One of the findings among the lower expressed miRNAs
are miRNA-142-3p and miRNA-34a. We obtained the same target gene,
HDAC1 of miRNA-142-3p and miRNA-34a, by target prediction analysis.
The finding indicates that miRNA-142-3p and miRNA-34a are capable
of reducing the expression level of their target gene, HDAC1. HDAC1
is a member of the histone deacetylase (HDAC) family. Proteins of
this family are critical cellular regulators that are involved in
fundamental cellular events, including cell cycle progression,
differentiation and tumorigenesis (23,24).
HDAC1 was predominantly detected in mature areas of differentiated
tumours (teratomas), rendering HDAC1 a potential novel biomarker
for benign teratomas (25). In the
present study, we demonstrated that miRNA-142-3p and miRNA-34a were
lowly expressed in mature ovarian teratomas, meaning that HDAC1 had
a relatively high expression in mature ovarian teratomas. Thus,
these results are consistent with previous findings.
In conclusion, the present study was the first to
demonstrate the differential profile of 16 miRNAs in mature ovarian
teratomas. An aberrant expression of miRNAs may be essential for
the pathogenesis of mature ovarian teratomas. Future studies
addressing the function of these miRNAs are required to provide
insights into their role in the development of ovarian
teratomas.
Acknowledgements
This study was supported by grants from the Nanjing
Science and Technology Agency (No. 200901084).
References
|
1
|
Matz MH: Benign cystic teratomas of the
ovary: a review. Obstet Gynecol Surv. 16:591–594. 1961. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lakkis WG, Martin MC and Gelfand MM:
Benign cystic teratoma of the ovary: a 6-year review. Can J Surg.
28:444–446. 1985.PubMed/NCBI
|
|
3
|
Ayhan A, Bukulmez O, Genc C, Karamursel BS
and Ayhan A: Mature cystic teratomas of the ovary: case series from
one institution over 34 years. Eur J Obstet Gynecol Reprod Biol.
88:153–157. 2000.PubMed/NCBI
|
|
4
|
Emin U, Tayfun G, Cantekin I, Ozlem UB,
Umit B and Leyla M: Tumor markers in mature cystic teratomas of the
ovary. Arch Gynecol Obstet. 279:145–147. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park SB, Kim JK, Kim KR and Cho KS:
Imaging findings of complications and unusual manifestations of
ovarian teratomas. Radiographics. 28:969–983. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shah AA, Meese E and Blin N: Profiling of
regulatory microRNA transcriptomes in various biological processes:
a review. J Appl Genet. 51:501–507. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Corney DC, Hwang CI, Matoso A, Vogt M,
Flesken-Nikitin A, Godwin AK, Kamat AA, Sood AK, Ellenson LH,
Hermeking H and Nikitin AY: Frequent downregulation of miR-34
family in human ovarian cancers. Clin Cancer Res. 16:1119–1128.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bhattacharya R, Nicoloso M, Arvizo R, Wang
E, Cortez A, Rossi S, Calin GA and Mukherjee P: MiR-15a and MiR-16
control Bmi-1 expression in ovarian cancer. Cancer Res.
69:9090–9095. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Miura K, Obama M, Yun K, Masuzaki H, Ikeda
Y, Yoshimura S, Akashi T, Niikawa N, Ishimaru T and Jinno Y:
Methylation imprinting of H19 and SNRPN genes in human benign
ovarian teratomas. Am J Hum Genet. 65:1359–1367. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lakshmipathy U, Love B, Goff LA, Jornsten
R, Graichen R, Hart RP and Chesnut JD: MicroRNA expression pattern
of undifferentiated and differentiated human embryonic stem cells.
Stem Cells. 16:1003–1016. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
von Frowein J, Pagel P, Kappler R, von
Schweinitz D, Roscher A and Schmid I: MicroRNA-492 is processed
from the keratin 19 gene and up-regulated in metastatic
hepatoblastoma. Hepatology. 53:833–842. 2011.PubMed/NCBI
|
|
15
|
Hu H, Du L, Nagabayashi G, Seeger RC and
Gatti RA: ATM is down-regulated by N-Myc-regulated microRNA-421.
Proc Natl Acad Sci USA. 107:1506–1511. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Otsuka M, Zheng M, Hayashi M, Lee JD,
Yoshino O, Lin S and Han J: Impaired microRNA processing causes
corpus luteum insufficiency and infertility in mice. J Clin Invest.
118:1944–1954. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vaksman O, Stavnes HT, Kærn J, Trope CG,
Davidson B and Reich R: miRNA profiling along tumor progression in
ovarian carcinoma. J Cell Mol Med. 15:1593–602. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Laios A, O’Toole S, Flavin R, Martin C,
Kelly L, Ring M, Finn SP, Barrett C, Loda M, Gleeson N, D’Arcy T,
McGuinness E, Sheils O, Sheppard B and O’Leary J: Potential role of
miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer.
7:352008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Q, Yu JJ, Mu C, Yunmbam MK, Slavsky D,
Cross CL, Bostick-Bruton F and Reed E: Association between the
level of ERCC-1 expression and the repair of cisplatin-induced DNA
damage in human ovarian cancer cells. Anticancer Res. 20:645–652.
2000.PubMed/NCBI
|
|
20
|
Teodoridis JM, Hall J, Marsh S, Kannall
HD, Smyth C, Curto J, Siddiqui N, Gabra H, McLeod HL, Strathdee G
and Brown R: CpG island methylation of DNA damage response genes in
advanced ovarian cancer. Cancer Res. 65:8961–8967. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Matsuoka S, Ballif BA, Smogorzewska A,
McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini
N, Lerenthal Y, Shiloh Y, Gygi SP and Elledge SJ: ATM and ATR
substrate analysis reveals extensive protein networks responsive to
DNA damage. Science. 316:1160–1166. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang L, Volinia S, Bonome T, Calin GA,
Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K,
et al: Genomic and epigenetic alterations deregulate microRNA
expression in human epithelial ovarian cancer. Proc Natl Acad Sci
USA. 105:7004–7009. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thiagalingam S, Cheng KH, Lee HJ, Mineva
N, Thiagalingam A and Ponte JF: Histone deacetylases: unique
players in shaping the epigenetic histone code. Ann N Y Acad Sci.
983:84–100. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee H, Rezai-Zadeh N and Seto E: Negative
regulation of histone deacetylase 8 activity by cyclic
AMP-dependent protein kinase A. Mol Cell Biol. 24:765–773. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Simboeck E and Di Croce L: HDAC1, a novel
marker for benign teratomas. EMBO J. 29:3992–4007. 2010. View Article : Google Scholar
|