Introduction
Colorectal cancer (CRC) is among the most common
types of cancer in both males and females and is associated with
high mortality, particularly at advanced stages (1). Markers for defining individual risk
signatures in CRC patients are of great clinical value, as they may
allow for targeted therapies to improve the outcomes of CRC
patients.
CRC arises through the accumulation of multiple
genetic and epigenetic changes. Somatic mutations in APC,
BRAF, KRAS, PIK3CA and TP53, as well as
in other genes, have been frequently observed in CRC and are
thought to drive colorectal tumorigenesis (2). We previously examined the mutation of
genes in the EGFR pathway and found that the PIK3CA mutation
is associated with poor prognosis in CRC patients (8,11).
In addition to genetic alterations, epigenetic
silencing is a prevalent mechanism by which abnormal gene
inactivation may occur in cancer and is involved in initiation and
promotion (3). A predominant mode
of epigenetic alteration in cancer is gene silencing via the
hypermethylation of the CpG island promoter. Hypermethylation
results in abnormal silencing of a number of tumor suppressors in
numerous human malignancies including CRC (3).
It has been suggested that epigenetic abnormalities
are able to cooperate with genetic alterations to cause aberrant
gene function and result in cancer (4). Thus, in the present study, we examined
KRAS and PIK3CA mutations and the methylation status
of BNIP3, p16 and hMLH1 and analyzed the
correlation between these molecular alterations and the
clinicopathological characteristics of CRC. We aimed to clarify
whether a combination of genetic and epigenetic alterations could
be used to classify CRC patients in relation to their
clinicopathological characteristics and outcomes.
Materials and methods
Patients and tissue samples
After providing informed consent, a total of 165 CRC
patients who underwent surgical resection at Tokyo Medical and
Dental University Hospital between March 2000 and April 2003 were
recruited in this study. This study was approved by the
institutional review board of the Tokyo Medical and Dental
University. The study population (Table
I) consisted of 62 females and 103 males, with a mean age of
64.5±10.3 years (range, 37–89). Sixty-two cancers were located in
the proximal site and 103 were located in the distal site,
including the rectum. Histological classification and tumor staging
were performed according to the International Union Against Cancer
Tumor-Node-Metastasis classification guidelines. No patient
received preoperative chemotherapy or radiotherapy. Following
surgery, patients with stage III CRC received oral or intravenous
5-fluorouracil (FU)-based adjuvant chemotherapy and patients with
stage IV tumors received 5-FU-based systemic chemotherapy without
radiotherapy. Patients were prospectively followed up following
surgery for a median period of 64 months (range, 0.07–107.6).
Resected specimens were fixed in 10% neutral buffered formalin and
embedded in paraffin. For all cases, archival hematoxylin and eosin
(H&E) slides of primary tumors were retrieved and reviewed to
confirm pathological features.
 | Table IClinicopathological characteristics
according to the mutation and methylation status of each gene. |
Table I
Clinicopathological characteristics
according to the mutation and methylation status of each gene.
| KRAS | PIK3CA | BNIP3 | p16 | hMLH1 |
|---|
|
|
|
|
|
|
|---|
|
Characteristics | Mut
52 (32%) | Wt
113 (69%) | P | Mut
20 (12%) | Wt
145 (88%) | P | Met
84 (58%) | Unm
61 (42%) | P | Met
48 (33%) | Unm
97 (67%) | P | Met
30 (21%) | Unm
115 (79%) | P |
|---|
| Age (years) | 64.6 (37–89) | 64.6 (41–88) | 0.898 | 67.8 (54–84) | 64.1 (37–89) | 0.145 | 65.0 (37–89) | 63.8 (41–88) | 0.614 | 66.2 (48–87) | 64.0 (37–89) | 0.292 | 65.7 (37–88) | 64.4 (41–89) | 0.496 |
| Gender | | | 0.216 | | | 0.811 | | | 0.456 | | | 0.661 | | | 0.288 |
| Female | 23 (37) | 39 (63) | | 8 (13) | 54 (87) | | 28 (54) | 24 (46) | | 17 (31) | 38 (69) | | 13 (24) | 42 (76) | |
| Male | 29 (28) | 74 (72) | | 12 (12) | 91 (88) | | 56 (60) | 37 (40) | | 31 (34) | 59 (64) | | 17 (19) | 73 (81) | |
| Location | | | 0.372 | | | 0.221 | | | 0.918 | | | 0.305 | | | 0.338 |
| Proximal | 22 (35) | 40 (65) | | 10 (16) | 52 (84) | | 31 (58) | 22 (42) | | 20 (38) | 32 (68) | | 13 (25) | 39 (75) | |
| Distal | 30 (29) | 73 (71) | | 10 (10) | 93 (90) | | 53 (58) | 39 (42) | | 28 (30) | 65 (70) | | 17 (18) | 76 (82) | |
|
Differentiation | | | 0.773 | | | 0.886 | | | 0.466 | | | 0.331 | | | 0.529 |
| Wel | 23 (34) | 44 (66) | | 9 (13) | 59 (87) | | 32 (53) | 28 (47) | | 22 (38) | 36 (62) | | 11 (19) | 47 (81) | |
| Mod | 24 (28) | 62 (72) | | 10 (12) | 74 (88) | | 43 (59) | 30 (41) | | 23 (31) | 51 (69) | | 15 (20) | 59 (80) | |
| Por | 4 (36) | 7 (64) | | 1 (8) | 11 (92) | | 8 (73) | 3 (27) | | 2 (17) | 10 (83) | | 4 (33) | 8 (67) | |
| T | | | 0.223 | | | 0.346 | | | 0.641 | | | 0.371 | | | 0.558 |
| Tis | 5 (56) | 4 (44) | | 0 (0) | 9 (100) | | 4 (67) | 2 (33) | | 1 (17) | 5 (83) | | 0 (0) | 6 (100) | |
| T1 | 2 (14) | 12 (86) | | 3 (23) | 10 (77) | | 8 (67) | 4 (33) | | 6 (55) | 5 (45) | | 1 (9) | 10 (91) | |
| T2 | 6 (33) | 12 (67) | | 1 (6) | 17 (94) | | 9 (64) | 5 (36) | | 6 (43) | 8 (57) | | 3 (21) | 11 (79) | |
| T3 | 22 (36) | 39 (64) | | 6 (10) | 55 (90) | | 35 (61) | 22 (39) | | 19 (33) | 38 (67) | | 14 (25) | 43 (75) | |
| T4 | 17 (27) | 47 (73) | | 10 (16) | 54 (84) | | 28 (50) | 28 (50) | | 16 (28) | 41 (72) | | 12 (21) | 45 (79) | |
| N | | | 0.325 | | | 0.570 | | | 0.295 | | | 0.404 | | | 0.592 |
| N0 | 28 (29) | 69 (71) | | 13 (13) | 84 (87) | | 45 (54) | 38 (46) | | 30 (36) | 53 (64) | | 16 (19) | 67 (81) | |
| N1-3 | 24 (36) | 43 (64) | | 7 (10) | 60 (90) | | 39 (63) | 23 (37) | | 18 (30) | 43 (70) | | 14 (23) | 47 (77) | |
| M | | | 0.242 | | | 0.098 | | | 0.290 | | | 0.503 | | | 0.0005 |
| M0 | 45 (33) | 92 (67) | | 14 (10) | 123 (90) | | 69 (56) | 54 (44) | | 39 (32) | 83 (68) | | 19 (16) | 103 (84) | |
| M1 | 6 (21) | 22 (79) | | 6 (21) | 22 (79) | | 15 (68) | 7 (32) | | 9 (39) | 14 (61) | | 11 (48) | 12 (52) | |
DNA extraction
Tissues were cut into 10-μm sections from
paraffin-embedded blocks. After specimens were deparaffinized and
washed, tumor tissue was manually dissected for comparison with
H&E slides. The same amount of dissected tissue was used for
each case. Genomic DNA was extracted using standard proteinase K
(Invitrogen, Carlsbad, CA, USA) digestion, phenol/chloroform
extraction, and ethanol precipitation, as previously described
(5).
Mutation analysis
Exon 1 of the KRAS gene and exons 9 and 20 of
the PIK3CA gene were selected for mutation analysis, since
mutations are known to cluster in these regions (6,7).
Primer sequences and PCR conditions have been reported previously
(8). Following the purification of
PCR products using a Microcon YM-100 Centrifugal Filter (Millipore
Corporation, Billerica, MA, USA) and Centri-Sep Columns (Princeton
Separations, Adelphia, NJ, USA), direct sequencing was carried out
using the Big Dye Terminator Cycle Sequencing kit (Applied
Biosystems, Foster City, CA, USA) and analyzed using an Applied
Biosystems 3130 Genetic Analyzer (Applied Biosystems). Somatic
mutations were further validated by independent PCR amplification
and sequencing and matched with normal mucosa.
MethyLight analysis
Sodium bisulfite conversion and DNA recovery was
performed using EpiTect Bisulfite (Qiagen, Hilden, Germany).
Following sodium bisulfite conversion, genomic DNA was analyzed
using the MethyLight technique, a fluorescence-based real time PCR
(Q-PCR) assay (9) and the ABI Prism
7300 Real Time PCR system (Taqman; Applied Biosystems). Four sets
of primers and probes designed specifically for bisulfite-converted
DNA were used. Three sets were used to detect methylation in the
gene of interest and the remaining set served as a reference for
β-actin (ACTB) to normalize for input DNA. Reference primers and
probes were designed in a region of the ACTB gene lacking
CpG dinucleotides, thus allowing for equivalent amplification
regardless of methylation levels. Primer and probe sequences were
previously reported (10,11). SssI-treated HCT-15 DNA was
used as a fully methylated positive control (100% methylation
ratio). Parallel TaqMan PCR reactions were performed using primers
specific for the bisulfite-converted methylated sequence of a
particular locus and with ACTB reference primers. In each
case, triplicate threshold cycle (Ct) values were obtained and
averaged and expression levels were evaluated using the
2−ΔΔCt method. As an internal standard, each sample was
normalized to its ACTB content and compared with the gene
expression level of SssI-treated HCT-15 DNA as positive
controls (calibration sample) as follows: 2ΔΔCt where
ΔΔCt=(Ct-target-Ct-reference) treated sample -
(Ct-target-Ct-reference) calibrator sample. We defined the
percentage of the fully methylated reference (PMR) to be
2ΔΔCt ×100%. To define p16 or hMLH1
methylation status, a PMR cut-off value of 4 was used based on
previously validated data (9).
Based on the distribution of PMR values in normal colon epithelial
tissue (10), a PMR cut-off value
of >0 was used to define positive methylation status for
BNIP3.
Statistical analysis
Statistical analysis was carried out using StatView
Software (version 5.0). To estimate the differences among the
groups, the Chi-square test, Fisher’s exact test, Student’s t-test
and log-rank test were used where appropriate. The Kaplan-Meier
method was used to estimate survival. Survival was calculated
beginning from the date of surgery. Prognostic factors were
estimated using multivariate analysis and the Cox proportional
hazards model. P<0.05 was considered to indicate a statistically
significant result.
Results
Mutation of KRAS and PIK3CA and
methylation of BNIP3, p16 and hMLH1
KRAS mutations in exon 1 were observed in 32%
of cases and PIK3CA mutations in exons 9 and 20 in 12% of
cases. Methylation frequencies of the examined genes were 58% for
BNIP3, 33% for p16 and 21% for hMLH1. Although
hMLH1 methylation was significantly associated with distant
metastasis (P=0.0005), other associations were not observed between
clinicopathological characteristics and the mutation or methylation
status of individual genes (Table
I).
Correlation between mutation and
methylation
Associations among gene mutations and gene
methylations are shown in Table
II. The KRAS mutation was significantly associated with
BNIP3 (P<0.0001) and p16 methylation (P=0.04) as
well as with the number of methylated genes (P=0.0082). p16
methylation was associated with BNIP3 methylation
(P<0.0001) and hMLH1 methylation (P=0.03).
 | Table IIConcordance between the mutation and
methylation status of each gene. |
Table II
Concordance between the mutation and
methylation status of each gene.
| PIK3CA | BNIP3 | p16 | hMLH1 | Methylationa |
|---|
|
|
|
|
|
|
|---|
| Mut | Wt | Met | Unm | Met | Unm | Met | Unm | High | Low |
|---|
| KRAS | P=0.918 | P<0.0001 | P=0.0402 | P=0.467 | P=0.0082 |
| Mut | 7 | 45 | 39 | 9 | 21 | 26 | 8 | 39 | 24 | 23 |
| Wt | 13 | 88 | 45 | 52 | 27 | 71 | 22 | 77 | 28 | 70 |
| PIK3CA | | | P=0.827 | P=0.982 | P=0.428 | P=0.775 |
| Mut | | | 10 | 8 | 6 | 12 | 5 | 13 | 7 | 11 |
| Wt | | | 74 | 53 | 42 | 85 | 25 | 102 | 45 | 82 |
| BNIP3 | | | | | P<0.0001 | P=0.541 | | |
| Met | | | | | 42 | 39 | 19 | 64 | | |
| Unm | | | | | 4 | 54 | 11 | 48 | | |
| p16 | | | | | | | P=0.0295 | | |
| Met | | | | | | | 15 | 33 | | |
| Unm | | | | | | | 15 | 81 | | |
Correlation between molecular markers and
outcomes
The correlation between the molecular parameters and
disease-specific survival (DSS) were analyzed (Table III). Patients with
hMLH1-methylated tumors had significantly shorter DSS
compared with those without methylation [hazards ratio (HR), 2.231;
P=0.05], but mutation or methylation of other single genes did not
affect DSS. Having mutations in both the KRAS and
PIK3CA genes did not correlate with DSS. By contrast, when
cases were divided into 2 groups, namely a low methylation group (0
or 1 methylated genes) and a high methylation group (2 or 3
methylated genes), the high methylation group had significantly
shorter DSS than the low methylation group (HR, 2.681; P=0.01).
Integration of the KRAS mutation and methylation status of
multiple genes did not reveal an association with DSS. However,
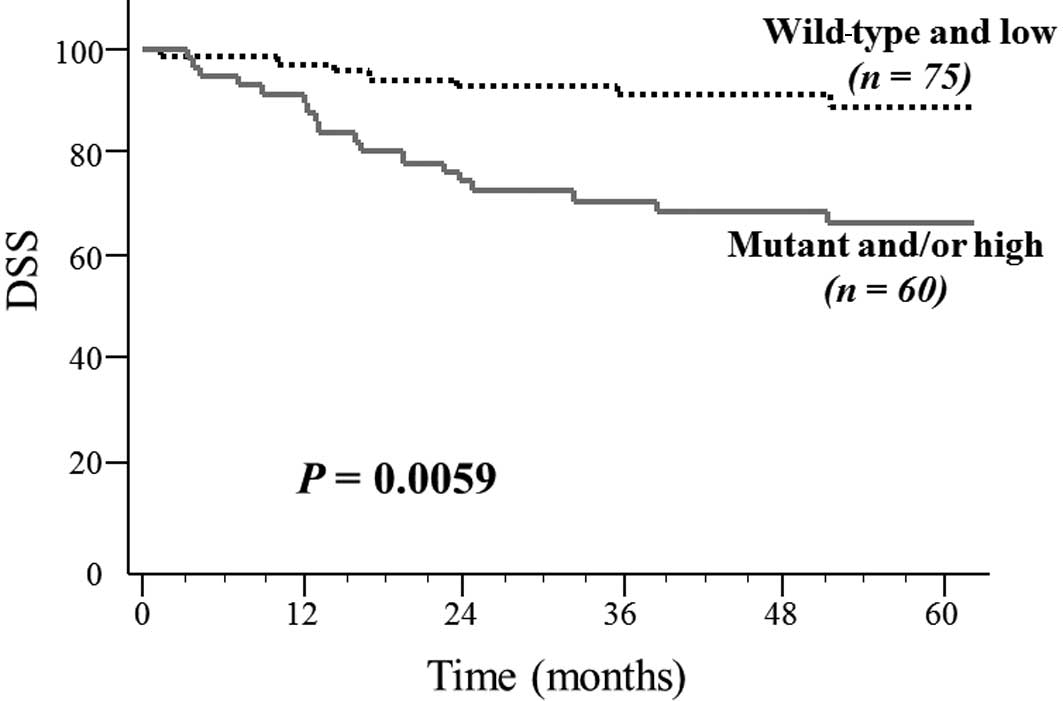
CRCs were divided into 4 groups: CRC containing i) PIK3CA
mutation and high methylation (5.2%); ii) PIK3CA mutation
and low methylation (8.1%); iii) no mutation and high methylation
(31.1%); and iv) no mutation and low methylation (55.6%). Patients
with the PIK3CA mutation and/or high methylation (groups i,
ii and iii) had a significantly poorer outcome in DSS compared with
those with wild-type PIK3CA and low methylation (group iv;
HR, 2.924; P=0.0087) (P=0.0059, log-rank test; Fig. 1).
 | Table IIICorrelation of molecular parameters
with DSS. |
Table III
Correlation of molecular parameters
with DSS.
| | HR | 95% CI | P |
|---|
| KRAS | (Mutant vs.
wild) | 0.930 | 0.418–2.071 | 0.8590 |
| PIK3CA | (Mutant vs.
wild) | 1.429 | 0.540–3.779 | 0.4721 |
| BNIP3 | (Methylated vs.
unmethylated) | 2.233 | 0.944–5.284 | 0.0675 |
| p16 | (Methylated vs.
unmethylated) | 1.950 | 0.912–4.169 | 0.0849 |
| hMLH1 | (Methylated vs.
unmethylated) | 2.231 | 1.000–4.977 | 0.0500 |
| Mutationa | (Mutant vs.
wild) | 1.001 | 0.468–2.139 | 0.9983 |
| Methylationb | (High vs. low) | 2.681 | 1.253–5.747 | 0.0110 |
|
Mutation/methylation | (Mutant and/or high
vs. wild and low) | 1.499 | 0.634–3.546 | 0.3569 |
|
KRAS/methylation | (Mutant and/or high
vs. others) | 1.675 | 0.766–3.663 | 0.1963 |
|
PIK3CA/methylation | (Mutant and/or high
vs. others) | 2.924 | 1.312–6.494 | 0.0087 |
Results of the univariate and multivariate analyses
using the Cox proportional hazards model are shown in Table IV. Univariate analysis revealed
that the PIK3CA mutation and/or high methylation was a
significant prognostic factor for DSS, when used with the depth of
the tumor, lymph node metastasis, distant metastasis, histological
differentiation, lymphatic invasion and vessel invasion. Variables
with P<0.05 in the univariate analysis were used for the
multivariate analysis. PIK3CA mutation and/or high
methylation, as well as distant metastasis and lymphatic invasion,
were found to be independent and significant prognostic factors for
DSS.
 | Table IVCox proportional hazards model of
prognostic factors of DSS. |
Table IV
Cox proportional hazards model of
prognostic factors of DSS.
| | Univariate
analysis | Multivariate
analysis |
|---|
| |
|
|
|---|
|
Characteristics | | HR | 95% CI | P | HR | 95% CI | P |
|---|
| Gender | (Female vs.
male) | 0.424 | 0.171–1.050 | 0.0636 | | | |
| Age | (≥65 vs.
<65) | 0.699 | 0.328–1.487 | 0.3526 | | | |
| Location | (Distal vs.
proximal) | 0.839 | 0.389–1.808 | 0.6539 | | | |
| T | (T1,2 vs.
T3,4) | 8.547 | 1.157–62.50 | 0.0354 | 2.451 | 0.270–22.20 | 0.4253 |
| N | (N− vs. N+) | 3.559 | 1.558–8.130 | 0.0026 | 1.368 | 0.504–3.717 | 0.5377 |
| M | (M0 vs. M1) | 7.874 | 3.448–17.86 | <0.0001 | 7.092 | 2.732–18.18 | <0.0001 |
|
Differentiation | (Por vs.
wel/mod) | 3.031 | 1.046–8.781 | 0.0411 | 0.611 | 0.172–2.172 | 0.4466 |
| Ly | (Ly0,1 vs.
ly2,3) | 4.831 | 2.247–10.31 | <0.0001 | 3.891 | 1.379–10.99 | 0.0102 |
| v | (v0,1 vs.
v2,3) | 4.049 | 1.399–11.76 | 0.0099 | 1.761 | 0.540–5.747 | 0.3482 |
|
PIK3CA/methylation | (Mutant and/or high
vs. others) | 2.924 | 1.312–6.494 | 0.0087 | 2.793 | 1.220–6.410 | 0.0151 |
Discussion
In this study, we examined mutations in KRAS
and PIK3CA, which comprise components of the EGFR pathway,
and the methylation status of BNIP3, p16 and
hMLH1 in CRCs to evaluate patients with the PIK3CA
mutation and/or 2 or more methylations with a poorer prognosis
compared with other patients. Our results suggest that a
combination of epigenetic and genetic alterations is a potent
biomarker for predicting CRC prognosis.
Integrated analysis of epigenetic and genetic
alterations is increasingly significant in cancer research. Since
CRC develops as a result of accumulating genetic and epigenetic
alterations, the combination of genetic and epigenetic profiles
could confer differential clinical phenotypes and potential
variability in survival. Ward et al reported that although
DNA methylation is associated with a worse outcome in CRC patients,
this adverse prognostic effect is lost in methylated tumors with
microsatellite instability (12).
By integrating genetic and epigenetic analyses, Shen et al
revealed that colon cancers correspond to 3 molecularly distinct
subclasses of disease, each of which is relatively homogeneous
(13). In the present study, CRCs
were divided into 4 groups according to PIK3CA mutations and
the number of methylated genes: CRCs with i) PIK3CA mutation
and 2 or 3 methylations (5.2%); ii) PIK3CA mutation and less
than 2 methylations (8.1%); iii) no mutation and 2 or 3
methylations (31.1%); and iv) no mutations and less than 2
methylations (55.6%). The last group was associated with better
prognosis than the former 3 groups. CRC patients with the
PIK3CA mutation and/or more than 1 methylation had worse
outcomes compared with the other groups. This finding suggests that
mutation analysis and determining methylation status may provide
significant predictive information regarding tumor behavior and
clinical outcome in CRC patients.
BNIP3 is a pro-apoptotic member of the Bcl-2 family
(14). The expression of BNIP3 is
induced by hypoxia, such as that which occurs during cardiac
ischemia and in the hypoxic regions of tumors, and it acts against
pro-survival proteins, including Bcl-2 and Bcl-xl (15–18).
Transcriptional silencing of the BNIP3 gene has recently
been observed in multiple human cancer cell lines, including
colorectal, gastric, pancreatic, and hematopoietic tumors (19). Murai et al reported that
aberrant hypermethylation of BNIP3 was detected in 66% of
primary colorectal and 49% of primary gastric cancers, but not in
normal tissue; these authors also revealed that BNIP3 is
silenced by DNA methylation (20).
Akada et al and Erkan et al revealed that loss of
BNIP3 expression correlates with poor prognosis and increased
chemoresistance in patients with pancreatic cancer (21,22).
Manka et al demonstrated that BNIP3 silencing induces
or increases the metastatic growth of breast cancer in distant
organs (23). Moreover, our
previous study also indicated that BNIP3 methylation is
associated with poor clinical outcome and chemoresistance in
primary CRC (10). These previous
studies suggest that reduced BNIP3 expression contributes to
carcinogenesis of the colon and rectum and may be a predictive
factor for the prognosis of CRC. Therefore, the BNIP3 gene,
as well as the hMLH1 and p16 genes, were tested in
our methylated gene sets for predicting the outcome of CRC.
Activation of the phosphatidylinositol 3-kinase
(PI3K)/AKT pathway is thought to be critical in CRC development and
clinical behavior (6,8,24,25).
PIK3CA encodes the p110α catalytic subunit of PI3K and is
mutated in 10–32% of CRCs (6,8,24,26–29).
PIK3CA mutations elevate kinase activity, thereby activating
the PI3K/AKT pathway and contributing to tumorigenesis through
decreased apoptosis, loss of contact inhibition and increased tumor
invasion (6,30); these factors have been associated
with poor outcomes in CRC patients (8,24,31).
Although previous studies have investigated the role of the
PIK3CA mutation and gene methylation in CRC, the data are
inconclusive. Certain studies reported a significantly higher
frequency of the PIK3CA mutation in the CpG island
methylator phenotype (CIMP)-high tumor compared with CIMP-negative
tumors and a significant correlation between the PIK3CA
mutation and RASSF2 gene methylation (29,32,33),
whereas other studies failed to observe this correlation (11,34).
In the present study, although CIMP status was not examined, the
PIK3CA mutation did not exhibit an association with the
methylation status of the genes examined. Moreover, few studies
have described the effect of the combination of the PIK3CA
mutation with gene methylation on patient outcome. Ogino et
al demonstrated that the effect of the PIK3CA mutation
on CRC patient survival is not significantly modified by CIMP
(31). Their result was
inconsistent with our finding that integration of the PIK3CA
mutation and methylation status may be used to predict outcomes for
CRC patients. It is possible that the methylated gene set used in
this study was different from that used in the Ogino study. The
BNIP gene included in our methylated set was not included in
CIMP markers used to date. Moreover, there have been no studies
regarding the influence of integrating the PIK3CA mutation
and methylation status of the BNIP3 gene on outcomes of CRC.
It would be useful to examine whether mutating other genes from the
EGFR pathway also affects CRC prognosis in cooperation with
BNIP3 methylation.
In conclusion, by integrating genetic and epigenetic
alterations, we revealed that CRC patients with poor prognosis
could be identified. Further studies are necessary to confirm our
finding that a combination of the PIK3CA mutation with gene
methylation may be useful for predicting outcomes of CRC patients
and to elucidate the underlying mechanism behind these
findings.
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics. CA Cancer J Clin. 60:277–300. 2010.
|
|
2
|
Wood LD, Parsons DW, Jones S, et al: The
genomic landscape of human breast and colorectal cancers. Science.
318:1108–1113. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
4
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Iida S, Akiyama Y, Nakajima T, Ichikawa W,
Nihei Z, Sugihara K and Yuasa Y: Alterations and hypermethylation
of the p14ARF gene in gastric cancer. Int J Cancer. 87:654–658.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Samuels Y, Wang Z, Bardelli A, et al: High
frequency of mutations of the PIK3CA gene in human cancers.
Science. 304:5542004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vogelstein B, Fearon ER, Hamilton SR, et
al: Genetic alterations during colorectal-tumor development. N Engl
J Med. 319:525–532. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kato S, Iida S, Higuchi T, et al: PIK3CA
mutation is predictive of poor survival in patients with colorectal
cancer. Int J Cancer. 121:1771–1778. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Eads CA, Danenberg KD, Kawakami K, et al:
MethyLight: a high-throughput assay to measure DNA methylation.
Nucleic Acids Res. 28:E32–00. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shimizu S, Iida S, Ishiguro M, et al:
Methylated BNIP3 gene in colorectal cancer prognosis. Oncol Lett.
1:865–872. 2010.PubMed/NCBI
|
|
11
|
Aoyagi H, Iida S, Uetake H, et al: Effect
of classification based on combination of mutation and methylation
in colorectal cancer prognosis. Oncol Rep. 25:789–794.
2011.PubMed/NCBI
|
|
12
|
Ward RL, Cheong K, Ku SL, Meagher A,
O’Connor T and Hawkins NJ: Adverse prognostic effect of methylation
in colorectal cancer is reversed by microsatellite instability. J
Clin Oncol. 21:3729–3736. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shen L, Toyota M, Kondo Y, et al:
Integrated genetic and epigenetic analysis identifies three
different subclasses of colon cancer. Proc Natl Acad Sci USA.
104:18654–18659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Reed JC: Bcl-2 family proteins. Oncogene.
17:3225–3236. 1998. View Article : Google Scholar
|
|
15
|
Labi V, Erlacher M, Kiessling S and
Villunger A: BH3-only proteins in cell death initiation, malignant
disease and anticancer therapy. Cell Death Differ. 13:1325–1338.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Strasser A: The role of BH3-only proteins
in the immune system. Nat Rev Immunol. 5:189–200. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Willis SN and Adams JM: Life in the
balance: how BH3-only proteins induce apoptosis. Curr Opin Cell
Biol. 17:617–625. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vande VC, Cizeau J, Dubik D, et al: BNIP3
and genetic control of necrosis-like cell death through the
mitochondrial permeability transition pore. Mol Cell Biol.
20:5454–5468. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mellor HR and Harris AL: The role of the
hypoxia-inducible BH3-only proteins BNIP3 and BNIP3L in cancer.
Cancer Metastasis Rev. 26:553–566. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Murai M, Toyota M, Suzuki H, et al:
Aberrant methylation and silencing of the BNIP3 gene in colorectal
cancer and gastric cancer. Clin Cancer Res. 11:1021–1027.
2005.PubMed/NCBI
|
|
21
|
Akada M, Crnogorac-Jurcevic T, Lattimore
S, et al: Intrinsic chemoresistance to gemcitabine is associated
with decreased expression of BNIP3 in pancreatic cancer. Clin
Cancer Res. 11:3094–3101. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Erkan M, Kleeff J, Esposito I, et al: Loss
of BNIP3 expression is a late event in pancreatic cancer
contributing to chemoresistance and worsened prognosis. Oncogene.
24:4421–4432. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Manka D, Spicer Z and Millhorn DE:
Bcl-2/adenovirus E1B 19 kDa interacting protein-3 knockdown enables
growth of breast cancer metastases in the lung, liver, and bone.
Cancer Res. 65:11689–11693. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Barault L, Veyrie N, Jooste V, et al:
Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH
kinase) signaling network correlate with poor survival in a
population-based series of colon cancers. Int J Cancer.
122:2255–2259. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Manning BD and Cantley LC: AKT/PKB
signaling: Navigating downstream. Cell. 129:1261–1274. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Velho S, Oliveira C, Ferreira A, et al:
The prevalence of PIK3CA mutations in gastric and colon cancer. Eur
J Cancer. 41:1649–1654. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Benvenuti S, Frattini M, Arena S, et al:
PIK3CA cancer mutations display gender and tissue specificity
patterns. Hum Mutat. 29:284–288. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Abubaker J, Bavi P, Al-Harbi S, et al:
Clinicopathological analysis of colorectal cancers with PIK3CA
mutations in Middle Eastern population. Oncogene. 27:3539–3545.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nosho K, Kawasaki T, Ohnishi M, et al:
PIK3CA mutation in colorectal cancer: relationship with genetic and
epigenetic alterations. Neoplasia. 10:534–541. 2008.PubMed/NCBI
|
|
30
|
Samuels Y, Diaz LA Jr, Schmidt-Kittler O,
et al: Mutant PIK3CA promotes cell growth and invasion of human
cancer cells. Cancer Cell. 7:561–573. 2005. View Article : Google Scholar
|
|
31
|
Ogino S, Nosho K, Kirkner GJ, et al:
PIK3CA mutation is associated with poor prognosis among patients
with curatively resected colon cancer. J Clin Oncol. 27:1477–1484.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nosho K, Yamamoto H, Takahashi T, et al:
Genetic and epigenetic profiling in early colorectal tumors and
prediction of invasive potential in pT1 (early invasive) colorectal
cancers. Carcinogenesis. 28:1364–1370. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Whitehall V, Rickman C, Bond C, et al:
Oncogenic PIK3CA mutations in colorectal cancers and polyps. Int J
Cancer. 2011.(Epub ahead of print). View Article : Google Scholar
|
|
34
|
Bruin SC, He Y, Mikolajewska-Hanclich I,
et al: Molecular alterations associated with liver metastases
development in colorectal cancer patients. Br J Cancer.
105:281–287. 2011. View Article : Google Scholar : PubMed/NCBI
|