Introduction
The incidence of esophageal cancer (EC) has been
high (68.88/100,000) among the Kazakh people living in the Xinjiang
Uygur Autonomous Region (Northwest of China) during the past 30
years. Nevertheless, early detection and treatment rates of EC
remain low, and it consequently has a poor prognosis. If early
diagnosis and treatment were possible, the 5-year survival rate
could be increased to above 90% as in other countries/regions that
have early detection programs (1–3).
Conventional pathological diagnosis plays a crucial
role in the diagnosis of EC, and provides important information on
tumor differentiation and the degree of morphological changes
(4,5). However, due to the limitations of
biopsy pathology, discrepancies between pathological diagnosis and
actual diagnosis occasionally occur, making clinical diagnosis and
treatment difficult. There is, therefore, a need to find a more
objective and quantitative method to distinguish benign from
malignant cells.
A number of studies suggest that the incidence and
evolution of EC involves a variety of chromosomal anomalies. During
carcinogenesis, a cell goes through molecular cytogenetic changes
prior to showing morphological changes. Nuclear chromosome
abnormality, which can be observed in cancer cells, is an early
event during the process of tumorigenesis, and it has become the
determining objective index of cancer cells.
Cell nuclear aneuploidy is one of the most common
features of a number of types of cancer, including EC (6,7).
Malignant cells are therefore capable of being diagnosed by
detecting aneuploidy, usually found in aneusomic nuclei.
Fluorescence in situ hybridization (FISH) technology is a
rapid and sensitive method for detecting aneusomy of a specific
chromosome and is widely used in the diagnosis of hematological
malignancies, lung, breast and kidney cancer, with high sensitivity
and specificity (8–11). The advantage of the FISH method has
been considered to lie in its objective and quantitative evaluation
of malignant cells.
Some studies have suggested that FISH has a certain
value in detecting a variety of cancer cells in conventional
cytology and early cancer diagnosis (12–14).
However, no comparative study on conventional pathology with FISH
using biopsy tissues for cancer cell detection has been reported
previously.
In this study, 50 Kazakh patients with suspected EC
underwent FISH examination and conventional pathological diagnosis
using biopsied samples, to analyze the value of the clinical
applications and prospective uses of the FISH method in the early
diagnosis of EC.
Patients and methods
Patients
This study was approved by the Ethics Committee of
the First Affiliated Hospital of Xinjiang Medical University,
China. Between March 2009 and December 2010, 50 Kazakh EC patients
were admitted to the Department of Thoracic Surgery (First
Affiliated Hospital of Xinjiang Medical University) and underwent
resection. The patients included 40 males and 10 females, with an
average age of 56.8 years (range 31–82).
Final pathological diagnosis confirmed esophageal
squamous cell carcinoma in 47 cases post-operatively, including
well differentiated tumors in 23 cases, moderately differentiated
tumors in 18 cases, and poorly differentiated tumors in 6 cases; as
well as poorly differentiated adenocarcinoma in 1 case, mucinous
adenocarcinoma in 1 case, and small cell carcinoma in 1 case
(Table I).
 | Table IPatient charateristics. |
Table I
Patient charateristics.
| Case | Gender | Age | Diagnosis |
|---|
| 1 | M | 55 | Sq |
| 2 | F | 63 | Sq |
| 3 | M | 50 | Sq |
| 4 | M | 48 | Sq |
| 5 | F | 65 | Sq |
| 6 | M | 56 | Sq |
| 7 | F | 52 | Sq |
| 8 | M | 63 | Sq |
| 9 | M | 66 | Scc |
| 10 | F | 59 | Sq |
| 11 | M | 54 | Sq |
| 12 | M | 47 | Sq |
| 13 | M | 63 | Sq |
| 14 | M | 82 | Sq |
| 15 | F | 59 | Sq |
| 16 | M | 31 | Sq |
| 17 | M | 51 | Sq |
| 18 | M | 50 | Sq |
| 19 | M | 64 | Sq |
| 20 | M | 46 | Sq |
| 21 | M | 44 | Sq |
| 22 | M | 56 | Sq |
| 23 | M | 42 | M-ad |
| 24 | M | 61 | Sq |
| 25 | F | 48 | Sq |
| 26 | M | 42 | Sq |
| 27 | M | 39 | Sq |
| 28 | M | 71 | Sq |
| 29 | F | 60 | Sq |
| 30 | M | 56 | Sq |
| 31 | M | 45 | Sq |
| 32 | M | 64 | Sq |
| 33 | M | 72 | Sq |
| 34 | M | 65 | Sq |
| 35 | M | 67 | Sq |
| 36 | M | 65 | Sq |
| 37 | M | 58 | Sq |
| 38 | F | 54 | Sq |
| 39 | M | 50 | Sq |
| 40 | F | 56 | Sq |
| 41 | M | 65 | Sq |
| 42 | M | 56 | Sq |
| 43 | M | 63 | Sq |
| 44 | M | 69 | Sq |
| 45 | M | 71 | Sq |
| 46 | M | 65 | Sq |
| 47 | M | 62 | Sq |
| 48 | M | 56 | Sq |
| 49 | F | 37 | Sq |
| 50 | M | 56 | Ad |
Methods
To determine the diagnosis pre-operatively, all
patients underwent esophagofiberscopic examination, and sites that
appeared to be suspicious for malignancy were biopsied using
standard biopsy forceps. After performing touch preparations of
cells on glass slides with the specimen, the same specimens were
used for conventional pathological diagnosis. Informed consent was
obtained from all 50 patients.
Samples were stained with hematoxylin and eosin
(H&E). Pathological evaluations were performed by three
qualified pathologists from the Department of Pathology.
Pathomorphological classification of biopsy
specimens were as follows: class I was mild grade squamous
epithelial hyperplasia; class II was mild dysplasia; class III was
moderate dysplasia, but without any malignant characteristics;
class IV was severe dysplasia, i.e., carcinoma in situ;
class V was typical cancer tissue. Classes IV and V were considered
to be EC.
Touch preparations of cells were made on glass
slides and air dried overnight at room temperature and then stored
at −80°C. Centromeric probes labeled with fluorochrome were used
for the visualization and enumeration of copy numbers. Spectrum
orange and green labeled probes were used to visualize centromeric
regions of chromosomes 3 and 17. Reagents were purchased from
Abbott Molecular, Inc. (Des Plaines, IL, USA).
Preparation of slides
Cells were denatured with 70% formamide and then
washed twice in standard saline citrate (SSC) at 74°C and at room
temperature, respectively, for 2 min in a water bath. Then, slides
were dehydrated through a graded ethanol series (70, 85 and 100%,
each for 2 min). We then applied 10 μl of hybridization solution
containing 1 μl of each of the DNA probes, 7 μl of hybridization
buffer and 1 μl of double distilled water. This was covered with a
cover slip and sealed with rubber cement. Following incubation for
16 h at 42°C in a humidity-controlled chamber, the slides were
washed with an SSC solution for 5 min at 74°C, and at room
temperature for 2 min. Diamidinophenylindole (DAPI, II) (5 μl) was
applied to each spot and covered with a cover slip. The slides were
observed under a fluorescence microscope that was connected to a
cooled charge-coupled device camera and an image analyzer system
(Leica Microsystems, Ltd., Germany).
FISH analysis
FISH signal analysis was performed as follows: all
cells, with the exception of damaged cells or those with
overlapping nuclei, were evaluated. We counted 100 nuclei from each
patient, and the total number of centromeric signals was recorded.
When the percentage of hyperdisomic nuclei with more than three
copies for at least one nucleus was >10%, we diagnosed
malignancy.
FISH diagnosis was made without knowing the result
of the conventional pathological diagnosis. Similarly, the results
of FISH analysis were not shown to the pathologists. Thus, the two
diagnoses were independently performed in a blind manner.
Statistical analysis
An IBM SPSS 16.0 statistical software package (IBM
Corporation, NY, USA) was used for statistical analysis. The
student's paired t-test was used to test the difference between the
number of countable centromere signals of chromosome 3 and 17.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Patients and FISH analysis
Biopsy pathology yielded a diagnosis of primary EC
in 45 patients, including squamous cell carcinoma in 42 cases,
adenocarcinoma in 1 case, mucinous adenocarcinoma in 1 case, and
small cell carcinoma in 1 case.
We classify the cases as follows: 3 cases were
classified as class II; 2 cases as class III; 4 cases as class IV;
41 cases as class V. Five cases were false-negative but there were
no false-positive cases. The sensitivity and specificity were 87.2
and 100%, respectively.
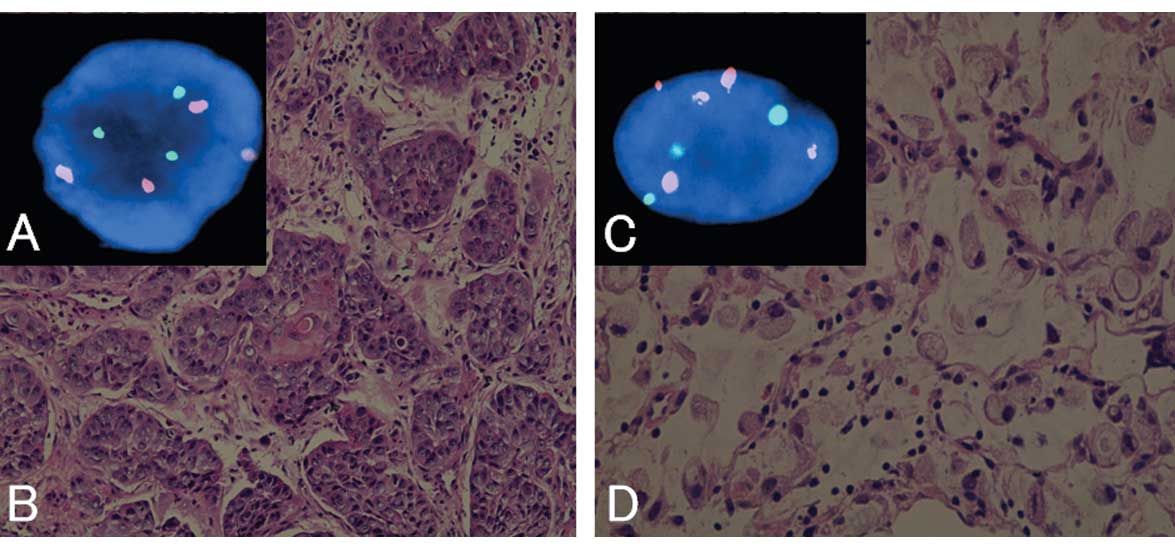
FISH analysis revealed that 48 cases had abnormal
copy numbers in either chromosome 3 or 17. Representative findings
of the pathology and FISH are shown in Fig. 1.
The FISH method yielded 2 false-negative and no
false-positive cases, with a sensitivity and specificity of 94.8
and 100%, respectively (Table II).
The copy numbers of centromeres in chromosome 3 were significantly
higher (P=0.0001) than the centromeres of chromosome 17.
 | Table IIResults of FISH, biopsy pathology and
final pathology. |
Table II
Results of FISH, biopsy pathology and
final pathology.
| Case | 3 copies
CEP3/CEP17 | 4 copies
CEP3/CEP17 | ≥5 copies
CEP3/CEP17 | Hyperdisomy
(%) | Biopsy
pathology | FISH | Pathology |
|---|
| 1 | 12/9 | 23/18 | 11/12 | 46/39 | Sq, V | Positive | pT2N2M0 |
| 2 | 18/17 | 17/13 | 23/23 | 58/53 | Sq, V | Positive | pT2N0M0 |
| 3 | 26/18 | 23/20 | 19/21 | 68/59 | Sq,V | Positive | pT4N3M0 |
| 4 | 14/15 | 19/15 | 18/16 | 51/46 | Sq,V | Positive | pT2N0M0 |
| 5 | 23/16 | 20/21 | 16/16 | 59/53 | Sq, IV | Positive | pT3N1M0 |
| 6 | 27/23 | 20/17 | 27/26 | 74/66 | Sq,V | Positive | pT2N1M0 |
| 7 | 15/17 | 25/19 | 25/20 | 65/56 | Sq, V | Positive | pT2N1M0 |
| 8 | 11/8 | 11/16 | 19/20 | 41/44 | Sq, V | Positive | pT3N2M0 |
| 9 | 31/28 | 25/20 | 33/32 | 89/80 | Scc, V | Positive | pT2N1M0 |
| 10 | 25/26 | 30/25 | 18/18 | 73/69 | Sq, V | Positive | pT3N1M0 |
| 11 | 22/20 | 27/28 | 20/23 | 69/71 | Sq, V | Positive | pT2N0M0 |
| 12 | 33/29 | 30/31 | 19/12 | 85/72 | Sq, V | Positive | pT4N1M0 |
| 13 | 3/1 | 1/2 | 1/0 | 5/3 | aNegative, II | aNegative | pT1N0M0 |
| 14 | 35/27 | 29/26 | 31/29 | 95/82 | Sq, V | Positive | pT3N2M0 |
| 15 | 19/13 | 22/25 | 22/19 | 63/57 | Sq, V | Positive | pT2N0M0 |
| 16 | 19/15 | 22/22 | 17/18 | 58/55 | Sq, V | Positive | pT3N1M0 |
| 17 | 24/20 | 30/26 | 14/17 | 68/63 | Sq, V | Positive | pT2N1M0 |
| 18 | 12/9 | 11/14 | 21/15 | 44/38 | Sq, V | Positive | pT3N1M0 |
| 19 | 23/17 | 23/20 | 21/21 | 67/58 | Sq, V | Positive | pT3N0M0 |
| 20 | 9/11 | 12/12 | 9/7 | 40/30 | Sq, IV | Positive | pT2N1M0 |
| 21 | 26/21 | 12/9 | 17/15 | 55/45 | Sq,V | Positive | pT3N2M0 |
| 22 | 17/14 | 29/23 | 19/11 | 65/48 | Sq, V | Positive | pT3N0M0 |
| 23 | 22/16 | 28/19 | 17/20 | 67/55 | M-ad,V | Positive | pT4N2M0 |
| 24 | 29/16 | 25/25 | 17/18 | 71/59 | Sq,V | Positive | pT2N0M0 |
| 25 | 2/2 | 0/2 | 1/0 | 3/4 | aNegative, II | aNegative | pT2N0M0 |
| 26 | 13/15 | 7/9 | 8/8 | 28/32 | Sq, V | Positive | pT3N1M0 |
| 27 | 17/15 | 10/15 | 5/5 | 32/35 | Sq, V | Positive | pT3N0M0 |
| 28 | 58/52 | 12/12 | 15/13 | 85/77 | Sq, V | Positive | pT3N1M0 |
| 29 | 30/24 | 22/16 | 20/21 | 72/61 | Sq, V | Positive | pT3N1M0 |
| 30 | 55/47 | 21/19 | 9/9 | 85/75 | Sq, V | Positive | pT4N2M0 |
| 31 | 45/39 | 9/13 | 7/2 | 61/54 | Sq, V | Positive | pT3N1M0 |
| 32 | 13/11 | 12/13 | 5/2 | 30/26 | aNegative, III | Positive | pT2N0M0 |
| 33 | 31/26 | 23/15 | 3/5 | 56/46 | Sq, V | Positive | pT3N1M0 |
| 34 | 48/38 | 24/19 | 11/12 | 83/69 | Sq, V | Positive | pT2N0M0 |
| 35 | 20/21 | 13/16 | 9/7 | 42/44 | Sq, V | Positive | pT2N1M0 |
| 36 | 19/21 | 8/5 | 12/12 | 39/38 | Sq, V | Positive | pT2N0M0 |
| 37 | 17/17 | 20/15 | 10/10 | 47/42 | Sq, V | Positive | pT3N1M0 |
| 38 | 21/22 | 9/6 | 3/1 | 33/29 | aNegative, III | Positive | pT1N0M0 |
| 39 | 25/20 | 19/16 | 8/7 | 52/43 | Sq, V | Positive | pT2N1M0 |
| 40 | 13/11 | 6/7 | 5/8 | 24/26 | aNegative, II | Positive | pT1N0M0 |
| 41 | 20/17 | 10/9 | 4/2 | 34/28 | Sq, IV | Positive | pT2N0M0 |
| 42 | 15/12 | 9/11 | 6/7 | 30/30 | Sq, IV | Positive | pT2N1M0 |
| 43 | 32/23 | 11/6 | 7/7 | 50/36 | Sq, V | Positive | pT2N0M0 |
| 44 | 51/47 | 16/12 | 9/9 | 76/68 | Sq, V | Positive | pT2N0M0 |
| 45 | 29/20 | 14/20 | 15/21 | 58/61 | Sq, V | Positive | pT3N2M0 |
| 46 | 44/38 | 19/13 | 10/7 | 73/58 | Sq, V | Positive | pT4N3M0 |
| 47 | 20/17 | 23/22 | 11/9 | 54/48 | Sq, V | Positive | pT2N0M0 |
| 48 | 49/41 | 20/15 | 13/6 | 82/62 | Sq, V | Positive | pT2N1M0 |
| 49 | 29/25 | 19/15 | 12/12 | 60/52 | Sq, V | Positive | pT3N0M0 |
| 50 | 38/32 | 25/19 | 13/5 | 76/56 | Ad, V | Positive | pT3N2M0 |
Discussion
The Xinjiang Uygur Autonomous Region is a
multi-ethnic area located in the Northwest of China and the Kazakh
ethnic group has a high incidence of EC. Improved early diagnosis
of EC among this ethnic group is likely to lessen the burden of EC
(1).
Aneuploidy is present in the nuclei of cancer cells.
It is a common molecular pathological characteristic in human
carcinoma (7,15,16). A
number of studies have suggested that using DNA probes for the
detection of aneuploidy in cancer cells may be a superior technique
to conventional pathological diagnosis (8–12,14).
Han et al examined 113 EC patients using specific centromere
DNA probes 3, 8, 10, 12, 17 and 20, and found that chromosomal
signal numbers and all chromosomes were found to have abnormal copy
numbers (12). In their study,
Fritcher et al analyzed esophageal adenocarcinoma using the
FISH method with the centromeric region probes C-MYC, P16, HER2 and
20q13 (9). These authors found that
the sensitivity of cytology is only 45% for the detection of
esophageal adenocarcinoma, but FISH yielded a detection rate of
100%. The same study used FISH analysis with centromeric probes 7,
11, 12, 17 and 18. Aneusomy was not found in the normal controls of
any chromosomes. By contrast, chromosomal abnormalities were found
in all carcinoma specimens (13).
Using FISH technology, the genetic analysis of
interphase nuclei closely reflected the real changes in
chromosomes. Simultaneous use of two or more different
fluorescent-labeled probes resulted in high sensitivity and
specificity for the detection of esophageal, lung and breast cancer
cells (9,11,14).
Therefore, we performed FISH analysis and conventional pathological
diagnosis for biopsied specimens in suspected EC patients to
compare their sensitivity and specificity in order to analyze
whether the early diagnosis of EC is possible or not and to obtain
its clinical value. Our results showed that using the molecular
pathological diagnostic method during the process of EC diagnosis
is more accurate than conventional pathological diagnosis. This is
consistent with the results of similar studies (8,12,14,17).
In our study, we selected the centromeres in
chromosome 3 and 17 probes, and set the cut-off value of the
percentage of hyperdisomic cells at 10%, whereas normal cells often
have less than 6%. This discrepancy is probably due to counting
sister chromatids as copies.
The sensitivity and specificity by biopsy pathology
were 87.2 and 100%, respectively. Using FISH analysis, the
sensitivity and specificity were 94.8 and 100%, respectively. These
results indicated that FISH is more sensitive than biopsy
pathology, the latter yielding 5 false-negative results: class II
in 3 cases, class III in 2 cases. Post-operative final pathological
diagnoses in all of these patients were esophageal squamous cell
carcinomas. FISH yielded 2 false-negative results, both of which
matched the 2 pathologically false-negative class II cases. This
finding was probably due to the shallowness of the biopsies.
FISH successfully detected cancer cells in 3 cases
in which biopsy pathology was false-negative. Post-operative
pathological staging confirmed stage IA in 2 cases, and stage IIA
in 1 case, suggesting that FISH is capable of detecting the
relevant chromosomal mutations in EC earlier. Therefore, FISH has
its own clinical detection value for the early diagnosis of EC, and
our findings have been supported by other studies (9,12,18).
FISH may provide objective information on malignant cells,
particularly in tissues with moderate or severe dysplasia.
Therefore, we recommend FISH as an ancillary test in cases with
moderate or severe dysplasia in order to avoid the misdiagnosis of
EC (19,20).
The FISH results showed that the copy numbers of
centromeres in chromosome 3 were significantly higher than those of
chromosome 17 (P=0.0001). The results may be associated with the
lifestyle of the Kazakh ethnic group, such as long-term excessive
consumption of smoked meat, fermented foods, alcohol and tobacco,
spicy foods, and lack of vegetables and foods rich in vitamins
(1,20). In the future, the most sensitive
probes should be selected to improve the early diagnosis of EC
using molecular pathological techniques (9,10,21).
In this study, centromeres of chromosome 3 and 17
copy numbers and degree of aneuploidy were suggested to be
correlated with the grade of tumor malignancy. These observations
should be proven in future studies.
In conclusion, FISH technology is more sensitive
than conventional pathology using biopsy specimens. Therefore,
using an ancillary FISH test during the pathological diagnosis of
cases with moderate or severe dysplasia may effectively improve the
early diagnosis of EC. In addition, the centromeres of the
chromosome 3 probe may be the most sensitive probe diagnostically
in Kazakh patients with EC.
Acknowledgements
This study was supported by a grant from the Chinese
postdoctoral fund of Xinjiang Medical University (20080-3014) and
the Xinjiang Uygur Autonomous Region key disciplines Fund. Thanks
are extended to the Department of Hematology, First Affiliated
Hospital of Xinjiang Medical University for the technical support.
We give thanks to Professor J. Patrick Barron, Professor and
Chairman, Department of International Medical Communications, Tokyo
Medical University for his review of this manuscript.
Abbreviations:
|
FISH
|
fluorescence in situ
hybridization
|
|
EC
|
esophageal cancer
|
|
SSC
|
standard saline citrate
|
References
|
1
|
Zhang YM: Distribution of esophageal
cancer in the Xinjiang. XMU Med. 11:139–145. 1988.
|
|
2
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Krasna MJ: Multimodality therapy for
esophageal cancer. Oncology. 24:1134–1138. 2010.PubMed/NCBI
|
|
4
|
Odze RD: Barrett esophagus: histology and
pathology for the clinician. Nat Rev Gastroenterol Hepatol.
6:478–490. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yerian L: Histology of metaplasia and
dysplasia in Barrett's esophagus. Surg Oncol Clin N Am. 18:411–422.
2009. View Article : Google Scholar
|
|
6
|
Duesberg P, Li R, Rasnick D, Rausch C,
Willer A, Kraemer A, Yerganian G and Hehlmann R: Aneuploidy
precedes and segregates with chemical carcinogenesis. Cancer Genet
Cytogenet. 119:83–93. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rajagopalan H and Lengauer C: Aneuploidy
and cancer. Nature. 432:338–341. 2004. View Article : Google Scholar
|
|
8
|
Halling KC and Kipp BR: Fluorescence in
situ hybridization in diagnostic cytology. Hum Pathol.
38:1137–1144. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fritcher EG, Brankley SM, Kipp BR, Voss
JS, Campion MB, Morrison LE, Legator MS, Lutzke LS, Wang KK, Sebo
TJ and Halling KC: A comparison of conventional cytology, DNA
ploidy analysis, and fluorescence in situ hybridization for the
detection of dysplasia and adenocarcinoma in patients with
Barrett's esophagus. Hum Pathol. 39:1128–1135. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Idiris A, Madiniyet N, Xie HZ, Hadeti B,
Zhang HY, Zhang Z, Ilyar S, Zhang CM, Zhang LW and Wen H: Genetic
diagnosis of patients with esophageal cancer using FISH. Oncol
Lett. 1:809–814. 2010.PubMed/NCBI
|
|
11
|
Murphy CG and Fornier M: HER2-positive
breast cancer: beyond Trastuzumab. Oncology. 24:410–415.
2010.PubMed/NCBI
|
|
12
|
Han QY, Shun H, Yu PW, Xiao CW, Ya LH and
Xin X: Application of multicolor fluorescence in situ hybridization
to early diagnosis of esophageal squamous cell carcinoma. Chin J
Can. 27:1137–1143. 2008.PubMed/NCBI
|
|
13
|
Yang Y, Fruehauf J, Xiang S and Li CJ:
Genomic instability in precancerous lesions before inactivation of
tumor suppressors p53 and APC in patients. Cell Cycle.
13:1443–1447. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nakamura H, Aute I, Kawasaki N, Taguchi M,
Ohira T and Kato H: Quantitative detection of lung cancer cells by
fluorescence in situ hybridization: comparison with conventional
cytology. Chest. 128:906–911. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dey P: Aneuploidy and malignancy: an
unsolved equation. J Clin Pathol. 57:1245–1249. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Albertson DG, Collins C, McCormick F and
Gray JW: Chromosome aberrations in solid tumors. Nat Genet.
34:369–76. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Falk GW, Skacel M, Gramlich TL, Casey G,
Goldblum JR and Tubbs RR: Fluorescence in site hybridization of
cytologic specimens from Barrett's esophagus: a pilot feasibility
study. Gastrointest Endosc. 60:280–284. 2004. View Article : Google Scholar
|
|
18
|
Fahmy M, Skacel M, Gramlich TL, Brainard
JA, Rice TW, Goldblum JR, Connor JT, Casey G, Legator MS, Tubbs RR
and Falk GW: Chromosomal gains and genomic loss of p53 and p16
genes in Barrett's esophagus detected by fluorescence in situ
hybridization of cytology specimens. Mod Pathol. 17:588–596. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fiegl M, Massoner A, Haun M, Sturm W,
Kaufmann H, Hack R, Krugmann J, Fritzer-Szekeres M, Grünewald K and
Gastl G: Sensitive detection of tumour cells in effusions by
combining cytology and fluorescence in situ hybridisation (FISH).
Br J Cancer. 91:558–563. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bahmanyar S and Ye W: Dietary patterns and
risk of squamous-cell carcinoma and adenocarcinoma of the esophagus
and adenocarcinoma of the gastric cardia: a population based
case-control study in Sweden. Nutr Cancer. 54:171–178. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Delektorskaya VV, Chemeris GY, Zavalishina
LE, Ryazantseva AA, Grigorchuk AY, Kononets PV and Davydov MI:
Squamous cell carcinoma of the esophagus: evaluation of the status
of epidermal growth factor receptors (EGFR and HER-2) by
immunohistochemistry and in situ hybridization. Bull Exp Biol Med.
149:615–620. 2010. View Article : Google Scholar : PubMed/NCBI
|