Introduction
The MIR125A gene is located on chromosome 19
in a cluster with MIR99B and MIR7E. Mature
hsa-miR-125a interacts with a conserved 8-nt binding element,
CUCAGGGA, located within the proximal 3′-UTR of ERBB2 and is
able to bifunctionally mediate ERBB2 transcript decay and
translational inhibition (1).
Besides ERBB2, HuR, Rock-1, KLF13 and
ARID3B mRNA levels are also diminished by hsa-miR-125a
(2,3). Decreased levels of hsa-miR-125a have
been detected in breast cancer and gastric cancer (4,5).
Furthermore, increased levels of miR-125 in breast cancer cells
inhibit cell growth via the suppression of cell proliferation and
by promotion of apoptosis (6). In
the nucleus, pri-miR-125a is transformed into mature hsa-miR-125a
by the Drosha system (7). This
process is obstructed by estrogen receptor (ER) α (8). Germline mutations in BRCA1/2,
ATM, PTEN and CHEK2 are common in familial
breast cancer, but they explain only one-quarter of the familial
risk (9). Thus, it is likely that
there are a number of unidentified genes which contain loci
encoding miRNAs that confer susceptibility to breast cancer. An SNP
(rs12975333) has been observed in the hsa-miR-125a miRNA precursor
sequence, which blocks the pri- to pre-miR-125a processing step
(10).
In our study, we hypothesized that three other known
genetic variants of pri-miR-125a (rs10404453, rs12976445 and
rs143525573) should correlate with levels of mature hsa-miR-125a in
breast cancer cells. Consequently, ERBB2 mRNA levels would be
increased in breast tumors with these genetic variants, suggesting
that genetic variants that influence hsa-miR-125a expression have
potential as genetic markers of breast cancer. In particular, these
variants may have predictive value in designing treatment with
drugs against HER2.
Materials and methods
Patients and samples
Tissue samples were obtained from 26 Polish patients
undergoing surgery for breast cancer in the Department of Surgery,
Chair of Oncology of Poznan University of Medical Sciences (PUMS;
see Table I for patient and tumor
characteristics). No preoperative radiotherapy or chemotherapy was
used. The study protocol was approved by the bioethics board of
PUMS. Tumor tissue and blood samples for comprehensive experiments
were collected after obtaining written informed consent from all
participants.
 | Table I.Groups of patients classified in the
TNM system and ER, PR and HER2 expression receptors. |
Table I.
Groups of patients classified in the
TNM system and ER, PR and HER2 expression receptors.
| Type | Number of
patients | Percentage |
|---|
| G1 | 5 | 19.2 |
| G2 | 12 | 46.2 |
| G3 | 9 | 34.6 |
| pT1 | 13 | 50.0 |
| pT2 | 12 | 46.2 |
| pT3 | 0 | 0.0 |
| pT4 | 1 | 3.8 |
| pN0 | 14 | 53.8 |
| pN1 | 11 | 42.3 |
| pN2 | 1 | 3.8 |
| pN3 | 0 | 0.0 |
| ER (−) | 8 | 30.8 |
| ER (+) | 18 | 69.2 |
| PR (−) | 9 | 34.6 |
| PR (+) | 17 | 65.4 |
| HER2
0/1+ | 16 | 61.5 |
| HER2
2+ | 4 | 15.4 |
| HER2
3+ | 6 | 23.1 |
Immediately after surgery, the tissue samples were
stored in liquid nitrogen. Formalin-fixed paraffin-embedded (FFPE)
tissue samples of breast tumors were collected separately.
Subsequently, the patients’ cases were classified according to the
TNM classification of tumors (7th edition). Prior to RNA
extraction, corresponding hematoxylin and eosin (HE) stained tumor
tissue sections were made and the percentage of cancer cells in the
sections was evaluated using a light microscope (Olympus BX41,
Olympus, Tokyo, Japan). In the present study, the average
percentage of tumor cells per section was 76%.
Blood sampling and measurements
Blood samples were obtained by puncture of the
antecubital vein, in the Department of Surgery, Chair of Oncology
at PUMS.
RNA and DNA extraction
miRNA and mRNA were extracted from frozen tissue
using the mirVana™ miRNA Isolation kit (Life Technologies,
Carlsbad, CA, USA). Total RNA and DNA from the paraffin-embedded
tissues was extracted using the RecoverAll™ Total Nucleic Acid
Isolation kit (Life Technologies).
DNA from frozen tissue and blood was extracted using
GenElute™ Mammalian Genomic DNA Miniprep kit (Sigma-Aldrich, St.
Luis, MO, USA). The quantity of obtained nucleic acid was assessed
using a BioPhotometer™ (Eppendorf, Hamburg, Germany).
PCR, sequencing and restriction
analysis
DNA specimens were amplified using standard PCR
protocols. The PCR primers corresponding to pri-pre-miR-125a used
for MIR125A sequencing were: 5′-TTTTGGTCTTTCTGTCTCTGG-3′ and
5′-TGGAGGAAGGGTATGAGGAGT-3′. The PCR products were purified with
the Gel-out purification kit (A&A Biotechnology, Gdynia,
Poland) and sequenced at the DNA Sequencing and Oligonucleotide
Synthesis Laboratory of the Institute of Biochemistry and
Biophysics of the Polish Academy of Sciences (Warsaw, Poland). The
sequencing results were analyzed using BioEdit Sequence Alignment
Editor. In addition to sequencing, the SNP (rs12976445) was
genotyped using the restriction enzyme BaeGI (New England
Biolabs, Ipswich, MA, USA). All results were in agreement.
Real-time PCR
To evaluate the MIR125A expression level,
TaqMan microRNA Assays (Life Technologies) for real-time RT-PCR
were used. Similar assays were also used for hsa-miR-206,
hsa-miR-125b, hsa-miR-17 and hsa-miR-27b; U6 RNA was used as a
reference gene. All samples were reverse transcribed using the
TaqMan MicroRNA Reverse Transcription kit and specific starters
from the TaqMan microRNA Assay. TaqMan Universal PCR Master Mix and
specific primers from TaqMan microRNA Assays were used to quantify
the samples in a Roche (Indianapolis, IN, USA) LC 480 cycler. The
relative amounts of all miRNAs were calculated using standard
curves and compared as ratios using the U6 reference gene.
ERBB2, ESR1 and PGR mRNA levels
were analyzed by reverse transcription (Life Technologies) and
TaqMan real-time PCR (Roche). HMBS and POL2 were used
as reference genes.
Statistical analysis
Statistical analysis was conducted using Instat
software. P≤0.05 was considered to indicate statistically
significant differences. A t-test was used to compare the
differences in mean expression levels between the groups of samples
from the real-time RT-PCR experiment. The mean of the
log10 of the ratio between target gene expression levels
and reference gene expression levels was also calculated.
Results
Frequency of the rs12976445 variant
A total of 26 surgically removed breast tumors were
analyzed with the aim of identifying genetic polymorphisms in the
gene encoding hsa-miR-125a. Of the samples, 21 were congealed in
liquid nitrogen and 5 were paraffin-embedded archival samples. The
group of 26 samples was analyzed routinely using histopathological
methods. The patients were classified by the TNM system and the
samples were also grouped into ER, PR and HER positive and negative
cases. The results are presented in Table I. DNA extracted from the samples was
amplified using primers spanning pri-, pre- and mature
hsa-miR-125a. There are four known SNPs in the amplified fragment
of MIR125A: rs12976445, rs10404453, rs12975333 and
rs143525573 (11). Subsequently all
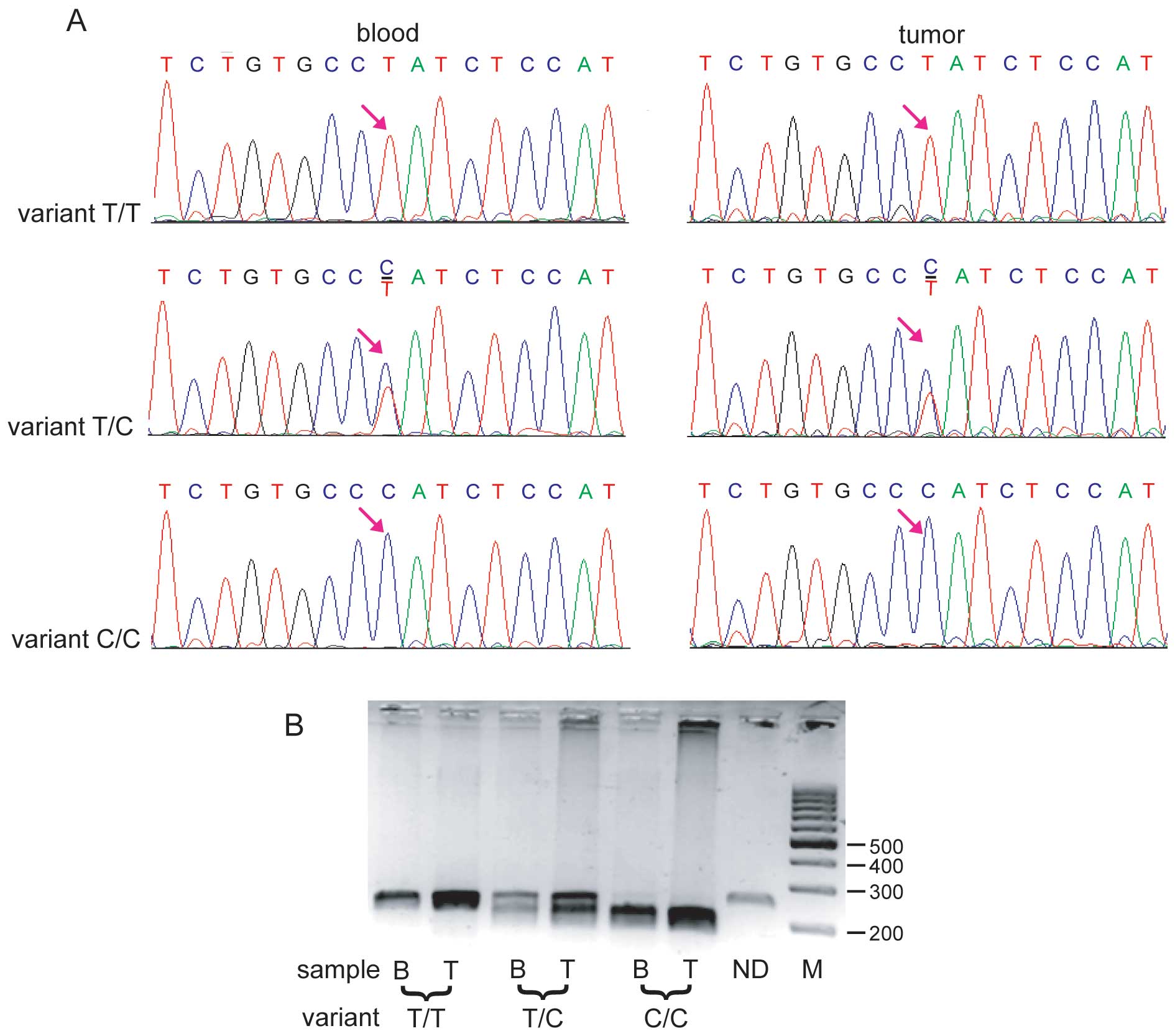
amplicons from the tumor samples were sequenced. A single
nucleotide change from T to C (variant rs12976445) was identified
in the sequence of pri-miR-125 in the breast cancer patients (15
nucleotides downstream from the start of pri-miR-125a, 54
nucleotides upstream from the start of pre-miR-125, 68 nucleotides
upstream of the miR-125a-5p). The three other variants were not
present in these samples.
To confirm the results of the MIR125A
sequencing and to establish the frequency of the variant, the DNA
extracted from the congealed tumors and paraffin-embedded tissues
was amplified using the same pair of primers as for the sequencing.
Subsequently the products were digested using the restriction
enzyme BaeGI (GKGCM^C). The restriction analysis revealed
that 8 out of 26 patients were T/T homozygotes (30.8%), 15 (57.7%)
were T/C heterozygotes and 3 were C/C homozygotes (11.5%; Fig. 1). For 14 tumors, corresponding blood
samples were available and the same genetic variant was
investigated using restriction analysis with BaeGI (Fig. 1B). The results revealed exactly the
same distribution of variants in the blood as in the corresponding
tumors.
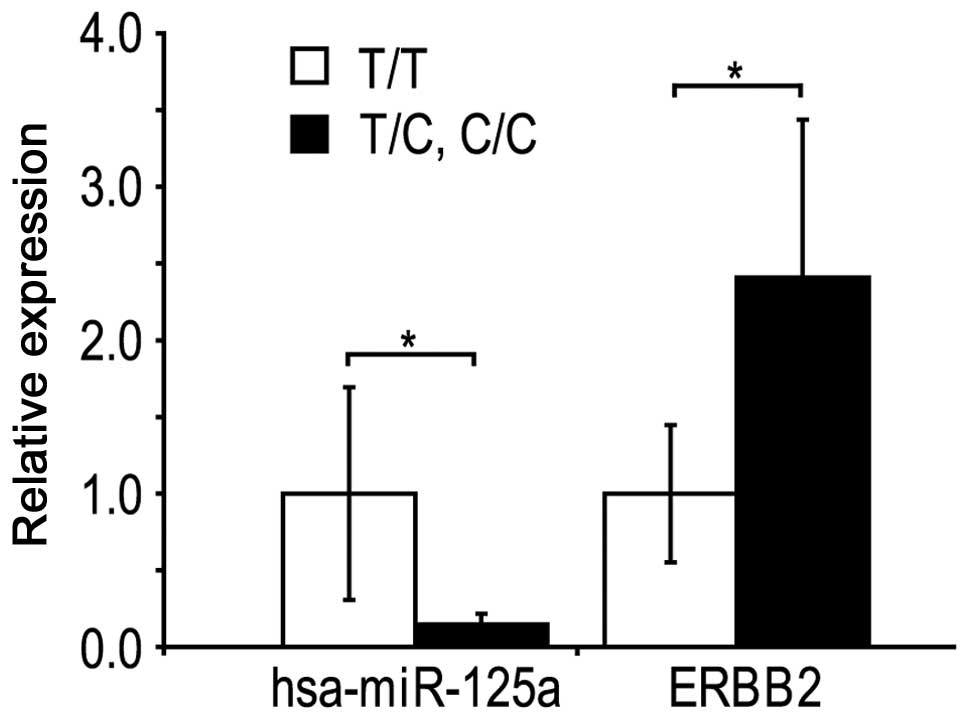
MiRNA level
The expression of MIR125A was analyzed in the
tumours using real-time PCR (Fig.
2). The hsa-miR-125a level was decreased by 85% (P=0.024) in
the samples with the variants CC and CT, compared with the TT
variant. When the T homozygous variant, heterozygotoes and the C
homozygotes were compared, a gradual decrease of hsa-miR-125a
levels was observed. In the present samples, no correlation was
observed between the expression of hsa-miR-125b, hsa-miR-27,
hsa-miR-17-5 or hsa-miR-206 and the rs12976445 variant. mRNA of the
ERBB2 gene was increased by 2.4-fold (P=0.044) in the tumor
samples with the variant C/C and C/T MIR125A variants,
compared with the T/T variant. ESR1 and PGR mRNA
levels did not correlate with the rs12976445 variant.
These findings suggest that the ERBB2 mRNA
increase may be caused by diminished levels of inhibitory
hsa-miR-125a.
Discussion
The present study shows that in breast cancers the
rs12976445 variant (in the T/C and C/C types as opposed to the T/T
type) located in pri-hsa-miR-125a correlated with lower levels of
hsa-miR-125a and that the mRNA levels of ERBB2 were
increased. ERBB2 encodes the HER2 breast cancer marker gene.
The normal HER2 level, designated 0, may be increased and so is
measured by histopathological methods which allocate results to a
scale from 1 to 3. The scale is used to test whether it may be
possible to treat the patient with trastuzumab (12).
Deregulation of ERBB2 and ERBB3,
singly or in combination, is able to induce malignant
transformation. Of all human breast cancers, ∼25% are associated
with amplification and overexpression of ERBB2. In
particular, overexpression of ERBB2 drives cell survival,
proliferation, motility and the invasion mechanisms characteristic
of this aggressive form of human breast cancer (1).
Decreased levels of hsa-miR-125a were observed in
correlation with the rs12976445 variant. Similar results were
described by Hu et al in correlation with recurrent
pregnancy loss (13). For the first
time, the present study shows the correlation between the
rs12976445 variant (T/C and C/C) and miR-125 levels in breast
tumors. The rs12976445 variant is located in the pri-miR-125a
sequence and disrupts a potential GATA-1 site. This suggests that
lower hsa-miR-125a levels in the T/C and C/C variants, compared
with the T/T variant, may be explained by diminished MIR125A
transcription. According to the data of Hu et al, the
fragment may potentially interact with undetermined nuclear
proteins. The mechanism of the rs12976445 variant’s effect on the
level of MIR125A remains undetermined and it is unknown
whether it is the transcriptional regulation effect, maturation of
hsa-miR-125a effect or transport of hsa-miR-125a. The obstructed
processing hypothesis is supported by the observation that
Drosha-mediated maturation of pri-miR-125a is inhibited by
p68/p72-dependent mechanisms upon stimulation of ERα (8). We suggest that the rs12976445 variant
may also reduce Drosha-mediated maturation, by disrupting the
Drosha interaction with pri-miRNA. No correlation was observed
between rs12976445 (T/T vs. T/C and C/C variants) and ESR1
mRNA levels in the present samples, however it may be that
estrogens increase the effect of the variant, resulting in a
hsa-miR-125a drop. It may be a matter of considerable interest to
measure the estradiol levels in tumor samples.
There are three other known SNP variants in the
amplified fragment containing has-miR-125a: rs10404453, rs12975333
and rs143525573 (11). None of
these variants were present in our samples. As determined by Li
et al, the rs12975333 (variant T/C) is rare and is observed
in only 8% of investigated breast cancers while it was absent in
controls (11). In the present
study rs12976445 (variants T/C and C/C) was observed in 69.2% of
tumors and in all the corresponding blood samples, where available.
From this we deduced that rs12976445 is not a somatic tumor-origin
mutation (11). We concluded that
the SNP rs12976445 in pri-miRNA may be an example of robustness in
miRNA evolution (14) since the
frequent genetic variant of this sequence does not cause marked
phenotypic effects. Duan et al reported that rs12975333, in
a +8-bp hairpin region of mature hsa-miR-125a, alters the
processing of miRNA (10). The
hairpin region of miRNAs appears to be more conservative than the
pri-miRNA flanking regions and, therefore, no rs12975333 variants
of hsa-mir-125a was detected in the present small-scale study
(n=26) and a very low frequency was described by Li et al in
larger group (n=72) (11).
The correlation between rs12976445 and the risk of
breast cancer remains to be confirmed. There may be several
possible explanations for the robustness of the cell in spite of
deregulated expression of MIR125A. The first explanation
originates from observations of miRNA gene knockouts. Regardless of
the large number of target genes predicted to be affected by miRNA,
a complete loss of function in gene-knockout experiments for
individual miRNAs has yielded no change in phenotype (15). The functional redundancy of miRNA is
postulated as a possible explanation for the phenomenon (15). The second explanation is based on
the hypothesis that certain genetic variations associated with the
rs12976445 variant, may compensate for the lower expression of
MIR125A.
Changes in the expression pattern of an miRNA
generate novel sets of signals, leading to the formation of novel
regulatory circuits (16). At
present, the complete regulatory circuit, called a feedforward loop
(FFL) and involving hsa-miR-125a, ERBB2 and a third unknown
component closing the loop, concomitantly regulated by ERBB2
and regulating miR125, has yet to be described (15). An elevated level of ERBB2
mRNA in the T/C and C/C variants in comparison with the T/T variant
was observed in the present study. This effect may be a consequence
of a decrease in the level of hsa-miR-125a, which promotes ERBB2
mRNA decay and inhibits translation (1). The increase of ERBB2 transcript
in tumors with the T/C and C/C variants was specific since neither
a positive or a negative correlation was observed for the
ESR1 and PGR transcripts. The patients with the T/C
and C/C variants may potentially respond better to trastuzumab,
though this response would need to be verified further in future
studies. The level of the ERBB2 transcript correlated with
the HER2 protein levels measured by immunohistochemical methods.
Independently of the unconfirmed and possibly low, correlation
between the rs12976445 variant and breast cancer risk, an awareness
of miR-SNPs in the patient genome may allow oncologists to predict
whether the patient is likely to respond to anti-HER2
chemotherapeutic drugs (17).
Acknowledgements
The authors would like to thank Ms.
Beata Raczak and Bogumiła Ratajczak M.Sc. for their indispensable
assistance during preparation of this manuscript. The study was
supported by a grant from the Polish Ministry of Science and Higher
Education (no. N N403 598538).
References
|
1.
|
Scott GK, Goga A, Bhaumik D, Berger CE,
Sullivan CS and Benz CC: Coordinate suppression of ERBB2 and ERBB3
by enforced expression of micro-RNA miR-125a or miR-125b. J Biol
Chem. 282:1479–1486. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Zhao X, Tang Y, Qu B, et al: MicroRNA-125a
contributes to elevated inflammatory chemokine RANTES levels via
targeting KLF13 in systemic lupus erythematosus. Arthritis Rheum.
62:3425–3435. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Cowden Dahl KD, Dahl R, Kruichak JN and
Hudson LG: The epidermal growth factor receptor responsive miR-125a
represses mesenchymal morphology in ovarian cancer cells.
Neoplasia. 11:1208–1215. 2009.PubMed/NCBI
|
|
4.
|
Nishida N, Mimori K, Fabbri M, et al:
MicroRNA-125a-5p is an independent prognostic factor in gastric
cancer and inhibits the proliferation of human gastric cancer cells
in combination with trastuzumab. Clin Cancer Res. 17:2725–2733.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Guo X, Wu Y and Hartley RS: MicroRNA-125a
represses cell growth by targeting HuR in breast cancer. RNA Biol.
6:575–583. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Kusenda B, Mraz M, Mayer J and Pospisilova
S: MicroRNA biogenesis, functionality and cancer relevance. Biomed
Pap Med Fac Univ Palacky Olomouc Czech Repub. 150:205–215. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Yamagata K, Fujiyama S, Ito S, et al:
Maturation of microRNA is hormonally regulated by a nuclear
receptor. Mol Cell. 36:340–347. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
van der Groep P, van der Wall E and van
Diest PJ: Pathology of hereditary breast cancer. Cell Oncol
(Dordr). 34:71–88. 2011.
|
|
10.
|
Duan R, Pak C and Jin P: Single nucleotide
polymorphism associated with mature miR-125a alters the processing
of pri-miRNA. Hum Mol Genet. 16:1124–1131. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Li W, Duan R, Kooy F, Sherman SL, Zhou W
and Jin P: Germline mutation of microRNA-125a is associated with
breast cancer. J Med Genet. 46:358–360. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Fiszman GL and Jasnis MA: Molecular
mechanisms of trastuzumab resistance in HER2 overexpressing breast
cancer. Int J Breast Cancer. 2011:3521822011. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Hu Y, Liu CM, Qi L, et al: Two common SNPs
in pri-miR-125a alter the mature miRNA expression and associate
with recurrent pregnancy loss in a Han-Chinese population. RNA
Biol. 8:861–872. 2011. View Article : Google Scholar
|
|
14.
|
Borenstein E and Ruppin E: Direct
evolution of genetic robustness in microRNA. Proc Natl Acad Sci U S
A. 103:6593–6598. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Ebert MS and Sharp PA: Roles for microRNAs
in conferring robustness to biological processes. Cell.
149:515–524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Berezikov E: Evolution of microRNA
diversity and regulation in animals. Nat Rev Genet. 12:846–860.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Zheng T, Wang J, Chen X and Liu L: Role of
microRNA in anti-cancer drug resistance. Int J Cancer. 126:2–10.
2010. View Article : Google Scholar
|