Introduction
Renal cell carcinoma (RCC) is the most common type
of cancer of the kidney. Metastasis frequently becomes evident at
the time of diagnosis or patients develop distant metastases
following removal of the primary tumor. The treatment of
metastatic, locally unresectable RCCs remains a challenge for
urologists (1). There is a great
need to understand the basic biology of RCC and develop better
therapeutic options.
The CCN3 (nephroblastoma overexpressed, NOV) gene
belongs to the CCN family of genes, which has five additional
members: cystein-rich protein 61 (Cyr61), connective tissue growth
factor (CTGF), Wnt-1-induced secreted protein (WISP)-1, WISP-2 and
WISP-3. CCN proteins are involved in fundamental biological
processes such as cell proliferation, attachment, migration,
differentiation, wound healing, angiogenesis and tumorigenesis
(2).
The CCN3 protein is detectable at varying levels in
the kidney, muscle, cartilage, brain, lung, ovary and heart
(3–5). The functions of CCN3 protein among
these different tissues are, however, different. CCN3 was
originally described as antiproliferative (3), and its expression was associated with
differentiation of Wilms' tumor (4), rhabdomyosarcomas (6), neuroblastomas (7), cartilaginous tumors (8), adrenocortical tumors (9) and with inhibition of the growth and
decrease in tumorigenicity of several tumor cell lines including
glioblastoma (10), choriocarcinoma
(11) and Ewing's sarcoma (12). Furthermore, CCN3 expression was
correlated with the increased proliferative index of 3T3 fibroblast
(13) and tissue samples of the
prostate (14). Although CCN3
reduced the growth rate of Ewing's sarcoma transfectants ex
vivo, CCN3 expression was associated with poor prognosis and
shown to increase cell motility, resulting in enhanced metastatic
potential (6,12).
In the present study, we confirm that the expression
of CCN3 protein is decreased in clear cell RCC tissues compared to
normal kidney tissues. To further investigate the role of CCN3 in
the biology of clear cell RCC cells, we transfected 786-O cells
with the CCN3 gene. The effects of CCN3 on cell proliferation,
adhesion, migration and invasion are described.
Materials and methods
Human tissues
RCC tissues were obtained from 32 patients with
clear cell RCC (median age, 65 years; range, 45–83 years) treated
by radical nephrectomy at Shandong Provincial Hospital (Jinan,
China). Samples were fixed in 4% buffered formalin, embedded in
paraffin, and routinely stained with hematoxylin and eosin
(H&E). Corresponding normal kidney tissues were also obtained
from these patients. Our study was approved by the Ethics Committee
of the Shandong Provincial Hospital.
Immunohistochemistry
Formalin-fixed, paraffin-embedded RCC and
corresponding normal kidney tissue sections (5 μm) were
deparaffinized with xylene, rehydrated and washed with
phosphate-buffered saline (PBS). Endogenous peroxidase activity was
blocked with 3% H2O2 for 20 min. Antigen
retrieval was performed in a 0.01 M sodium citrate buffer in a
microwave oven. Slides were blocked with 10% bovine serum for 1 h,
followed by incubation with primary antibodies at 4°C overnight.
The primary antibodies used were: rabbit polyclonal to CCN3 (Abcam,
Cambridge, UK, 1/2000) and rabbit polyclonal to Ki67 (Abcam, 1/50).
After washing in 0.01 M PBS, the slides were incubated with
biotinylated goat anti-rabbit IgG (1:50) for 30 min, then stained
with 0.03% diaminobenzidine for 10 min. Slides were counterstained
with hematoxylin. The stain was evaluated by two independent
assessors. The staining intensity for CCN3 was expressed as
follows: −, no stain; +, weak; ++, moderate; and +++, strong. After
staining with Ki67 antibody, cells were enumerated by counting the
number of positive cells in 10 fields over the total cells at ×400
magnification.
Cell lines and transfections
The RCC cell line (786-O cell line) was purchased
from the cell bank of the Chinese Academy of Sciences. We
constructed a plasmid based on the PCMV-NOVH plasmid which contains
human CCN3 under the control of a constitutive or inducible
promoter (kindly provided by Dr Cecile Martinerie, Hospital
Saint-Antoine, France). To monitor the transfection rate by
fluorescence microscopy, EGFP and Neo, which encode enhanced green
fluorescence protein and Neomycin resistance protein, respectively,
were inserted to make the construct (pEGFP-C1-CCN3), and the
plasmid was verified by sequencing. The 786-O cell line was
transfected with this vector and Lipofectamine 2000 (Invitrogen,
Carlsbad, CA, USA). A fluorescence microscope was used to monitor
the transfection rate. Stable transfectants were selected in G418
(800 μg/ml, Invitrogen) and characterized for CCN3 expression level
by real-time reverse transcriptase-polymerase chain reaction
(RT-PCR) analysis and western blotting, respectively. Among the
clones obtained, one line with significantly higher levels of CCN3
expression was selected and compared with the parental 786-O cell
line. Cells transfected with the empty vector pEGFP-C1 were used as
negative controls.
Real-time RT-PCR analysis
Quantitative real-time RT-PCR was carried out using
SYBR-Green according to the standard protocol (Roche Diagnostics
GmbH, Mannheim, Germany). Briefly, total RNA was isolated, then
cDNA was reverse transcribed from 1 μg of RNA by RT-PCR standard
methods. Real-time PCR reactions were carried out in triplicate on
an ABI 7500 machine using gene expression assays (Applied
Biosystems, Foster City, CA, USA). The real-time PCR primers for
CCN3 were 5′-GCATCTGCACGGCGGTAG-3′ (forward) and
5′-CACTGGAATTTGCAGCTTGG-3′ (reverse). PCR products were detected by
measuring the emitted fluorescence at the end of each reaction step
(reaction cycle). The threshold cycle (Ct) corresponds with the
cycle number required to detect a fluorescence signal above the
baseline. We used the housekeeping gene glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) as an endogenous control and the 786-O cell
line as a calibrator. A negative control without a cDNA template
was run with each assay. Data analysis was carried out using SDS
version 1.3.0.
Western blot analysis
To determine the expression of CCN3, proteins were
extracted from cells via suspension in RIPA buffer. Samples were
centrifuged at 10,000 × g at 4°C for 10 min and the supernatant was
used for analysis. The protein concentration was determined using
the Bradford protein method and the BCA protein assay kit (Sigma,
St Louis, MO, USA). Protein (50 μg) was loaded onto a pre-cast
Bis-Tris polyacrylamide gel (8%) and subsequently transferred to a
PVDF membrane, then blotted with anti-CCN3 rabbit polyclonal
antibodies, rabbit anti-actin (Abcam) and (HRP)-conjugated
secondary antibodies (Abcam). Immunoblots were visualized by
enhanced chemiluminescence (LAS4000).
Growth curve
Cells were trypsinized, and 1.5×104 cells
were plated in each well of 24-well plates containing RPMI-1640
with FBS at a concentration of 10%. Every 24 h, the medium was
removed, adherent cells were trypsinized, and total adherent cells
in each well were quantified using a hematocytometer. Cell counts
of 3 wells per time point per group were averaged and the data were
used to draw growth curves.
WST-1 assay for cell proliferation
For WST-1 proliferation assays (WST-1 cell
proliferation and cytotoxicity assay kit; Beyotime, Institute of
Biotechnology, Haimen, China), cells in an exponential phase of
growth were harvested and seeded in 96-well plates at a density of
2,000 cells per well, and cultured for 24, 48, and 72 h at 37°C
with 5% CO2. Following the culture, 10 μl WST-1 kit was
added into each well. Plates were then incubated for an additional
1 h period at 37°C. Subsequently, absorbance was determined in a
microplate reader (EL340 Bio-TEK instruments, Hopkinton, MA, USA)
at 450 nm.
Cell adhesion assays
The effect of CCN3 expression on the adhesive
properties of RCC cells was analyzed using different ECM
(extracellular matrix) substrates, such as laminin (LN) and
fibronectin (FN). Cell adhesion to laminin and fibronectin was
tested on tissue culture plates, which were coated overnight at 4°C
with 0.2 μg/cm2 human placental laminin and fibronectin
(Sigma), then blocked with 1% bovine serum albumin at room
temperature for 1 h. Cells (105/ml × 200 μl) were plated
and incubated at 37°C for 90 min, non-adherent cells were rinsed
off with RPMI, and adherent cells were fixed with methanol for 15
min and stained with 0.1% crystal violet. Plates were washed by
immersion in tap water and then dried, and absorbance at 550 nm was
measured following cell solubilization with a 50:50 mixture of 0.1
mol/l NaH2PO (pH 4.5) in 50% ethanol. The experiment was
performed in triplicate.
Transwell migration assay
Transwell chambers (Costar, Cambridge, UK) with 8-μm
pore size polycarbonate filter inserts for 24-well plates were used
according to the manual. In brief, 1×105 cells in
serum-free medium (RPMI-1640) were plated in the upper well of the
transwell chambers, whereas medium with 10% fetal bovine serum was
added to the lower well. Following a 12-h incubation, the cells on
the upper side of the inserts were removed using a cotton swab. The
inserts were fixed in methanol and stained with H&E. The number
of migrated cells attached to the other side of the insert was
counted under a light microscope in 8 random fields at a
magnification of ×200. The experiment was performed in
triplicate.
Matrigel invasion assay
Invasion assays were performed using Matrigel (BD
Bioscience, Bedford, MA, USA)-coated transwell chambers. Briefly,
the transwell inserts were pre-coated with 40 μl of 1 mg/ml
Matrigel matrix. Cells (1×105) in serum-free medium were
plated in the upper well, whereas medium with 10% fetal bovine
serum was added to the lower well. After incubating for 24 h, the
cells on the Matrigel side of the chambers were removed with a
cotton swab. The inserts were fixed in methanol and stained by
H&E staining. The number of invaded cells attached to the other
side of the inserts was counted under a light microscope in 8
random fields at a magnification of ×200. The experiment was
performed in triplicate.
Statistical analysis
Results are presented as the means ± SD. Statistical
analyses included the Student's t-test, performed using the
Statistical Package for Social Science (SPSS for Windows package
release 10.0; SPSS Inc., Chicago, IL, USA) as indicated in Results
and figure legends. P<0.05 was considered to represent a
statistically significant result.
Results
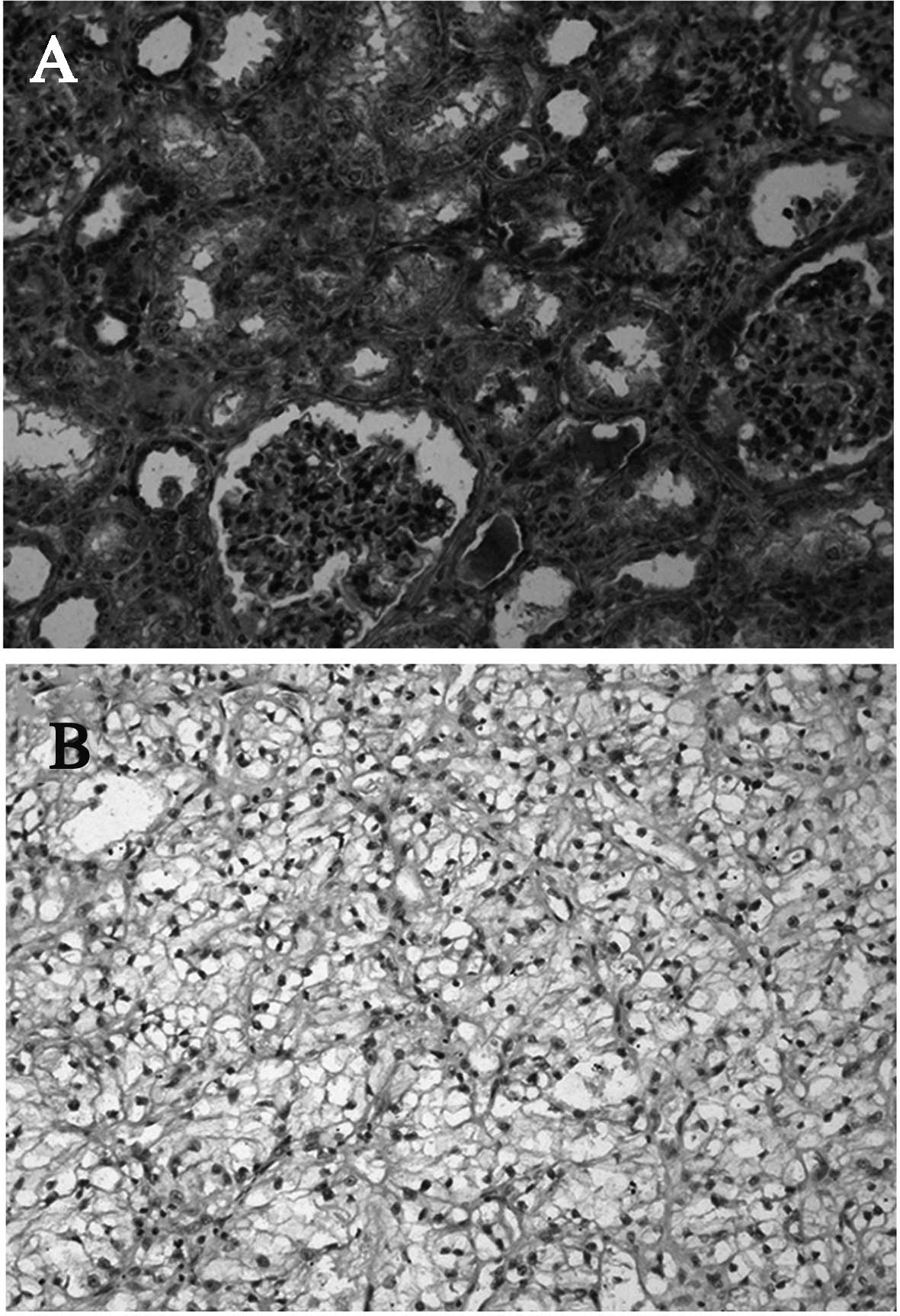
Immunohistochemistry
In all cases, the adjacent normal kidney tissue
demonstrated intense CCN3 immunostaining. CCN3 was localized in the
cytoplasm of the renal tubules and parietal epithelial cells of the
glomeruli (Fig. 1A). CCN3 staining
was observed in 21/32 (65.6%) of clear cell RCCs. The intensity of
CCN3 staining was generally less pronounced than in the
corresponding normal kidney tissue. The majority of cell membrane
staining was observed in tumor cell RCCs (Fig. 1B). The proportion of Ki67-positive
tumor cells was significantly higher in the CCN3-negative tumors
(3.1±1.7%) compared to the CCN3-positive tumors (1.7±1.2%)
(P<0.05) (Table I). However, no
obvious correlations were observed between the intensity of
staining for CCN3 and tumor stage or grade.
 | Table IImmunostaining intensity of CCN3 in
RCC tissues. |
Table I
Immunostaining intensity of CCN3 in
RCC tissues.
| No. | Gradea | T stageb | CCN3 | Ki67 (%) |
|---|
| 1 | 2 | 1 | + | 2.4 |
| 2 | 2 | 2 | − | 0.5 |
| 3 | 3 | 2 | − | 5.6 |
| 4 | 1 | 2 | + | 1.5 |
| 5 | 1 | 1 | − | 2.2 |
| 6 | 2 | 2 | + | 4.2 |
| 7 | 2 | 1 | + | 2.8 |
| 8 | 2 | 1 | ++ | 0.6 |
| 9 | 3 | 2 | − | 3.3 |
| 10 | 2 | 2 | + | 2.7 |
| 11 | 1 | 1 | ++ | 0.2 |
| 12 | 3 | 2 | + | 2.8 |
| 13 | 2 | 1 | − | 2.4 |
| 14 | 2 | 1 | + | 1.7 |
| 15 | 2 | 1 | − | 2.8 |
| 16 | 1 | 2 | + | 0.5 |
| 17 | 2 | 2 | ++ | 2.8 |
| 18 | 1 | 2 | + | 0.3 |
| 19 | 2 | 3 | + | 0.5 |
| 20 | 2 | 2 | ++ | 2.9 |
| 21 | 2 | 3 | +++ | 3.1 |
| 22 | 2 | 2 | − | 1.7 |
| 23 | 3 | 3 | ++ | 2.4 |
| 24 | 1 | 3 | + | 0.5 |
| 25 | 2 | 3 | ++ | 0.4 |
| 26 | 2 | 3 | − | 5.9 |
| 27 | 3 | 3 | − | 4.5 |
| 28 | 3 | 3 | ++ | 0.8 |
| 29 | 2 | 2 | ++ | 1.6 |
| 30 | 3 | 3 | +++ | 1.2 |
| 31 | 2 | 2 | − | 1.5 |
| 32 | 2 | 2 | − | 3.2 |
CCN3 expression changed by plasmid
transfection
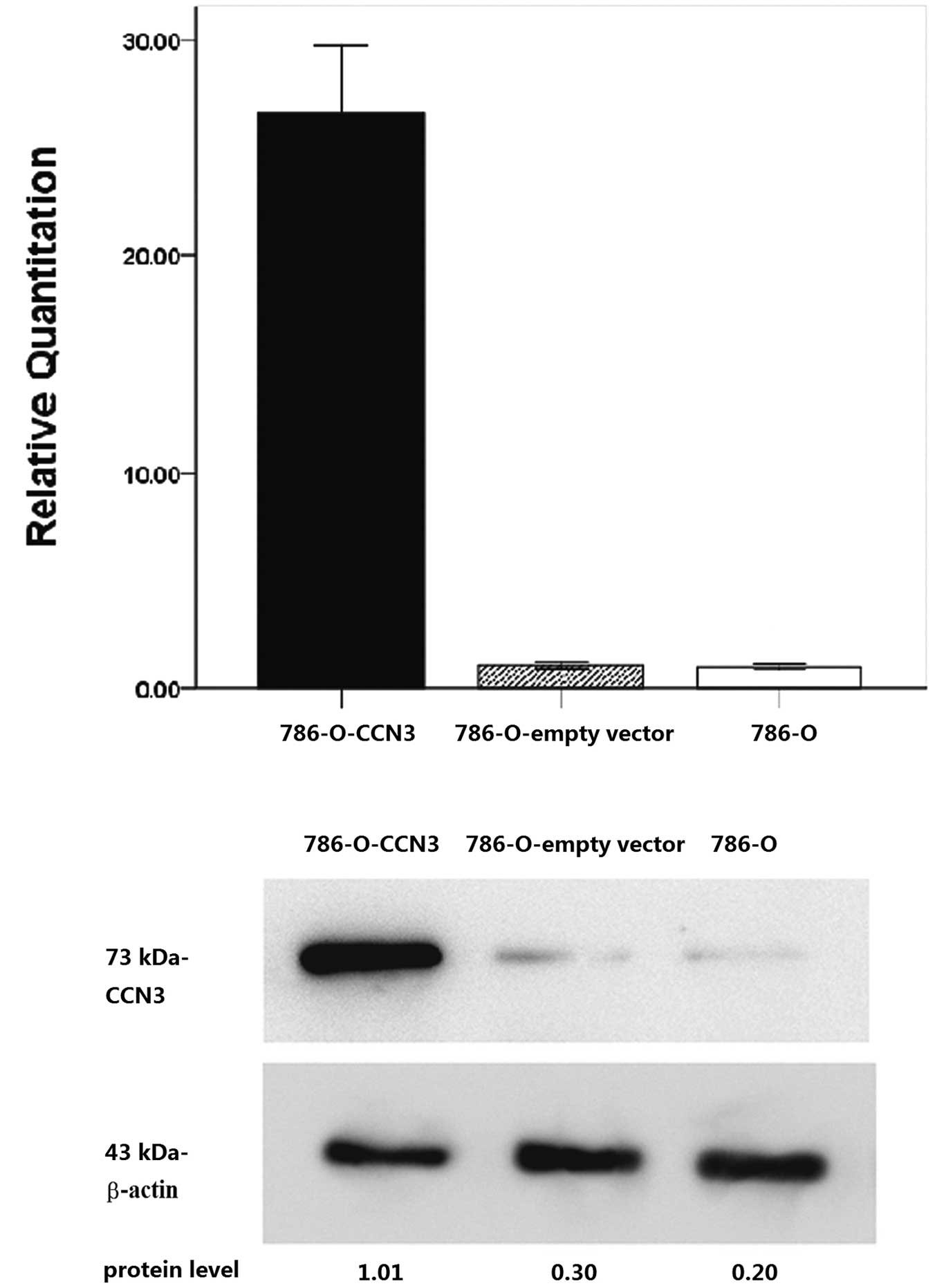
To determine the CCN3 expression level in 786-O
cells with CCN3 plasmid transfection, quantitative real-time RT-PCR
analysis and western blot analysis of CCN3 were measured in
CCN3-transfected 786-O cells, mock transfectants and parental 786-O
cells. The expression of CCN3 mRNA and protein was found to be
significantly increased in CCN3-transfected 786-O cells compared to
the parental cells (Fig. 2).
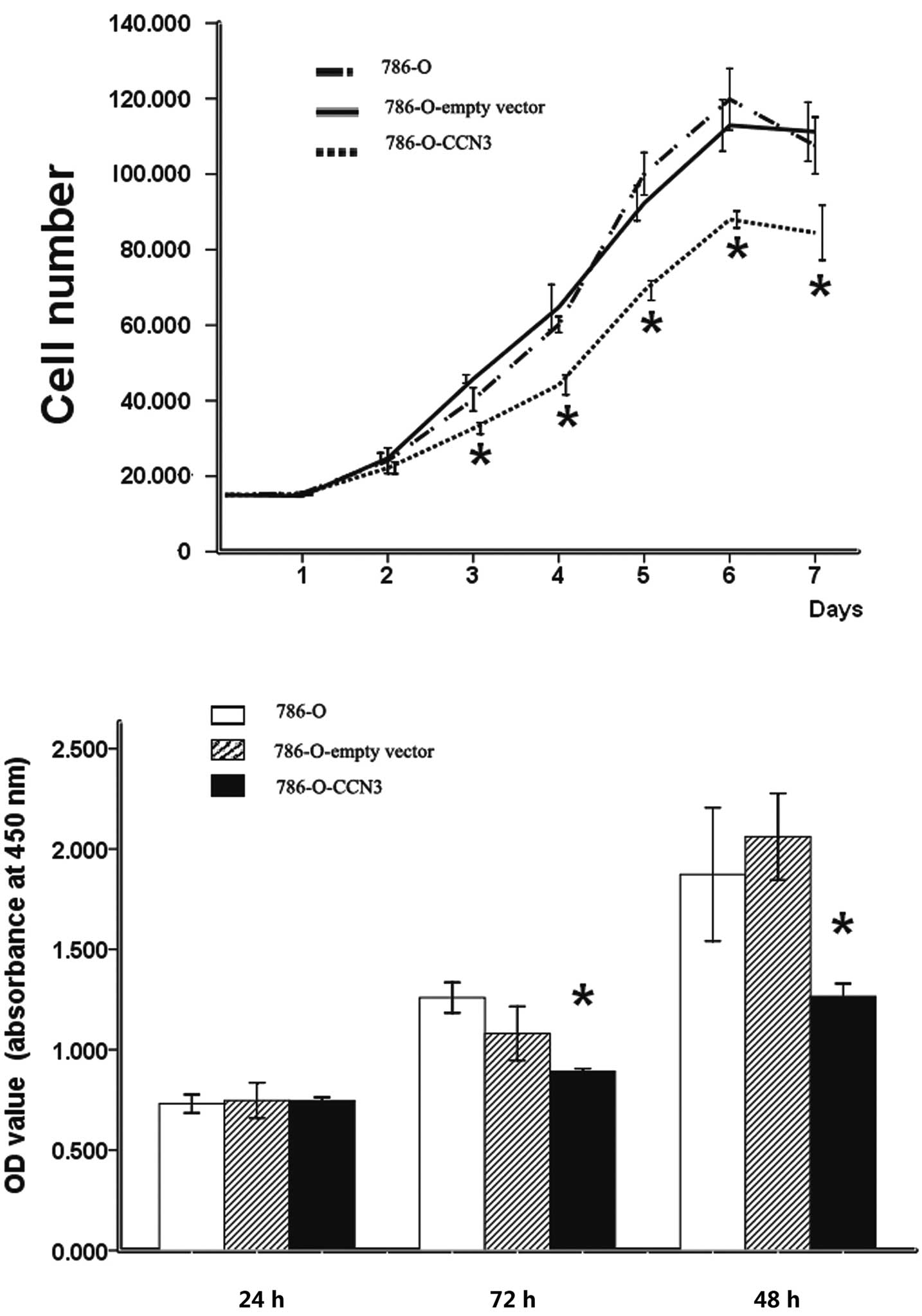
In vitro growth features of
CCN3-transfected clones
Our data show that a high expression of CCN3
significantly decreased the growth rate of CCN3-transfected 786-O
cells, with a 22.07±1.97 to 28.6±3.34% reduction at different time
points compared to their parental cells, evident after three days
of culture (P<0.05) (Fig.
3A).
The WST-1 proliferation assay was employed to test
the proliferation of CCN3-transfected cells. The proliferation of
CCN3-transfected cells was significantly decreased at 48 h
(P<0.05; percent inhibition, 17.45±1.23%) and 72 h (P<0.05;
percent inhibition, 36.12±6.92%), compared to the parental cells
(Fig. 3B).
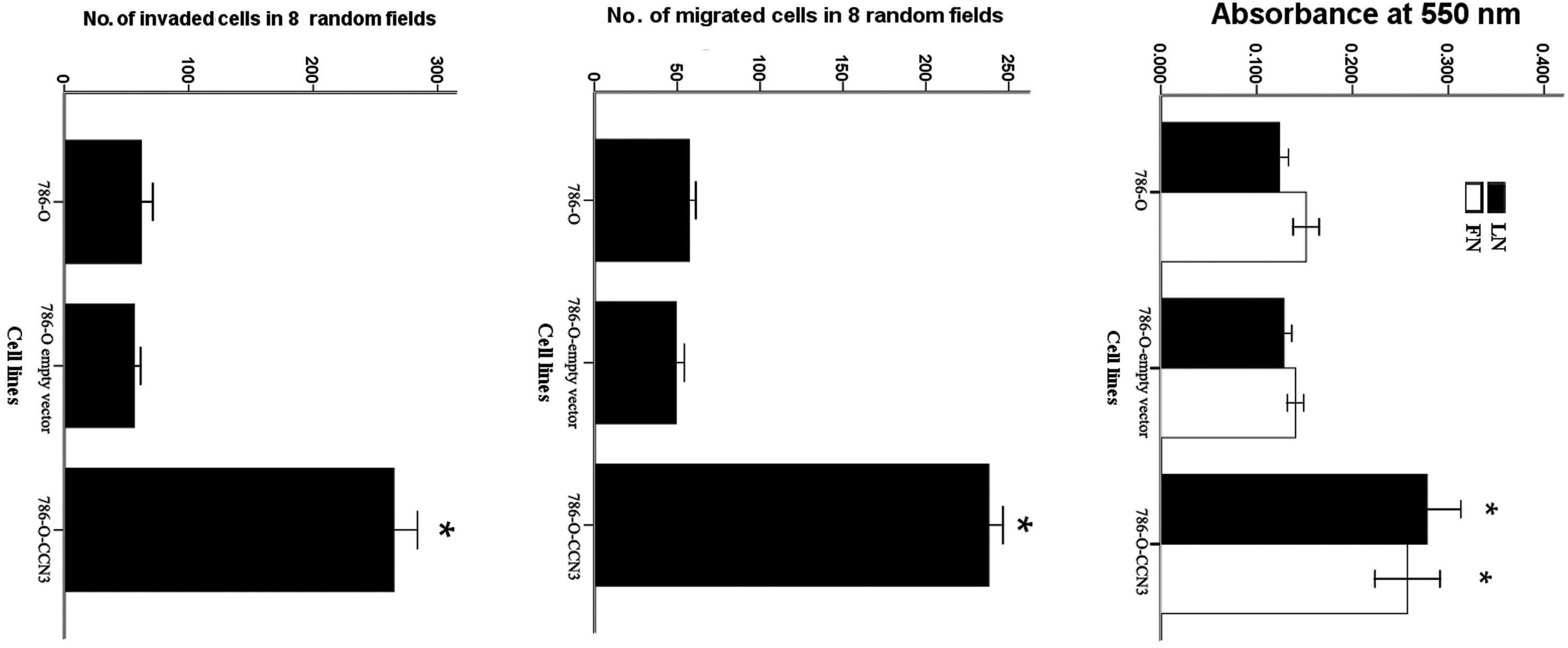
Effects of CCN3 expression on the
adhesive, migratory and invasive capabilities of 786-O cells
The effect of CCN3 expression on the adhesive
properties of 786-O cells was analyzed with respect to the
different ECM substrates, such as laminin and fibronectin. The
results indicate increased adhesion of CCN3-transfected cells to
fibronectin and laminin compared to the parental cells (Fig. 4A).
The migration of 786-O cells was measured in
transwell chambers. CCN3-transfected cells exhibited a significant
increase in cell migration ability compared to the parental cells
(P<0.01) (Fig. 4B).
We further tested the ability of 786-O cells to
invade through a reconstituted membrane barrier (Matrigel).
Similarly to the migration assay, CCN3-transfected cells exhibited
a significant increase in the invasive ability of these cells
compared to the parental cells (P<0.01, Fig. 4C).
Discussion
The expression of the CCN3 gene has been found to be
regulated temporally and spatially during chicken kidney
development since high amounts of CCN3 were detected in embryonic
kidneys, whereas much lower amounts were detected in the kidneys of
birds after hatching, suggesting that CCN3 is related to the
development of the kidney (15). In
human kidneys, CCN3 expression is implicated in the differentiation
of glomerular podocytes during normal nephrogenesis (4). High levels of CCN3 expression have
been associated with heterotypic tumor differentiation in human
nephroblastomas. Additionally, it has been suggested that
overexpression of the CCN3 protein may be associated with tumor
cell growth arrest (4). In the
present study, we observed that the expression of CCN3 was lower in
all clear cell RCC tissues compared to their corresponding normal
kidney tissues using immunohistochemical staining. This finding is
in line with previous findings obtained with quantitative RT-PCR in
a panel of sporadic RCC tissues (16). In addition, we observed that the
expression of CCN3 was inversely correlated with the Ki67 index in
clear cell RCC tissues. Therefore, CCN3 may play an inhibitory role
in clear cell RCC.
Since CCN3 was observed to be downregulated in clear
cell RCC tissues, we transfected a clear cell RCC cell line (786-O)
and obtained stable clones expressing the CCN3 gene to further
study the role of CCN3 in the biology of clear cell RCC. We showed
that the overexpression of CCN3 significantly inhibited the
proliferation of clear cell RCC cells in vitro. This finding
further indicated that CCN3 may be involved in the negative control
of clear cell RCC cell growth.
However, CCN3 overexpression was found to promote
clear cell RCC cell adhesion to ECM proteins, and clear cell RCC
cell migration and invasion were increased through Matrigel. Our
results are in line with previous findings obtained in Ewing's
sarcoma (12). The expression of
CCN3 by stable transfection significantly reduced the proliferative
activity of Ewing's sarcoma cells, while increasing cell adhesion
to ECM proteins and increased migration and invasion through
Matrigel. Moreover, in a study by Manara et al (6), the expression of CCN3 was only
observed in approximately 30% of cases of primary Ewing's sarcoma,
but it was associated with a significantly higher risk of
developing lung and bone metastasis. These findings supported the
hypothesis that while an increase in proliferation is important for
the initiation and maintenance of primary tumors, growth inhibition
may be crucial for the survival of cancer cells in the circulation
and secondary organs, thereby leading to the development of a more
malignant phenotype (17). It is
therefore conceivable that CCN3 plays a role in the metastasis of
RCC.
Purified CCN3 has been shown to support adhesion and
migration in endothelial cells and fibroblasts via integrins and
heparan sulfate proteoglycans (18,19).
Despite the absence of an RGD sequence motif, CCN3 is a direct
ligand of integrins αvβ3 and α5β1 as demonstrated by solid phase
binding assays (18). In Ewing's
sarcoma, CCN3-transfected tumor cells exhibited a decreased
expression of α2β1 integrin receptor, while cells lacking α2β1 are
less less stably anchored to the ECM in Ewing's sarcoma tissue,
resulting in increased migratory abilities (12). In melanoma, CCN3-transfected cells
exhibited an increased expression of laminin and vitronectin
integrin receptors α7β1 and ανβ5, resulting in increased cell
adhesion (20). Integrin subunits
α3, β1, αν, α2 and α5 were highly expressed by 786-O cells. The
failure of 786-O cells to organize β1 fibrillar adhesions may be
associated with their highly migratory phenotype (21). It is therefore conceivable that
integrins may also be involved in CCN3-enhanced 786-O cell adhesion
and migration. Further investigation is required to confirm the
correlation between CCN3 and integrins in clear cell RCC.
In conclusion, our data indicate that CCN3 may play
a significant role in clear cell RCC biology. The molecule exhibits
anti-proliferative effects on tumor cells, while it promotes the
adhesion, migration and invasion of clear cell RCC cells.
Acknowledgements
This study was supported by the Shandong Provincial
Natural Science Foundation, China. We are grateful for the help and
support of Dr Cecile Martinerie.
References
|
1
|
Rini BI and Atkins MB: Resistance to
targeted therapy in renal-cell carcinoma. Lancet Oncol.
10:992–1000. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Perbal B: NOV (nephroblastoma
overexpressed) and the CCN family of genes: structural and
functional issues. Mol Pathol. 54:57–79. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Joliot V, Martinerie C, Dambrine G, et al:
Proviral rearrangements and overexpression of a new cellular gene
(nov) in myeloblastosis-associated virus type 1-induced
nephroblastomas. Mol Cell Biol. 12:10–21. 1992.PubMed/NCBI
|
|
4
|
Chevalier G, Yeger H, Martinerie C, et al:
novH: differential expression in developing kidney and Wilms'
tumors. Am J Pathol. 152:1563–1575. 1998.PubMed/NCBI
|
|
5
|
Perbal B: The CCN family of cell growth
regulators: a new family of normal and pathologic cell growth and
differentiation regulators: lessons from the first international
workshop on CCN gene family. Bull Cancer. 88:645–649.
2001.PubMed/NCBI
|
|
6
|
Manara MC, Perbal B, Benini S, et al: The
expression of ccn3(nov) gene in musculoskeletal tumors. Am J
Pathol. 160:849–859. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Perbal B: The CCN3 protein and cancer. Adv
Exp Med Biol. 587:23–40. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu C, Le AT, Yeger H, Perbal B and Alman
BAY: NOV (CCN3) regulation in the growth plate and CCN family
member expression in cartilage neoplasia. J Pathol. 201:609–615.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Doghman M, Arhatte M, Thibout H, et al:
Nephroblastoma overexpressed/cysteine-rich protein 61/connective
tissue growth factor/nephroblastoma overexpressed gene-3
(NOV/CCN3), a selective adrenocortical cell proapoptotic factor, is
down-regulated in childhood adrenocortical tumors. J Clin
Endocrinol Metab. 92:3253–3260. 2007. View Article : Google Scholar
|
|
10
|
Gupta N, Wang H, McLeod TL, et al:
Inhibition of glioma cell growth and tumorigenic potential by CCN3
(NOV). Mol Pathol. 54:293–299. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gellhaus A, Dong X, Propson S, et al:
Connexin43 interacts with NOV: a possible mechanism for negative
regulation of cell growth in choriocarcinoma cells. J Biol Chem.
279:36931–36942. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Benini S, Perbal B, Zambelli D, et al: In
Ewing's sarcoma CCN3 (NOV) inhibits proliferation while promoting
migration and invasion of the same cell type. Oncogene.
24:4349–4361. 2005.
|
|
13
|
Liu C, Liu XJ, Crowe PD, et al:
Nephroblastoma overexpressed gene (NOV) codes for a growth factor
that induces protein tyrosine phosphorylation. Gene. 238:471–478.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Maillard M, Cadot B, Ball RY, et al:
Differential expression of the ccn3 (nov) proto-oncogene in human
prostate cell lines and tissues. Mol Pathol. 4:275–280. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Perbal B: Contribution of MAV-1-induced
nephroblastoma to the study of genes involved in human Wilms' tumor
development. Crit Rev Oncog. 5:589–613. 1994.PubMed/NCBI
|
|
16
|
Niu Z, Ito M, Awakura Y, et al: The
expression of NOV and WT1 in renal cell carcinoma: a quantitative
reverse transcriptase-polymerase chain reaction analysis. J Urol.
174:1460–1462. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Evdokimova V, Tognon C, Ng T and Sorensen
PH: Reduced proliferation and enhanced migration: two sides of the
same coin? Molecular mechanisms of metastatic progression by YB-1.
Cell Cycle. 8:2901–2906. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin CG, Leu SJ, Chen N, et al: CCN3 (NOV)
is a novel angiogenic regulator of the CCN protein family. J Biol
Chem. 278:24200–24208. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin CG, Chen CC, Leu SJ, Grzeszkiewicz TM
and Lau LF: Integrin-dependent functions of the angiogenic inducer
NOV (CCN3): implication in wound healing. J Biol Chem.
280:8229–8237. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fukunaga-Kalabis M, Martinez G, Telson SM,
et al: Downregulation of CCN3 expression as a potential mechanism
for melanoma progression. Oncogene. 27:2552–2560. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Esteban-Barragán MA, Avila P,
Alvarez-Tejado M, et al: Role of the von Hippel-Lindau tumor
suppressor gene in the formation of beta1-integrin fibrillar
adhesions. Cancer Res. 62:2929–2936. 2002.PubMed/NCBI
|