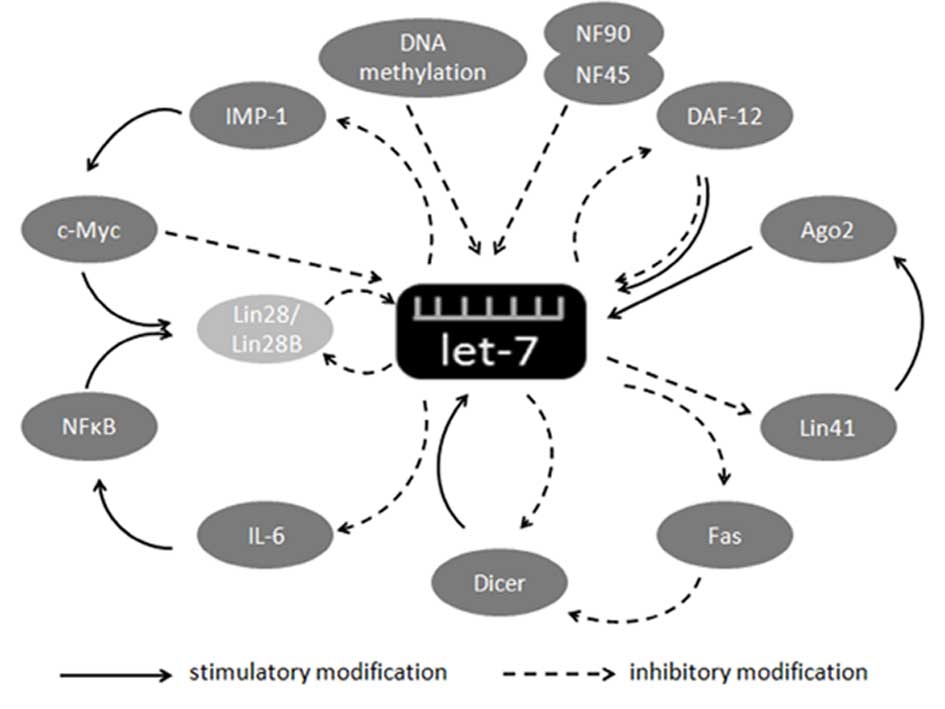
|
1
|
Nilsen TW: Mechanisms of microRNA-mediated
gene regulation in animal cells. Trends Genet. 23:243–249. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hutvágner G, McLachlan J, Pasquinelli AE,
Bálint E, Tuschl T and Zamore PD: A cellular function for the
RNA-interference enzyme Dicer in the maturation of the let-7 small
temporal RNA. Science. 293:834–838. 2001.PubMed/NCBI
|
|
3
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Roush S and Slack FJ: The let-7 family of
microRNAs. Trends Cell Biol. 18:505–516. 2008. View Article : Google Scholar
|
|
7
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
et al: Reduced expression of the let-7 microRNAs in human lung
cancers in association with shortened postoperative survival.
Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang HH, Wang XJ, Li GX, Yang E and Yang
NM: Detection of let-7a microRNA by real-time PCR in gastric
carcinoma. World J Gastroenterol. 13:2883–2888. 2007.PubMed/NCBI
|
|
9
|
Akao Y, Nakagawa Y and Naoe T: Let-7
microRNA functions as a potential growth suppressor in human colon
cancer cells. Biol Pharm Bull. 29:903–906. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sampson VB, Rong NH, Han J, Yang Q, Aris
V, Soteropoulos P, Petrelli NJ, Dunn SP and Krueger LJ: MicroRNA
let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt
lymphoma cells. Cancer Res. 67:9762–9770. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Grosshans H, Johnson T, Reinert KL,
Gerstein M and Slack FJ: The temporal patterning microRNA let-7
regulates several transcription factors at the larval to adult
transition in C. elegans. Dev Cell. 8:321–330. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sokol NS, Xu P, Jan YN and Ambros V:
Drosophila let-7 microRNA is required for remodeling of the
neuromusculature during metamorphosis. Genes Dev. 22:1591–1596.
2008. View Article : Google Scholar
|
|
13
|
Caygill EE and Johnston LA: Temporal
regulation of metamorphic processes in Drosophila by the
let-7 and miR-125 heterochronic microRNAs. Curr Biol. 18:943–950.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thomson JM, Parker J, Perou CM and Hammond
SM: A custom microarray platform for analysis of microRNA gene
expression. Nat Methods. 1:47–53. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Landgraf P, Rusu M, Sheridan R, Sewer A,
Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M,
et al: A mammalian microRNA expression atlas based on small RNA
library sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
17
|
Chang TC and Mendell JT: MicroRNAs in
vertebrate physiology and human disease. Annu Rev Genomics Hum
Genet. 8:215–239. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Viswanathan SR, Daley GQ and Gregory RI:
Selective blockade of microRNA processing by Lin28. Science.
320:97–100. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Newman MA, Thomson JM and Hammond SM:
Lin-28 interaction with the let-7 precursor loop mediates regulated
microRNA processing. RNA. 14:1539–1549. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Piskounova E, Viswanathan SR, Janas M,
LaPierre RJ, Daley GQ, Sliz P and Gregory RI: Determinants of
microRNA processing inhibition by the developmentally regulated
RNA-binding protein Lin28. J Biol Chem. 283:21310–21314. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Heo I, Joo C, Cho J, Ha M, Han J and Kim
VN: Lin28 mediates the terminal uridylation of let-7 precursor
microRNA. Mol Cell. 32:276–284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hagan JP, Piskounova E and Gregory RI:
Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in
mouse embryonic stem cells. Nat Struct Mol Biol. 16:1021–1025.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho
J, Yeom KH, Han J and Kim VN: TUT4 in concert with Lin28 suppresses
microRNA biogenesis through pre-microRNA uridylation. Cell.
138:696–708. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lehrbach NJ, Armisen J, Lightfoot HL,
Murfitt KJ, Bugaut A, Balasubramanian S and Miska EA: LIN-28 and
the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in
Caenorhabditis elegans. Nat Struct Mol Biol. 16:1016–1020.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
26
|
Ehrlich M: DNA methylation in cancer: too
much, but also too little. Oncogene. 21:5400–5413. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Esteller M: Relevance of DNA methylation
in the management of cancer. Lancet Oncol. 4:351–358. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lujambio A, Ropero S, Ballestar E, Fraga
MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A,
Sanchez-Cespedes M, Git A, et al: Genetic unmasking of an
epigenetically silenced microRNA in human cancer cells. Cancer Res.
67:1424–1429. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Saito Y, Liang G, Egger G, Friedman JM,
Chuang JC, Coetzee GA and Jones PA: Specific activation of
microRNA-127 with downregulation of the proto-oncogene BCL6 by
chromatin-modifying drugs in human cancer cells. Cancer Cell.
9:435–443. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yoshitomi T, Kawakami K, Enokida H,
Chiyomaru T, Kagara I, Tatarano S, Yoshino H, Arimura H, Nishiyama
K, Seki N and Nakagawa M: Restoration of miR-517a expression
induces cell apoptosis in bladder cancer cell lines. Oncol Rep.
25:1661–1668. 2011.PubMed/NCBI
|
|
31
|
Brueckner B, Stresemann C, Kuner R, Mund
C, Musch T, Meister M, Sültmann H and Lyko F: The human let-7a-3
locus contains an epigenetically regulated microRNA gene with
oncogenic function. Cancer Res. 67:1419–1423. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lu L, Katsaros D, de la Longrais IA,
Sochirca O and Yu H: Hypermethylation of let-7a-3 in epithelial
ovarian cancer is associated with low insulin-like growth factor-II
expression and favorable prognosis. Cancer Res. 67:10117–10122.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sakamoto S, Aoki K, Higuchi T, Todaka H,
Morisawa K, Tamaki N, Hatano E, Fukushima A, Taniguchi T and Agata
Y: The NF90-NF45 complex functions as a negative regulator in the
micro RNA processing pathway. Mol Cell Biol. 29:3754–3769. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Iliopoulos D, Hirsch HA and Struhl K: An
epigenetic switch involving NF-kappaB, Lin28, Let-7 microRNA, and
IL6 links inflammation to cell transformation. Cell. 139:693–706.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Boyerinas B, Park SM, Shomron N, Hedegaard
MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB and Peter ME:
Identification of let-7-regulated oncofetal genes. Cancer Res.
68:2587–2591. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ioannidis P, Mahaira LG, Perez SA,
Gritzapis AD, Sotiropoulou PA, Kavalakis GJ, Antsaklis AI,
Baxevanis CN and Papamichail M: CRD-BP/IMP1 expression
characterizes cord blood CD34+ stem cells and affects c-myc and
IGF-II expression in MCF-7 cancer cells. J Biol Chem.
280:20086–20093. 2005.PubMed/NCBI
|
|
37
|
Chang TC, Zeitels LR, Hwang HW, Chivukula
RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, et al:
Lin-28B transactivation is necessary for Myc-mediated let-7
repression and proliferation. Proc Natl Acad Sci USA.
106:3384–3389. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dangi-Garimella S, Yun J, Eves EM, Newman
M, Erkeland SJ, Hammond SM, Minn AJ and Rosner MR: Raf kinase
inhibitory protein suppresses a metastasis signalling cascade
involving LIN28 and let-7. EMBO J. 28:347–358. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chang TC, Yu D, Lee YS, Wentzel EA, Arking
DE, West KM, Dang CV, Thomas-Tikhonenko A and Mendell JT:
Widespread microRNA repression by Myc contributes to tumorigenesis.
Nat Genet. 40:43–50. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hammell CM, Karp X and Ambros V: A
feedback circuit involving let-7-family miRNAs and DAF-12
integrates environmental signals and developmental timing in
Caenorhabditis elegans. Proc Natl Acad Sci USA.
106:18668–18673. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Diederichs S and Haber DA: Dual role for
argonautes in microRNA processing and posttranscriptional
regulation of microRNA expression. Cell. 131:1097–1108. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rybak A, Fuchs H, Hadian K, Smirnova L,
Wulczyn EA, Michel G, Nitsch R, Krappmann D and Wulczyn FG: The
let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin
ligase for the miRNA pathway protein Ago2. Nat Cell Biol.
11:1411–1420. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Forman JJ, Legesse-Miller A and Coller HA:
A search for conserved sequences in coding regions reveals that the
let-7 microRNA targets Dicer within its coding sequence. Proc Natl
Acad Sci USA. 105:14879–14884. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jakymiw A, Patel RS, Deming N,
Bhattacharyya I, Shah P, Lamont RJ, Stewart CM, Cohen DM and Chan
EK: Overexpression of dicer as a result of reduced let-7 microRNA
levels contributes to increased cell proliferation of oral cancer
cells. Genes Chromosomes Cancer. 49:549–559. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yonehara S, Ishii A and Yonehara M: A
cell-killing monoclonal antibody (anti-Fas) to a cell surface
antigen co-downregulated with the receptor of tumor necrosis
factor. J Exp Med. 169:1747–1756. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Peter ME, Budd RC, Desbarats J, Hedrick
SM, Hueber AO, Newell MK, Owen LB, Pope RM, Tschopp J, Wajant H,
Wallach D, et al: The CD95 receptor: apoptosis revisited. Cell.
129:447–450. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Barnhart BC, Legembre P, Pietras E, Bubici
C, Franzoso G and Peter ME: CD95 ligand induces motility and
invasiveness of apoptosis-resistant tumor cells. EMBO J.
23:3175–3185. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gordon N and Kleinerman ES: Aerosol
therapy for the treatment of osteosarcoma lung metastases:
targeting the Fas/FasL pathway and rationale for the use of
gemcitabine. J Aerosol Med Pulm Drug Deliv. 23:189–196. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang S, Tang Y, Cui H, Zhao X, Luo X, Pan
W, Huang X and Shen N: Let-7/miR-98 regulate Fas and Fas-mediated
apoptosis. Genes Immun. 12:149–154. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Geng L, Zhu B, Dai BH, Sui CJ, Xu F, Kan
T, Shen WF and Yang JM: A let-7/Fas double-negative feedback loop
regulates human colon carcinoma cells sensitivity to Fas-related
apoptosis. Biochem Biophys Res Commun. 408:494–499. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liang D, Meyer L, Chang DW, Lin J, Pu X,
Ye Y, Gu J, Wu X and Lu K: Genetic variants in microRNA
biosynthesis pathways and binding sites modify ovarian cancer risk,
survival, and treatment response. Cancer Res. 70:9765–9776. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Permuth-Wey J, Kim D, Tsai YY, Lin HY,
Chen YA, Barnholtz-Sloan J, Birrer MJ, Bloom G, Chanock SJ, Chen Z,
et al: LIN28B polymorphisms influence susceptibility to epithelial
ovarian cancer. Cancer Res. 71:3896–3903. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen AX, Yu KD, Fan L, Li JY, Yang C,
Huang AJ and Shao ZM: Germline genetic variants disturbing the
Let-7/LIN28 double-negative feedback loop alter breast cancer
susceptibility. PLoS Genet. 7:e10022592011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lytle JR, Yario TA and Steitz JA: Target
mRNAs are repressed as efficiently by microRNA-binding sites in the
5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 104:9667–9672.
2007.PubMed/NCBI
|
|
55
|
Nottrott S, Simard MJ and Richter JD:
Human let-7a miRNA blocks protein production on actively
translating polyribosomes. Nat Struct Mol Biol. 13:1108–1114. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Beilharz TH, Humphreys DT, Clancy JL,
Thermann R, Martin DI, Hentze MW and Preiss T: MicroRNA-mediated
messenger RNA deadenylation contributes to translational repression
in mammalian cells. PLoS One. 4:e67832009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: Ras is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Nelson HH, Christensen BC, Plaza SL,
Wiencke JK, Marsit CJ and Kelsey KT: KRAS mutation, KRAS-LCS6
polymorphism, and non-small cell lung cancer. Lung Cancer.
69:51–53. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Chin LJ, Ratner E, Leng S, Zhai R, Nallur
S, Babar I, Muller RU, Straka E, Su L, Burki EA, et al: A SNP in a
let-7 microRNA complementary site in the KRAS 3′ untranslated
region increases non-small cell lung cancer risk. Cancer Res.
68:8535–8540. 2008.PubMed/NCBI
|
|
60
|
Paranjape T, Heneghan H, Lindner R, Keane
FK, Hoffman A, Hollestelle A, Dorairaj J, Geyda K, Pelletier C,
Nallur S, et al: A 3′-untranslated region KRAS variant and
triple-negative breast cancer: a case-control and genetic analysis.
Lancet Oncol. 12:377–386. 2011.
|
|
61
|
Lee ST, Chu K, Oh HJ, Im WS, Lim JY, Kim
SK, Park CK, Jung KH, Lee SK, Kim M and Roh JK: Let-7 microRNA
inhibits the proliferation of human glioblastoma cells. J
Neurooncol. 102:19–24. 2010.PubMed/NCBI
|
|
62
|
He XY, Chen JX, Zhang Z, Li CL, Peng QL
and Peng HM: The let-7a microRNA protects from growth of lung
carcinoma by suppression of k-Ras and c-Myc in nude mice. J Cancer
Res Clin Oncol. 136:1023–1028. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Sgarra R, Rustighi A, Tessari MA, Di
Bernardo J, Altamura S, Fusco A, Manfioletti G and Giancotti V:
Nuclear phosphoproteins HMGA and their relationship with chromatin
structure and cancer. FEBS Lett. 574:1–8. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Sarhadi VK, Wikman H, Salmenkivi K, Kuosma
E, Sioris T, Salo J, Karjalainen A, Knuutila S and Anttila S:
Increased expression of high mobility group A proteins in lung
cancer. J Pathol. 209:206–212. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Meyer B, Loeschke S, Schultze A, Weigel T,
Sandkamp M, Goldmann T, Vollmer E and Bullerdiek J: HMGA2
overexpression in non-small cell lung cancer. Mol Carcinog.
46:503–511. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Abe N, Watanabe T, Suzuki Y, Matsumoto N,
Masaki T, Mori T, Sugiyama M, Chiappetta G, Fusco A and Atomi Y: An
increased high-mobility group A2 expression level is associated
with malignant phenotype in pancreatic exocrine tissue. Br J
Cancer. 89:2104–2109. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Miyazawa J, Mitoro A, Kawashiri S, Chada
KK and Imai K: Expression of mesenchyme-specific gene HMGA2 in
squamous cell carcinomas of the oral cavity. Cancer Res.
64:2024–2029. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Belge G, Meyer A, Klemke M, Burchardt K,
Stern C, Wosniok W, Loeschke S and Bullerdiek J: Upregulation of
HMGA2 in thyroid carcinomas: a novel molecular marker to
distinguish between benign and malignant follicular neoplasias.
Genes Chromosomes Cancer. 47:56–63. 2008. View Article : Google Scholar
|
|
69
|
Borrmann L, Wilkening S and Bullerdiek J:
The expression of HMGA genes is regulated by their 3'UTR. Oncogene.
20:4537–4541. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Motoyama K, Inoue H, Nakamura Y, Uetake H,
Sugihara K and Mori M: Clinical significance of high mobility group
A2 in human gastric cancer and its relationship to let-7 microRNA
family. Clin Cancer Res. 14:2334–2340. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Shi G, Perle MA, Mittal K, Chen H, Zou X,
Narita M, Hernando E, Lee P and Wei JJ: Let-7 repression leads to
HMGA2 overexpression in uterine leiomyosarcoma. J Cell Mol Med.
13:3898–3905. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Mayr C, Hemann MT and Bartel DP:
Disrupting the pairing between let-7 and Hmga2 enhances oncogenic
transformation. Science. 315:1576–1579. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Albihn A, Johnsen JI and Henriksson MA:
Myc in oncogenesis and as a target for cancer therapies. Adv Cancer
Res. 107:163–224. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ruggero D: The role of Myc-induced protein
synthesis in cancer. Cancer Res. 69:8839–8843. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Schultz J, Lorenz P, Gross G, Ibrahim S
and Kunz M: MicroRNA let-7b targets important cell cycle molecules
in malignant melanoma cells and interferes with
anchorage-independent growth. Cell Res. 18:549–557. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Johnson CD, Esquela-Kerscher A, Stefani G,
Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J,
Shingara J, et al: The let-7 microRNA represses cell proliferation
pathways in human cells. Cancer Res. 67:7713–7722. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Leeman RJ, Lui VW and Grandis JR: STAT3 as
a therapeutic target in head and neck cancer. Expert Opin Biol
Ther. 6:231–241. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Bowman T, Garcia R, Turkson J and Jove R:
STATs in oncogenesis. Oncogene. 19:2474–2488. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Achcar RO, Cagle PT and Jagirdar J:
Expression of activated and latent signal transducer and activator
of transcription 3 in 303 non-small cell lung carcinomas and 44
malignant mesotheliomas: possible role for chemotherapeutic
intervention. Arch Pathol Lab Med. 131:1350–1360. 2007.
|
|
80
|
Ma XT, Wang S, Ye YJ, Du RY, Cui ZR and
Somsouk M: Constitutive activation of Stat3 signaling pathway in
human colorectal carcinoma. World J Gastroenterol. 10:1569–1573.
2004.PubMed/NCBI
|
|
81
|
Wang Y, Lu Y, Toh ST, Sung WK, Tan P, Chow
P, Chung AY, Jooi LL and Lee CG: Lethal-7 is down-regulated by the
hepatitis B virus x protein and targets signal transducer and
activator of transcription 3. J Hepatol. 53:57–66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Bronner C, Achour M, Arima Y, Chataigneau
T, Saya H and Schini-Kerth VB: The UHRF family: oncogenes that are
drugable targets for cancer therapy in the near future? Pharmacol
Ther. 115:419–434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Robinson PA and Ardley HC:
Ubiquitin-protein ligases. J Cell Sci. 117:5191–5194. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Arima Y, Hirota T, Bronner C, Mousli M,
Fujiwara T, Niwa S, Ishikawa H and Saya H: Down-regulation of
nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage
checkpoint signals contributes to cell cycle arrest at G1/S
transition. Genes Cells. 9:131–142. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
He X, Duan C, Chen J, Ou-Yang X, Zhang Z,
Li C and Peng H: Let-7a elevates p21(WAF1) levels by targeting of
NIRF and suppresses the growth of A549 lung cancer cells. FEBS
Lett. 583:3501–3507. 2009. View Article : Google Scholar : PubMed/NCBI
|