Introduction
Nasopharyngeal carcinoma (NPC) is one of the most
common types of malignant cancer, with a high occurrence rate in
the Chinese population compared with the western population
(1). The genetic mechanisms of
oncogenesis include the inactivation of tumor suppression genes,
such as p53, Rb and p16, as well as the overexpression of
oncogenes, such as bcl-2, ras and Cyclin D1. However, the mutations
of these genes are rare in NPC patients. One possibility is that
the regulation of gene expression through promoter methylation is
changed in NPC cases (2,3). The present study aimed to examine the
differences in the mRNA and protein expression levels of the Syk
gene as well as its promoter methylation in NPC cell lines with
different differentiation levels, to understand the potential
contribution of promoter methylation to NPC genesis.
Materials and methods
Materials
The NPC cell lines CNE-1, CNE-2 and NP69 were
purchased from Yingrun Biotechnol Co. (Shanghai, China). The
following reagents and instruments were used: Q-RT-PCR machine
(ABI, Applied Biosystems, Carlsbad, CA, USA), Q-RT-PCR kit
(Tiangen, Shanghai, China), keratinocyte-sfm culture medium
(Invitrogen, Carlsbad, CA, USA), Genomic DNA extraction and
purification kit, rabbit anti-β-actin, goat anti-rabbit secondary
antibody, PVDF membrane, ECL kit (Kaiji Biotechnology, Shanghai,
China).
Cell culture
The cell culture of the CNE-1, CNE-2 and NP69 cell
lines was performed with complete RPMI-1640 medium at 37°C in a 5%
CO2 incubator. The cells were digested with 0.25%
trypsin protease-EDTA.
MS-PCR
Following the DNA extraction, 8 μg total DNA with
ddH2O in a total volume of 18 μl was heated at 95°C and
then immediately placed into an ice water bath. NaOH (3 M) was
added for denaturation at 42°C for 20 min and the solution was then
mixed with freshly prepared hydroquinone and sodium bisulfite.
Liquid paraffin (200 μl) was added and incubated at 50°C for 16 h.
The liquid paraffin was then removed. The water phase was mixed
with a purified solution of TBE buffer before de-salting with
Wizard DNA purification system. An isopropanol wash was performed
twice and DNA was eluted into 50 μl ddH2O, then
centrifuged before being incubated with 3 M NaOH at 37°C for 15 min
for denaturation. Then 33 μl of 10 M ammonium acetate was added to
buffer the NaOH before the ethanol was added to precipitate the DNA
at -70°C for 1 h. Then the tube was centrifuged at 4°C at 10,000 ×
g for 20 min. The DNA was washed with 70% ethanol after removing
the supernatant. Finally the DNA was diluted in 50 μl TE buffer and
stored at -20°C. For the MS-PCR detection, methylation-specific
primers contained 9 CpG islands and non-methylation-specific
primers contained 8 CpG islands. The reaction was performed as
follows: methylation system: denaturation at 95°C for 5 min, then
50 cycles of 95°C for 10 sec, annealing at 60°C for 15 sec and
extension at 72°C for 25 sec; non-methylation system: denaturation
at 95°C for 5 min, then 50 cycles of 95°C for 30 sec, annealing at
60°C for 15 sec and extension at 72°C for 25 sec; with a final step
at 72°C for 10 min. PCR products (5 μl) were used for 2% agarose
gel electrophoresis and the image was analyzed for
semi-quantitative absorbance values in order to calculate the
methylation ratio [methylated primer band absorbance
value/(methylated primer extension increase in absorbance values +
non-methylated primer band absorbance) × 100%]. The sequences of
the primers are listed in Table
I.
 | Table ISequences of methylation and
non-methylation primers for the Syk gene promoter. |
Table I
Sequences of methylation and
non-methylation primers for the Syk gene promoter.
| Primer | Sequences | Length (bp) |
|---|
| Methylation | F:
5′-TTTCGGGTTTATGGGCGCGG-3′
R: 5′-ACGAAAACGAACGCAACGCGAA-3′ | 202 |
| Non-methylation | F:
5′GTTTTAGTTGATTTTTGTTTAGTTTTG-3′
R: 5′-ACCACCCACTCCTCCTCACT-3′ | 202 |
Q-RT-PCR
RNA extraction was performed using the TRIzol
approach with suspended cells and the purity was tested with the UV
spectrophotometer A260/A280 ratio between 1.8 and 2.1. The 18 S and
28 S integrity were analyzed with agarose gel electrophoresis. For
the cDNA synthesis reaction system, 2 μg RNA was mixed with 10× RT
mix, dNTP mix, 2 μl Oligo-dT15, 1 μl Quant Reverse Transcriptase
and RNase Free ddH2O to a final volume of 20 μl. The
mixture was incubated at 37°C for 60 min. The SYBR-Green
fluorescence dye was used during the real-time PCR. The 20-μl
reaction system included 9 μl 2.5× Real Master mix/20× SYBR
solution mixed reaction solution, 2 μl cDNA solution, 2 μl upstream
and downstream primers (100 nM) and ddH2O was added to
make a final volume of 20 μl. Each sample was repeated 3 times. The
reaction conditions were: pre-denaturation at 95°C for 2 min, then
40 cycles of 95°C for 15 sec, 62°C for 30 sec, 68°C for 60 sec;
final reaction at 95°C for 15 sec and 60°C for 1 min, then 95°C for
15 sec. The fluorescence signals were collected for the melting
curve analysis. The Ct value was recorded in the three repeated
experiments and averaged, then calculated using the comparative
ΔΔCt method. The sequences of primers are listed in Table II.
 | Table IISequences of Syk gene and GAPDH gene
primers. |
Table II
Sequences of Syk gene and GAPDH gene
primers.
| Gene | Sequences | Length (bp) |
|---|
| Syk | F: 5′-CAT GTC AAG GAT
AAG AAC ATC ATA GA-3′
R: 5′-AGT TCA CCA CGT CAT AGT AGT AAT T-3′ | 514 |
| GAPDH | F: 5′-ATG GCC TTC CGT
GTC CCC ACT G-3′
R: 5′-TGA GTG TGG CAG GGA CTC CCC A-3′ | 398 |
Western blot analysis
The total protein samples were transferred to PVDF
membranes (positively charged Heni dragon film) following SDS-PAGE.
The blocking was performed with 5% non-fat milk powder and the
primary antibody was incubated at 4°C overnight. The incubation
with the secondary antibody was performed at room temperature for 1
h prior to washing and ECL treatment and finally the X-ray film was
developed. GADPH was used as an internal control. The image
analysis system was used to find the absorbance value ratio between
Syk and GADPH.
Statistical analysis
The SPSS 17.0 software was used for statistical
analysis and the data are presented as mean ± standard deviation.
The single factor analysis of variance was used to compared the Syk
promoter methylation rate and mRNA and protein expression
differences among the cell lines. P<0.05 was considered to
indicate a statistically significant result.
Results
Promoter methylation in different cell
lines
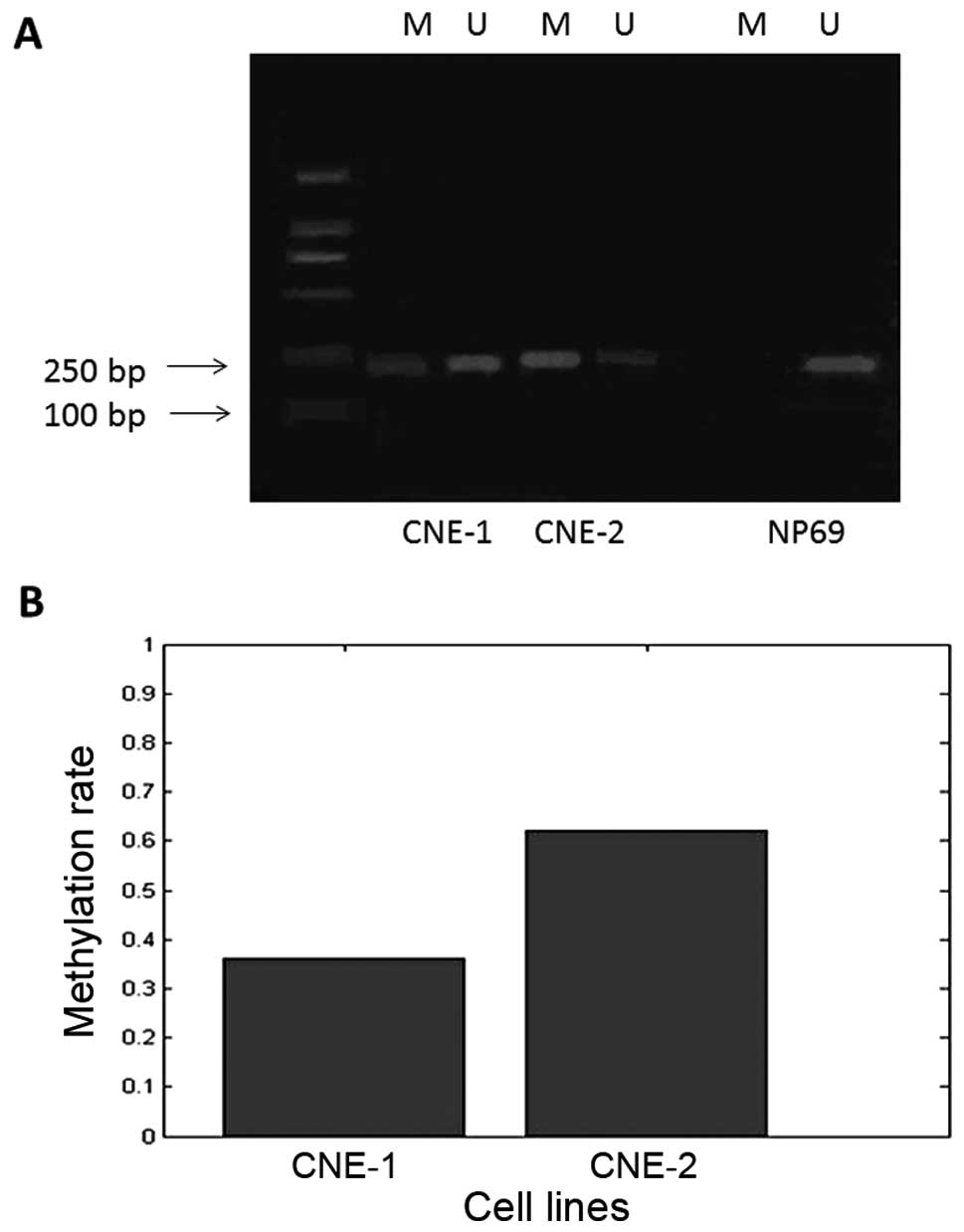
The electrophoresis results of the MS-PCR products
suggest that CNE-1 and CNE-2 cells showed incomplete Syk promoter
methylation (amplification with both groups of primers), while the
NP69 cell line showed no promoter methylation (Fig. 1A). The methylation rates in CNE-1
and CNE-2 cells were statistically different at 36 and 62%,
respectively (Fig. 1B;
P<0.01).
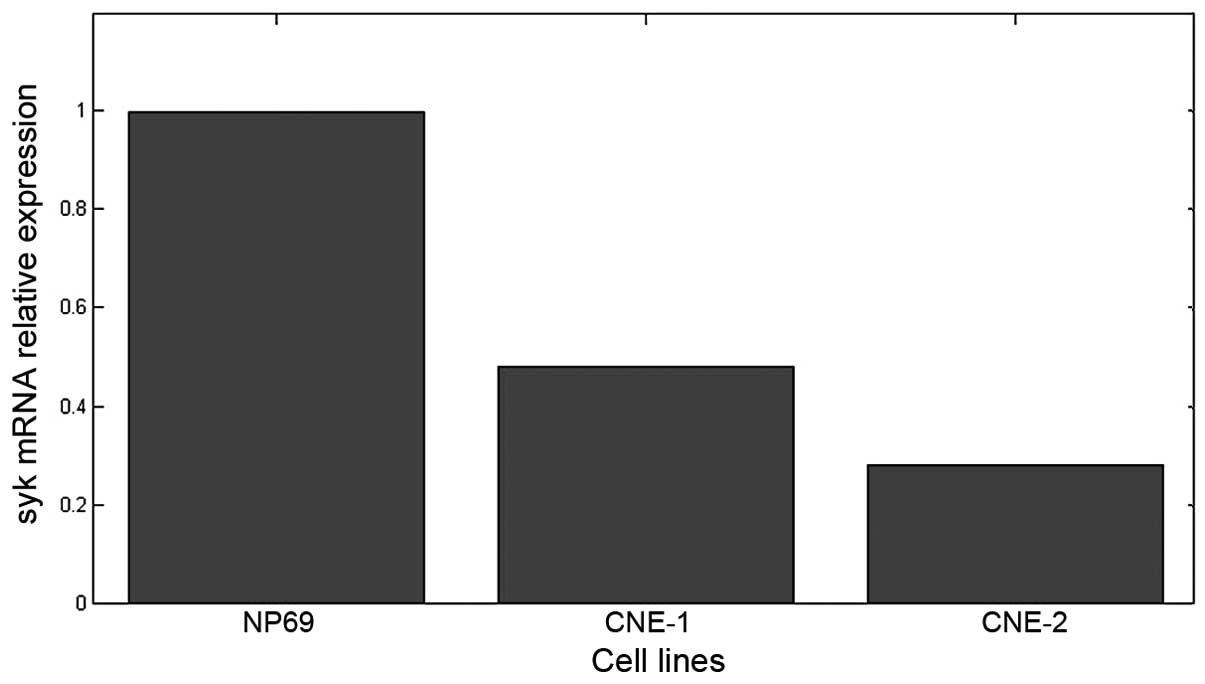
Syk mRNA expression in different cell
lines
The melting curves of the Syk and GAPDH genes had
single peaks at 80.57°C and 88.73°C, respectively, suggesting high
primer specificity. The results of quantitative study of Syk gene
expression are shown in Fig. 2. The
mRNA was expressed in all three cell lines. The mRNA levels in
CNE-1 and CNE-2 cells were lower, 42±3.5 and 28±2% of that in NP69,
respectively; the differences between the groups were statistically
significant (P<0.01).
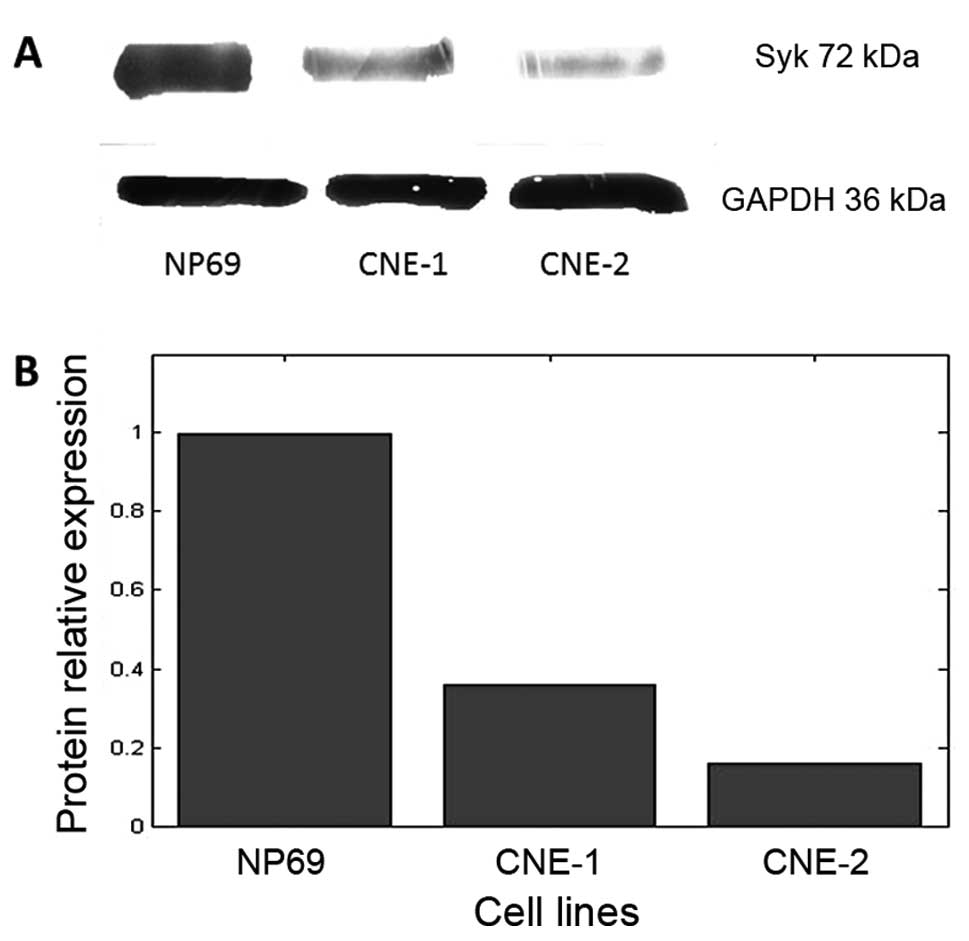
Syk protein expression in different cell
lines
The results of the western blot analysis of Syk
protein expression are shown in Fig.
3A. The protein was expressed in all three cell lines. The
protein levels in CNE-1 and CNE-2 cells were lower, 36±4.5 and
16±2.5% of that in NP69 (Fig. 3B),
respectively; the differences between the groups were statistically
significant (P<0.01).
Discussion
The epigenetic theory of cancer has emerged as a hot
spot in recent years, especially for tumor suppressor genes
(4,5). The present study used NPC as the model
to investigate the epigenetic changes of tumor suppressor genes.
Syk is a non-receptor tyrosine kinase which was first identified in
1991 (6) and locates to the q22
region of human chromosome 9. The protein contains 629 amino acids
and has a molecular weight of 72 kDa. Syk is a significant molecule
in the B cell antigen-receptor signaling pathway, and is considered
to be a candidate tumor suppressor gene, the loss of which results
in the retarded development and maturation of immune cells or even
severe combined immunodeficiency (SCID) (7,8). This
may lead to the failure of monitoring cancer cell development and
finally tumor growth.
The present study used NPC cells lines with varied
levels of differentiation with NP69 as the control to examine the
changes in the expression of the Syk gene and correlated changes in
epigenetic control. The results revealed that the mRNA expression
level in NP69 cells was higher than that in the NPC cell lines
(P<0.01) and that in CNE-1 cells the mRNA expression level was
higher than that in CNE-2 cells (P<0.01), suggesting a positive
correlation between mRNA expression and the differentiation level;
similar results were also observed for Syk protein expression.
Finally, the methylation level of the Syk gene promoter was
negatively correlated with the differentiation level.
Previous studies have suggested that the tumor
suppression function of Syk is due to its inhibition of cell
proliferation and migration, with the downregulation of its
expression in breast, gastric, colorectal and liver cancer, among
others (8–10). The detailed roles of Syk in NPC
growth and metastasis are not clear. Recent studies have
highlighted the roles of epigenetic changes of tumor suppressor
genes in oncogenesis (2,4,11,12).
Our results are consistent with this theory, and the low
differentiation level was associated with a low expression level of
Syk and high methylation level of the promoter region. Further
study is needed to elucidate the mechanism by which promoter
methylation contributes to the altered expression of the Syk gene
in NPC cell lines.
References
|
1
|
Tao Q and Chan AT: Nasopharyngeal
carcinoma: molecular pathogenesis and therapeutic developments.
Expert Rev Mol Med. 9:1–24. 2007.PubMed/NCBI
|
|
2
|
Li LL, Shu XS, Wang ZH, Cao Y and Tao Q:
Epigenetic disruption of cell signaling in nasopharyngeal
carcinoma. Chin J Cancer. 30:231–239. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lo KW and Huang DP: Genetic and epigenetic
changes in nasopharyngeal carcinoma. Semin Cancer Biol. 12:451–462.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Niller HH, Wolf H and Minarovits J:
Epigenetic dysregulation of the host cell genome in Epstein-Barr
virus-associated neoplasia. Semin Cancer Biol. 19:158–164. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Buchholz TA and Wazer DE: Molecular
biology and genetics of breast cancer development: a clinical
perspective. Semin Radiat Oncol. 12:285–295. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Taniguchi T, Kobayashi T, Kondo J, et al:
Molecular cloning of a porcine gene syk that encodes a 72-kDa
protein-tyrosine kinase showing high susceptibility to proteolysis.
J Biol Chem. 266:15790–15796. 1991.PubMed/NCBI
|
|
7
|
Medves S and Demoulin JB: Tyrosine kinase
gene fusions in cancer: translating mechanisms into targeted
therapies. J Cell Mol Med. 16:237–248. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stewart ZA and Pietenpol JA: Syk: a new
player in the field of breast cancer. Breast Cancer Res. 3:5–7.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Efremov DG and Laurenti L: The Syk kinase
as a therapeutic target in leukemia and lymphoma. Expert Opin
Investig Drugs. 20:623–636. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Page TH, Smolinska M, Gillespie J,
Urbaniak AM and Foxwell BM: Tyrosine kinases and inflammatory
signalling. Curr Mol Med. 9:69–85. 2009. View Article : Google Scholar
|
|
11
|
Simons MJ: Nasopharyngeal carcinoma as a
paradigm of cancer genetics. Chin J Cancer. 30:79–84. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hutajulu SH, Indrasari SR, Indrawati LP,
et al: Epigenetic markers for early detection of nasopharyngeal
carcinoma in a high risk population. Mol Cancer. 10:482011.
View Article : Google Scholar : PubMed/NCBI
|