Introduction
Colorectal cancer (CRC) is the second most common
form of cancer in developed countries, only surpassed by prostate
cancer in men and breast cancer in women (1). In Jordan, it is the most common type
of cancer among men and the second most common among women
(2). The reasons for this are
unknown and may include both genetic and environmental factors. CRC
can be cured by relatively simple colorectal procedures if detected
early. However, distant metastasis is the main cause of mortality
in CRC patients. Studies have shown that depending on the stage of
the primary tumor, liver metastases occur in 20–70% of patients and
lung metastases occur in 10–20% of patients (3).
Significant advances have been made in the treatment
and outcome of CRC over the last decade. An improved understanding
of the molecular pathways involved in the development and
progression of CRC has made it possible to provide prognoses for
patients with metastases, as well as the development of new
therapeutic strategies. The epidermal growth factor receptor (EGFR)
is a transmembrane tyrosine kinase receptor. It is expressed in
epithelial tissues and acts as a cell growth promoter. According to
literature, EGFR contributes to the development and progression of
several types of cancer, including CRC where it is overexpressed in
50–80% of colorectal tumors, making it a suitable target for
anticancer therapies (4). Abnormal
activation of the EGFR pathway may be caused by EGFR overexpression
or mutational activation of the downstream elements (5).
Currently, two strategies to attenuate EGFR
signaling are in use: monoclonal antibodies that bind to the
ligand-binding domain and inhibit the binding of specific ligands
(cetuximab and panitumumab), or small EGFR tyrosine kinase
inhibitor molecules that bind to the intracellular domain of EGFR
and compete for binding with ATP, inhibiting tyrosine
phosphorylation (gefitinib and erlotinib). These inhibitors of EGFR
have emerged as an important treatment for metastatic colorectal
cancer (mCRC) (6,7). To optimize the benefits and reduce the
risks of anti-EGFR therapies, EGFR as well as the molecules
involved in its signaling pathway have been evaluated as potential
markers for predicting therapy outcomes. Anti-EGFR therapies are
only effective in a subset of patients with CRC. A number of
clinical trials have demonstrated that EGFR-targeted therapies are
not effective in patients whose tumors have a mutation in the
oncogene Kirsten ras (KRAS) (8–10).
The v-Ki-ras2 Kirsten rat sarcoma (KRAS) gene
is a member of the Ras gene family that encodes small G proteins
with intrinsic GTPase activity. KRAS is a downstream
component of the EGFR signaling pathway. It acts as an
intracellular signal transducer by coupling the signal from the
cell surface receptors to intracellular targets, which regulate
significant functions for tumor progression, including
proliferation, differentiation and apoptosis (4). Mutations in the KRAS gene
(typically point mutations) result in constitutive guanine
triphosphotase activity, which continuously activates signaling
pathways in the absence of any upstream stimulation of EGFR/HER
receptors (11). Thus, patients
with KRAS mutations have poor responses to therapy with
anti-EGFR inhibitors. KRAS mutations are thought to be a
fairly early event in carcinogenesis and range from 35–45% in CRC
(12). KRAS mutations also
occur frequently in non-small-cell lung and pancreatic carcinomas
(13). The most common mutations
identified in CRC occur in exon 2 and to a lesser extent in exon 3
(14). Tumors that have a mutation
in codon 12 or 13 of the KRAS gene will not respond to
treatment with EGFR inhibitors, including cetuximab or
panitumumab.
The mutation status of the KRAS gene provides
diagnostic, prognostic and predictive information for several types
of cancer. The precise frequency and genotyping of KRAS
mutations in the Jordanian population have not been determined. The
high incidence and mortality among CRC Jordanian patients indicates
the need to determine whether specific ethnic, geographical,
dietary or lifestyle factors may possibly be correlated. This study
investigated the general incidence of KRAS mutations in CRC
in Jordan and the incidence of specific mutation types. The
possible correlation of molecular results with clinical and
histopathological data was also analyzed.
Materials and methods
Ethics statement
This was an observational study. All patients were
managed in accordance with normal clinical practice. The
Institutional Review Board at King Hussein Cancer Center in Amman,
Jordan, approved the current study.
Tissue attainment
Colorectal carcinoma specimens from 100 consecutive
patients who underwent surgical resection or colonoscopic biopsies
of colorectal tumors and had developed metastatic disease were
studied. Biopsies or resected tumors were reviewed for their
histological diagnosis and quantification of neoplastic cellularity
(>20%). Using hematoxylin and eosin-stained slides, areas with
>50% tumor cells were delimited.
All tumors were histologically confirmed to be
colorectal adenocarcinomas. In addition, medical records were
reviewed for information on the tumor site, pathological stage,
presence or absence of metastasis and outcome in patients prior to
anti-EGFR therapy. The formalin-fixed paraffin-embedded (FFPE)
tissues were previously processed according to routine
practices.
DNA extraction
DNA was extracted from paraffin blocks that best
represented each tumor, previously selected from hematoxylin-eosin
stained slides. To prevent cross contamination from tissues with
flakes of paraffin, disposable scalpel blades were used. Tumor
areas were carefully scraped from tissue blocks by macro-dissection
using a sterile scalpel blade and then transferred to a
microcentrifuge tube. Tissues were deparaffinized with three baths
of xylene for 10 minutes followed by three baths of 100% ethanol
solution for 5 minutes. Following this, tissues were digested with
Proteinase K and genomic DNA was isolated using the QIAamp DNA
Extraction kit (Qiagen, Crawley, UK) according to the
manufacturer’s instructions.
Samples of isolated genomic DNA were analysed by
0.8% agarose gel electrophoresis to evaluate the DNA quality. The
DNA quantity was assessed by using NanoDrop 1000 (Thermo
Scientific, DE, USA) and the purity was evaluated by calculating
the 260/280 ratio.
KRAS mutational analysis
KRAS mutations were detected by an
hybridization-based strip assay (ViennaLab® Diagnostics
GmbH, Vienna, Austria) as well as a real-time PCR-based mutation
assay (DxS KRAS Mutation Test kit, DxS Ltd; Manchester, UK)
according to the manufacturer’s instructions. The first assay is
based on reverse-hybridization of biotinylated PCR products to a
parallel array of allele-specific oligonucleotides immobilized on
membrane strips. The detection of specifically bound mutant
KRAS alleles is visible by an enzymatic color reaction which
can be compared to specific controls. This assay detects 10
mutations located in codons 12 and 13. The second assay designed by
DxS Diagnostic Innovations, combines allele-specific PCR
Amplification Refractory Mutation System (ARMS) with real-time PCR
to detect the seven most common mutations at KRAS codons 12
and 13 (p.Gly12Ala, p.Gly12Asp, p.Gly12Arg, p.Gly12Cys, p.Gly12Ser,
p.Gly12Val and p.Gly13Asp). Mutation detection was performed with a
Rotor Gene Q Real-Time PCR System (Corbett Robotics, Brisbane,
Australia).
Random samples were selected for confirmation by
standard Sanger sequencing using BigDye® terminator v3.1
(Applied Biosystems, Foster City, CA, USA). PCR was performed to
amplify codons 12 and 13 of exon 1 in KRAS using specific
primers under the PCR conditions described previously (15). The efficiency and quality of the
amplification PCR were confirmed by running the PCR products on a
2% agarose gel. A negative control containing all the components of
the PCR except the template was included in each PCR. DNA amplified
products were purified using a QIAquick DNA clean up kit (Qiagen),
according to the manufacturer’s instructions. Amplification
products were subjected to direct sequencing using the same primers
and all mutations were confirmed by sequences originating from both
the upstream and downstream primers on ABI 3130 Genetic Analyser
(Applied Biosystems, Foster City, CA, USA). The presence of a
mutation was accepted when its chromatographic peak height was 25%
or higher than the peak of the wild-type reference.
Statistical differences were analyzed using a
student’s t-test and P<0.05 was considered to indicate a
statistically significant difference.
Results
Patient characteristics
Specimens from 100 tumors were retrospectively
analyzed for the presence of KRAS mutations in codons 12 and
13. The study included almost equal numbers of males and females
(Table I). The median age at
diagnosis was 55 years. The most common metastatic site was the
liver (70% of patients). The primary tumor site was the colon NOS
(not otherwise specified) in 58% of patients, the rectum in 22% and
20% were considered to be rectosigmoidal. All cancers were
adenocarcinomas and were graded according to WHO criteria.
 | Table I.Characteristics of the 100 patients
enrolled in the study. |
Table I.
Characteristics of the 100 patients
enrolled in the study.
| Characteristic | Value |
|---|
| Total number of
tumors | 100 |
| Gender | |
| Female | 45 |
| Male | 55 |
| Median age at
diagnosis (range) | 55 (22–74
years) |
| Primary tumor
site | |
| Colon, NOS | 58 |
| Rectum | 22 |
| Rectosigmoid | 20 |
| Metastatic site at
diagnosis | |
| Liver | 70 |
| Lung | 17 |
| None | 13 |
| TNM stage at
diagnosis | |
| 1 | 0 |
| 2 | 5 |
| 3 | 8 |
| 4 | 87 |
Prevalence of KRAS subtype mutations in
Jordan
Of the 100 tumors included in this study, 44%
harbored KRAS mutations in either codon 12 or 13 (Fig. 1). Of the majority of KRAS
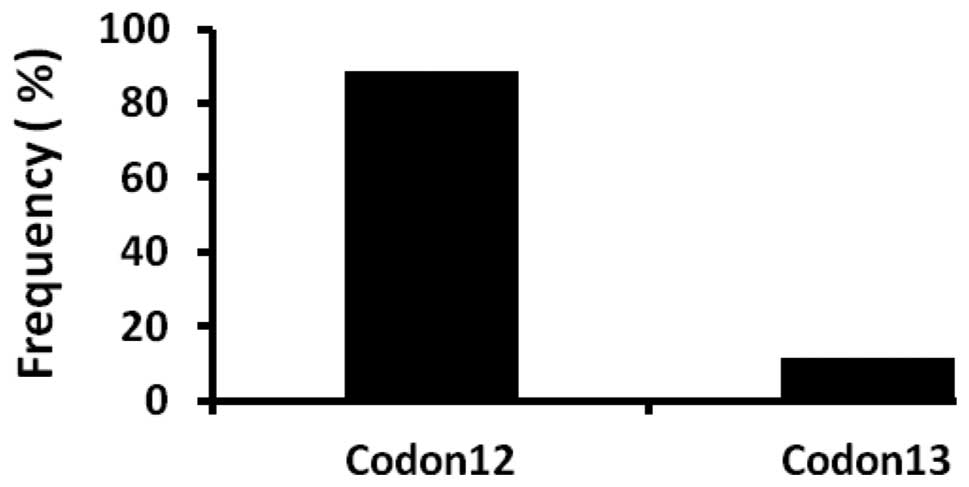
mutations, 39 (89%) were identified in codon 12, while codon 13 was
involved in 5 (11%) tumors (Fig.
2). Of the 39 mutations in codon 12 (wild-type GGT), 25 (62.5%)
were transition mutations, of which GAT (55%) was the most common
and 15 (37.5%) were transversion mutations, of which GTT (14%) was
the most frequent; in codon 13 (wild-type GGC), only GAC
transitions were present (Fig. 3).
In one tumor, two mutations were identified in codon 12, each with
a transition and transversion mutational type (Gly to Asp and Gly
to Cys). Several positive samples were randomly selected to confirm
the detected mutation(s) by sequencing. A summary of all molecular
types of KRAS mutations is shown in Table II.
 | Table II.Spectrum of KRAS mutations in
100 colorectal cancers. |
Table II.
Spectrum of KRAS mutations in
100 colorectal cancers.
| KRAS subtype
mutation | No. | % |
|---|
| pGly12Asp; codon 12
GGT>GAT | 24 | 54.5 |
| pGly12Val; codon 12
GGT>GTT | 6 | 13.6 |
| pGly12Cys; codon 12
GGT>TGT | 5 | 11.4 |
| pGly12Ala; codon 12
GGT>GCT | 2 | 4.5 |
| pGly12Arg; codon 12
GGT>CGT | 2 | 4.5 |
| pGly12Ser; codon 12
GGT>AGT | 1 | 2.3 |
| pGly12Leu; codon 12
GGT>CTT | 0 | 0 |
| pGly12Ile; codon 12
GGT>ATT | 0 | 0 |
| pGly13Asp; codon 13
GGC>GAC | 5 | 11.4 |
| pGly13Cys; codon 13
GGC >TGT | 0 | 0 |
Correlation of molecular findings with
clinical and demographic data
KRAS mutations (codon 12 or 13) did not show
any significant correlation with tumor location, stage, age at
onset or gender of the patient (Table
III).
 | Table III.Correlation between KRAS
mutational status and tumoral variables. |
Table III.
Correlation between KRAS
mutational status and tumoral variables.
| Characteristic | Mutated KRAS
| wt KRAS
| P-value |
|---|
| No. | % | No. | % |
|---|
| Gender | | | | | 1.000 |
| Female | 22 | 50 | 23 | 41 | |
| Male | 22 | 50 | 33 | 59 | |
| Age (years) | | | | | 1.000 |
| >60 | 11 | 25 | 22 | 39 | |
| <60 | 33 | 75 | 34 | 61 | |
| Tumor location | | | | | 1.000 |
| Colon, NOS | 58 | 58 | 20 | 67 | |
| Rectum | 22 | 22 | 5 | 17 | |
| Rectosigmoid | 20 | 20 | 5 | 16 | |
| Primary tumor
stage | | | | | 1.000 |
| I | 0 | 0 | 0 | 0 | |
| II | 4 | 9 | 3 | 5 | |
| III | 5 | 11 | 11 | 20 | |
| IV | 35 | 80 | 42 | 75 | |
KRAS mutations and prognosis
The response to standard therapy was documented for
only 51 patients in the KHCC Cancer Registry. This is a small pool
of data; however, findings are shared even if statistical analysis
was challenging. In the distribution of patients with either
wild-type or mutated KRAS, there is no association between
response to therapy and gene mutational status (Table IV). A complete response was observed
in one patient out of 25 with wild-type KRAS. Patients
(7) with partial responses included
3 KRAS wt and 4 KRAS mutants. A similar pattern was
also observed for patients with stable or progressive disease. Of
the patients with the mutated KRAS gene, 73% had progressive
disease compared with 68% of patients with the wild-type gene.
 | Table IV.Association of KRAS mutational
status with treatment outcomes. |
Table IV.
Association of KRAS mutational
status with treatment outcomes.
| Response to
therapy | Mutated KRAS
| wt KRAS
|
|---|
| No. | % | No. | % |
|---|
| Complete
response | 0 | 0 | 1 | 4 |
| Partial
response | 4 | 15.5 | 3 | 12 |
| Stable disease | 3 | 11.5 | 4 | 16 |
| Progressive
disease | 19 | 73 | 17 | 68 |
Prevalence of KRAS mutations in Jordan
and other countries
A review of studies published from various countries
concerning the prevalence of KRAS mutations in colorectal
tumors is shown in Table V
(16–21). In most countries, the KRAS
mutation rate ranged from 18–47%, as identified in Egypt and the
United States, respectively. Thus, the prevalence rate identified
in Jordan (44%) is at the higher end of this range. In addition,
the prevalence of KRAS mutations in Jordanian patients was
markedly higher than its two closest neighbors, Saudi Arabia and
Turkey (28 and 37.5%, respectively). However, the fraction of
mutations revealed in codon 12 and 13 of Jordanian CRC patients was
similar to all other countries, with the exception of the United
States.
 | Table V.Prevalence of KRAS mutations
in Jordan and other countries. |
Table V.
Prevalence of KRAS mutations
in Jordan and other countries.
| Jordan | Saudi Arabia | Egypt | USA | Brazil | Turkey | Spain | Slovenia |
|---|
| KRAS mutated
tumors (%) | 44 | 28 | 18 | 47 | 35 | 37.5 | 34 | 45.5 |
| Mutated in codon 12
(%) | 89 | 81 | NA | 96 | 85 | 82 | 84 | 81 |
| Mutated in codon 13
(%) | 11 | 19 | NA | 4 | 15 | 18 | 16 | 19 |
Discussion
CRC is one of the leading causes of mortality due to
cancer in Jordan. The poor prognosis of this disease would be
ameliorated if curative surgery was performed in its early stages.
This study analyzed KRAS mutations in CRCs of Jordanian
patients, in whom CRC incidence and mortality is one of the highest
in the Middle East and is still increasing (2). The incidence of KRAS mutations
in Jordan was 44% and these mutations were predominantly observed
in codon 12 (89%). These results, from a series of 100 patients,
are in accordance with a modern series conducted worldwide.
KRAS codon 12 and 13 mutations cover 98% of the entire
KRAS mutation spectrum in CRC (22,23).
Thus, this analysis did not include other codons, including 61
(exon 3) or 146 (exon 4) due to their infrequency in CRC.
To assess the specificity of both the TheraScreen
DxS KRAS Mutation Kit and Vienna Lab methods used to analyze
KRAS status in this study, the concordance of test results
was evaluated for retrospective samples with the results of the
Sanger sequencing method. The real-time PCR results (DxS kit) and
allele-specific oligonucleotide hybridization results (Vienna Lab)
were in 100% concordance when compared with the Sanger sequencing
method. A number of comparative studies have evaluated the
performance of the various methods used to accurately characterize
KRAS gene status (24,25).
The majority of these studies agree that the DxS and Vienna Lab
kits are equivalent and more reliable due to their higher
sensitivity than Sanger sequencing (24). The DxS kit tests for the seven most
common mutations in codon 12 and 13 of KRAS and Vienna Lab
detects 10 mutations. A significant number (44%) of KRAS
mutations were detected using these two kits which may account for
a large number of mutations in the Jordanian population. Thus, the
detection methods utilized in this study are considered to produce
an accurate frequency of KRAS mutations.
The frequency and spectrum of KRAS mutations
did not differ when compared with those of most other studies
(16–21). The distribution of the seven tested
mutations (p.Gly12Asp, 54.5%; p.Gly12Val, 13.6%; p.Gly12Cys, 11.4%;
p.Gly12Ala, 4.5%; p.Gly12Arg, 4.5%; pGly12Ser, 2.3% and p.Gly13Asp,
11.4%) among the mutated KRAS patients is in accordance with
published data (21). The findings
of this study suggest that the frequency of KRAS mutations
in Jordan is similar to those in European countries and the United
States. However, Jordanian KRAS mutation data contrasted
sharply with the neighboring countries of Saudi Arabia and Egypt. A
study concerning KRAS mutation status in Saudi Arabia
reported a significantly lower frequency (28%) than its neighboring
Jordan (19). An Egyptian study
reported that mutations of the KRAS proto-oncogene is
uncommon (18%) in Egyptian CRC in contrast to Western cases and was
not identified in any patients under the age of 40 years (26). Although Arabic countries share
certain cultural background and environmental exposures, these
findings reveal possible molecular genetic determinants playing a
role in KRAS gene mutation. Thus, ethnicity and geographical
differences should be considered in designing future clinical
trials. Overall, the frequency and spectrum of KRAS
mutations did not differ when compared to the majority of other
reports, possibly due to the nature of mutations in the KRAS
gene giving the tumor cell a growth advantage leading to clonal
selection.
In this study, no statistically significant
difference between KRAS positivity rate and the
clinicopathological findings was observed. Certain studies have
reported a higher frequency of KRAS mutations in females
compared with males (18,27). This study had an almost equal ratio
of males to females but identified no statistical difference in
KRAS mutation with respect to gender. Tumor stage revealed
no correlation with mutation status, which is in agreement with
published studies (17,28).
The association between KRAS mutational
status and prognosis remains controversial for patients with
metastatic CRC that have not been treated with anti-EGFR
antibodies. While certain studies reported a link between
KRAS mutations and poor prognosis (15,17),
others have identified no association (29). The biggest clinical trial designed
to analyze the prognostic value of KRAS status was the
RASCAL study, which revealed that a glycine-to-valine mutation in
codon 12 increased the risk of recurrence and mortality by 30%,
irrespective of the type of therapy administered (29). A smaller scale study from Spain
published findings that agreed with the RASCAL study on the poor
prognosis for patients with KRAS-mutated primary tumors.
However, the Spanish group revealed contrasting results to the
RASCAL study by declaring that the mutation type did not affect
prognosis (17). One limitation in
the current study was the unavailability of information concerning
treatment outcomes since the study was in retrospect. Only 51
patients had available information concerning response to therapy
documented in the KHCC Cancer Registry and of those only 26 were
KRAS-mutated patients. Thus, we were unable to perform
analysis to deduce whether an association existed among
disease-free, progression-free or overall survival rates and
KRAS mutational status or type. However, the available data
for the group of patients with stable disease provide inconclusive
evidence as the KRAS mutational status is negative for
almost half of the patients. Additionally, among the 36 patients
who were progressing and did not respond to therapy, 17 were
KRAS-wt and 19 harbored KRAS mutations. A similar
pattern of results was also reported by Licar et al
suggesting other molecular elements of response require
identification (21).
Information from a larger scale future study in
Jordan will either confirm or refute that the presence of
activating mutations in the KRAS gene reveals a poor
prognostic group and non-responding patients to anti-EGFR
antibodies. Further investigation will be directed at how Food and
Drug Administration agencies handle KRAS mutational analysis
prior to targeted drug administration to CRC patients.
Increasing efforts are exerted to assess individual
specific molecular alterations for personalized diagnosis,
prognosis and/or treatment. Future studies should be of larger
sizes and any existing concordance between KRAS mutations of
primary and metastatic tumors from patients with CRC requires
identification. A number of previous studies have reported a high
degree of concordance in KRAS mutational status between
primary tumors and their related liver metastases (17,30–32).
The liver and lung are common sites of CRC metastases, however,
this study revealed liver metastases to be more common (70%).
Determining the degree of concordance is critical from three
aspects. First, the frequency of KRAS mutations in primary
and secondary tumors in patients needs to be compared with
published studies to determine whether the acquired data are in
agreement. Second, evidence is required to further support previous
studies that KRAS mutations occur early in carcinogenesis
(33). Third, evaluation of the
mutational status of KRAS may be performed from a metastatic
site in the case of a primary tumor sample being unavailable.
In summary, KRAS mutation and subtyping
analysis of CRCs in Jordanian patients confirmed the data from
other studies but also yielded potentially new recommendations. The
results of this study will have a major impact on disease
management as the cost of treating metastatic CRC will be reduced
by millions of Jordanian Dinars a year, if all patients were tested
for KRAS mutations. Using KRAS testing to restrict
the use of EGFR-inhibitor therapy to patients with wild-type
KRAS tumors would avoid the administration of unnecessary,
ineffective and toxic treatments to patients with KRAS
mutations who would not benefit from them.
Acknowledgements
The authors would like to thank Selena
Audeh, Roubi Abu Obeid, Firas Zaitar and Nadim Musleh for their
kind assistance.
References
|
1.
|
A JemalR SiegelE WardY HaoJ XuT MurrayMJ
ThunCancer statistics, 2008CA Cancer J
Clin587196200810.3322/CA.2007.0010
|
|
2.
|
M TarawnehO NimriK ArkoobM Al ZhagalCancer
Incidence in Jordan annual report, 2009
|
|
3.
|
C PennaB NordlingerColorectal metastasis
(liver and lung)Surg Clin North
Am8210751090200210.1016/S0039-6109(02)00051-812507210
|
|
4.
|
JH Van KriekenA JungT KirchnerKRAS
mutation testing for predicting response to anti-EGFR therapy for
colorectal carcinoma: proposal for an European quality assurance
programVirchows Arch453417431200818802721
|
|
5.
|
K FransėnM KlintenäsA OsterströmMutation
analysis of the BRAF, ARAF and RAF-1 genes in human colorectal
adenocarcinomasCarcinogenesis25527533200414688025
|
|
6.
|
DJ JonkerCJ O’CallaghanCS
KarapetisCetuximab for the treatment of colorectal cancerN Engl J
Med35720402048200710.1056/NEJMoa07183418003960
|
|
7.
|
E Van CutsemS SienaY HumbletAn open-label,
single-arm study assessing safety and efficacy of panitumumab in
patients with metastatic colorectal cancer refractory to standard
chemotherapyAnn Oncol199298200817785764
|
|
8.
|
RG AmadoM WolfM PeetersE Van
CutsemWild-type KRAS is required for panitumumab efficacy in
patients with metastatic colorectal cancerJ Clin
Oncol2616261634200810.1200/JCO.2007.14.711618316791
|
|
9.
|
S Khambata-FordCR GarrettNJ
MeropolExpression of epiregulin and amphiregulin and K-ras mutation
status predict disease control in metastatic colorectal cancer
patients treated with cetuximabJ Clin
Oncol2532303237200710.1200/JCO.2006.10.543717664471
|
|
10.
|
CS KarapetisS Khambata-FordDJ JonkerK-ras
mutations and benefit from cetuximab in advanced colorectal cancerN
Engl J Med35917571765200810.1056/NEJMoa080438518946061
|
|
11.
|
M ZenkerK LehmannAL SchulzExpansion of the
genotypic and phenotypic spectrum in patients with KRAS germline
mutationsJ Med Genet44131135200710.1136/jmg.2006.04630017056636
|
|
12.
|
R WongD CunninghamUsing predictive
biomarkers to select patients with advanced colorectal cancer for
treatment with epidermal growth factor receptor antibodiesJ Clin
Oncol2656685670200810.1200/JCO.2008.19.502419001346
|
|
13.
|
T MinamotoM MaiZ RonaiK-ras mutation:
early detection in molecular diagnosis and risk assessment of
colorectal, pancreas, and lung cancers - a reviewCancer Detect
Prev24112200010757118
|
|
14.
|
D CalistriC RengucciI SeymourMutation
analysis of p53, K-ras, and BRAF genes in colorectal cancer
progressionJ Cell Physiol204484488200510.1002/jcp.2031015702478
|
|
15.
|
A LièvreJB BachetD Le CorreKRAS mutation
status is predictive of response to cetuximab therapy in colorectal
cancerCancer Res6639923995200616618717
|
|
16.
|
S EdkinsS O’MearaA ParkerRecurrent KRAS
codon 146 mutations in human colorectal cancerCancer Biol
Ther5928932200610.4161/cbt.5.8.325116969076
|
|
17.
|
P CejasM López-GómezC AguayoKRAS mutations
in primary colorectal cancer tumors and related metastases: a
potential role in prediction of lung metastasisPLoS
One4e8199200910.1371/journal.pone.000819920020061
|
|
18.
|
CG FerreiraI Zalcberg-RenaultFM VieiraMH
BonaminoM ZalisAnalysis of KRAS mutations in colorectal cancer
(CRC) patients by gender in a Brazilian cohort of 3,346 patientsJ
Clin Oncol2815s2010
|
|
19.
|
J AbubakerP BaviW Al-HaqawiPrognostic
significance of alterations in KRAS isoforms KRAS-4A/4B and KRAS
mutations in colorectal carcinomaJ
Pathol219435445200910.1002/path.262519824059
|
|
20.
|
M ConzelmannU LinnemannMR BergerK-ras
codon 12 and 13 mutations are correlated with differential patterns
of tumor cell dissemination in colorectal cancer patientsInt J
Oncol2415371544200415138598
|
|
21.
|
A LicarP CerkovnikJ OcvirkS NovakovicKRAS
mutations in Slovene patients with colorectal cancer: frequency,
distribution and correlation with the response to treatmentInt J
Oncol3611371144201020372787
|
|
22.
|
ER FearonMolecular genetic studies of the
adenoma-carcinoma sequenceAdv Intern Med3912314719948140953
|
|
23.
|
P ShawS TardyE BenitoOccurrence of Ki-ras
and p53 mutations in primary colorectal
tumorsOncogene62121212819911945416
|
|
24.
|
M Herreros-VillanuevaG AggarwalKRAS assay
selection: sensitivity and accuracy in clinical applicationMol Biol
Rep3924672470201210.1007/s11033-011-0997-621656377
|
|
25.
|
P PintoP RochaI VeigaComparison of
methodologies for KRAS mutation detection in metastatic colorectal
cancerCancer
Genet20443946201110.1016/j.cancergen.2011.07.00321962894
|
|
26.
|
AS SolimanML BondySA El-BadawyContrasting
molecular pathology of colorectal carcinoma in Egyptian and Western
patientsBr J Cancer8510371046200110.1054/bjoc.2001.183811592777
|
|
27.
|
T NagasakaH SasamotoK NotoharaColorectal
cancer with mutation in BRAF, KRAS, and wild-type with respect to
both oncogenes showing different patterns of DNA methylationJ Clin
Oncol2245844594200410.1200/JCO.2004.02.15415542810
|
|
28.
|
N SharmaM SaifoI TamaskarR BhuvaneswariT
MashtareM FakihKRAS status and clinical outcome in metastatic
colorectal cancer patients treated with first-line FOLFOX
chemotherapyJ Gastrointest Oncol19096201022811812
|
|
29.
|
HJ AndreyevAR NormanD CunninghamKirsten
ras mutations in patients with colorectal cancer: the ‘RASCAL II’
studyBr J Cancer856926962001
|
|
30.
|
D SantiniF LoupakisB VincenziHigh
concordance of KRAS status between primary colorectal tumors and
related metastatic sites: implications for clinical
practiceOncologist1312701275200810.1634/theoncologist.2008-018119056857
|
|
31.
|
MC Etienne-GrimaldiJL FormentoM
FrancoualK-Ras mutations and treatment outcome in colorectal cancer
patients receiving exclusive fluoropyrimidine therapyClin Cancer
Res1448304835200810.1158/1078-0432.CCR-07-4906
|
|
32.
|
S ArtaleA Sartore-BianchiSM
VeroneseMutations of KRAS and BRAF in primary and matched
metastatic sites of colorectal cancerJ Clin
Oncol2642174219200810.1200/JCO.2008.18.728618757341
|
|
33.
|
B VogelsteinER FearonSR HamiltonGenetic
alterations during colorectal-tumor developmentN Engl J
Med319525532198810.1056/NEJM1988090131909012841597
|