Introduction
Dimerization is essential for transmembrane KIT
receptor activity (1–6). KIT is a receptor tyrosine kinase (RTK)
which belongs to the class III RTK family. This subclass is
characterized by an extracellular region consisting of five
immunoglobulin (Ig)-like domains, a single transmembrane domain and
an intracellular tyrosine kinase domain split in two by an insert
region (7). There are two forms of
KIT receptor: wild-type (145 kDa) and mutant-type (125 kDa)
(8–10). Once interactions between KIT
receptors and specific stromal ligands (stem cell factor, SCF)
occur, the formation of various homodimers or heterodimers and
receptor cross-talk govern the activation of specific intracellular
signaling pathways, including mitogen-activated protein kinase
(MAPK) and phosphatidylinositol 3-kinase (PI3K)/Akt (11,12).
Dysregulation of the complex KIT signaling network is known to be
associated with malignant transformation and cancer progression
(13–16). Previous studies (16–19)
have demonstrated that KIT receptor dimerization mediates
intracellular signaling events, leading to cancer cell
proliferation, survival and resistance to anticancer therapy in
tumor cells associated with KIT dysregulation.
Although gastrointestinal stromal tumors (GISTs)
exhibit a spectrum of biological behaviors from benign to
malignant, the molecular mechanism of tumor progression has not
been fully clarified. Previous studies have reported that the
phosphorylation of KIT is one of the key steps in the growth and
metastasis of GISTs. The phosphorylation of KIT is induced by KIT
dimerization. The KIT-dimer plays a major role in promoting GISTs
(16–19). However, there has been no study
concerning the associations between KIT-dimer expression,
clinicopathological features, c-kit mutation types and SCF
expression. In a previous study, Native-PAGE was used to detect
KIT-dimers to preserve the noncovalent binding, however, this
failed to obtain the bands obtained by SDS gel electrophoresis,
where separation was based on size alone. In order to further
confine the KIT-dimers in tissue, modifications were made to
Native-PAGE and correlations between the clinicopathological
factors and KIT-dimers were analyzed further.
Materials and methods
Human tissues
A total of 49 GIST patients who underwent surgery at
Changhai Hospital (Shanghai, China) were examined. The GIST
diagnosis was confirmed by two pathologists (Bai CG and Ma DL). No
patients had received imatinib prior to surgical resection of the
tumor. The use of all human tissues was approved by the
Institutional Committee for Human Research of the hospital and
informed consent was obtained from all subjects. Demographic data
and clinical and histological features of all GISTs analyzed in
this study are summarized in Table
I.
 | Table I.Correlation between KIT-dimer
expression and clinicopathological features, including c-kit
mutations of GISTs. |
Table I.
Correlation between KIT-dimer
expression and clinicopathological features, including c-kit
mutations of GISTs.
| GIST KIT-dimer
expression
| |
|---|
| Variable | Positive | Negative | P-value |
|---|
| Total | 15 | 34 | |
| Mean age
(years) | 59.21 ± 13.84 | 61.27 ± 12.61 | 0.621 (T-test) |
| Gender | | | |
| Male | 8/15 | 20/34 | 0.480 |
| Female | 7/15 | 14/34 | |
| Tumor diameter
(cm) | | | |
| <5 | 4/15 | 16/34 | <0.001 |
| 5–<10 | 3/15 | 17/34 | |
| ≥10 | 8/15 | 1/34 | |
| Localization of
primary tumor | | | |
| Stomach | 9/15 | 25/34 | 0.208 (sto vs.
smbo) |
| Small bowel | 5/15 | 6/34 | |
| Others | 1/15 | 3/34 | |
| Mitotic rate (/50
HPF) | | | |
| <10 | 7/15 | 30/34 | 0.040 |
| ≥10 | 8/15 | 4/34 | |
| Ki-67 index
(%) | | | |
| <10 | 8/15 | 28/34 | 0.041 |
| ≥10 | 7/15 | 6/34 | |
| Risk of
malignancy | | | |
| M+L+VL | 5/15 | 28/34 | 0.001 (M+L+VL vs.
H) |
| H | 10/15 | 6/34 | |
| SCF expression | | | |
| Positive | 11/15 | 24/34 | 0.566 |
| Negative | 4/15 | 10/34 | |
| Mutation | | | |
|
c-kit-positive | 10/15 | 25/33 | 0.373 |
|
c-kit-negative | 5/15 | 8/33 | |
| Exon 11 | 9/10 | 20/25 | 0.436 |
| Exon 9 | 1/10 | 5/25 | |
Immunohistochemical analysis
Immunohistochemical staining of anti-SCF (rabbit
monoclonal, Abcam, Cambridge, MA, USA; 1:50 dilution), anti-KIT
(rabbit polyclonal, Dako, Glostrup, Denmark; 1:500 dilution) and
anti-ki-67 (mouse monoclonal, Dako; 1:200 dilution) was performed
using EnVision™ systems (Dako) according to the manufacturer’s
instruction.
The results of the ki-67 labeling index
immunohistochemical staining were evaluated as follows:
ki-67-positive was defined as positive staining of ≥5% of the tumor
cell nucleus. An assessment was performed by estimating the rate of
positive cells in 5 consecutive fields (magnification: x400). The
intensity of tissue staining was graded semi-quantitatively on a
four-point scale (−, +, ++, +++) and the proportion of stained
cells was calculated on a four-tier scale (<5%, 5–<10%,
10–<15% and ≥15%). In brief, 4-μm-tissue sections were
deparaffinized and boiled in 10% citric acid buffer solution (pH
6.0) for 20 min for antigen retrieval. After blocking the
endogenous peroxidase activity with 3% hydrogen peroxide, the
sections were incubated with primary antibodies and then with
secondary antibodies conjugated with peroxidase-labeled dextran
polymer. Staining was detected with diaminobenzidine (DAB) as
chromogen and counterstained with hematoxylin prior to
coverslipping. The results of the routine immunohistochemical
diagnostics were included in the statistical analysis.
C-kit mutation analysis
The genomic DNA of fresh tissues was extracted using
standard protenase-K digestion/phenol-chloroform procedures. Exons
11 and 9 of the c-kit gene were amplified by PCR using 2 μl
DNA solution, 25 μl 2X Taq PCR MasterMix, 1 μl each
primer (10 μmol/l) in a final volume of 50 μl. C-kit
exons 9 and 11 were amplified using the following designed primer
sequences and annealing temperatures: exon 9 (forward,
5′-GCCACATCCCAAGTGTTTTATG-3′ and reverse,
5′-GAGCCTAAACATCCCCTTAAATTG-3′ at 60°C), exon 11 (forward,
5′-CCAGAGTGCTCTAATGACTG-3′ and reverse, 5′-AGCCCCTGTTTCATACTGAC-3′
at 56°C). A total of 38 amplification cycles, consisting of
denaturation at 9°C for 30 sec, annealing at 60 or 56°C for 30 sec
and extension at 72°C for 1 min, were performed in PCR systems
(Thermo Scientific, Barrington, IL, USA). PCR products were used
directly for sequencing analysis.
Qualification and detection of
KIT-dimers
In a pilot study, total frozen GIST samples were
calibrated and homogenized in lysis buffer (20 mM Tris, 150 mM
NaCl, 1 mM othovanadate, 10 mM NaF, 1 mM PMSF, 0.5 μg/ml
leupeptine, 1 μg/ml pepstatine, 10 K IU/ml aprotinine, 1%
triton X-100). Lysates were rocked at 4°C for 30 min and then
centrifuged at 12,000 rpm for 15 min. The supernatant protein
concentration was measured using a BAC protein assay kit (Merck
KGaA, Darmstadt, Germany). Each sample was prepared in triplicate.
One portion already in solution was diluted accordingly with
ordinary 5X sample buffer (10% SDS and 5% 2-mercaptothiazoline
obtained) and then analyzed by SDS-PAGE. Another portion was
diluted accordingly with 5X sample buffer (SDS and
2-mercaptothiazoline omitted) and then analyzed by Native-PAGE. The
third potion was diluted accordingly with 5X sample buffer (SDS and
2-mercaptothiazoline omitted) and then analyzed by SDS-PAGE.
Proteins were separated by 6% SDS-PAGE and transferred to a
nitrocellulose membrane, which was then blocked for 15 h at 4°C
temperature with 5% BSA and then reacted with anti-KIT primary
antibody (Dako) for 2 h at 37°C. Peroxidase-labeled anti-rabbit IgG
was used as the second antibody for 1 h at 37°C. The western
lightening chemiluminescence reagent (Santa Cruz Biotechnology,
Inc., Santa Cruz, CA, USA) was used for the detection of
proteins.
Statistical analysis
An exploratory data analysis was performed using
SPSS 17.0 (SPSS Inc., Chicago, IL, USA) and Excel 2007 (Microsoft
Corporation, Redmond, WA, USA). All criteria were rated equally
significant, without the adjustment of P-values for multiple
testing. Tumor size and age were considered to be continuous
variables, while positivity for immunohistochemical markers, the
mitotic rate and gender were treated as categorical variables.
P<0.05 was considered to indicate statistically significant
diferences (α= 0.05). No correction for multiple testing was
performed.
Results
Clinicopathological findings
A total of 49 GIST patients (28 men and 21 women)
underwent surgical resection for GISTs. Patients ranged in age from
29 to 84 years (mean, 63.3±11.8; median, 60). The tumors were
located in the stomach (34 cases, 69%), small intestine (11 cases,
22%), large intestine (1 case, 2%), omentum (1 case, 2%),
enterocoelia (1 case, 2%) and rectum (1 case, 2%). The tumors
varied between 0.2 and 32.0 cm (mean: 12.7 ± 8.4; median: 5.0) in
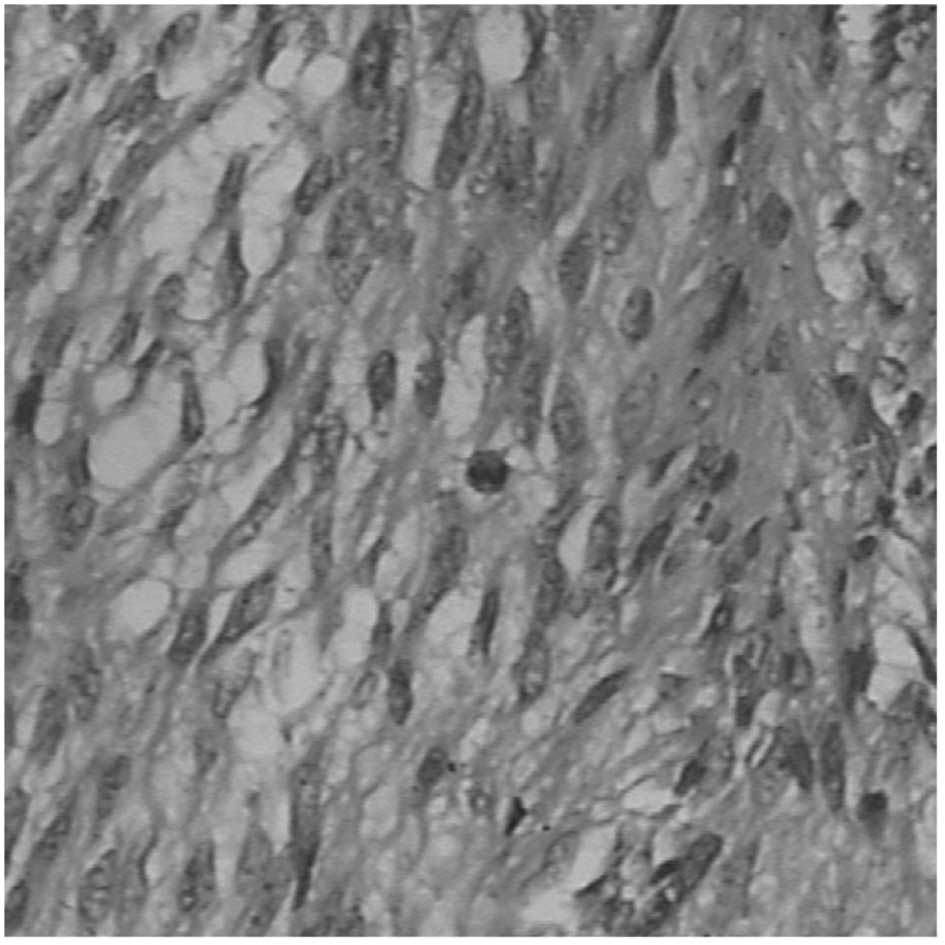
size. Histologically, 13 tumors were of the spindle-cell type,
seven tumors were of the epithelioid-cell type and the remaining 29
were of the mixed spindle- and epithelioid-cell type (Fig. 1). Mitotic counts in 24 cases were
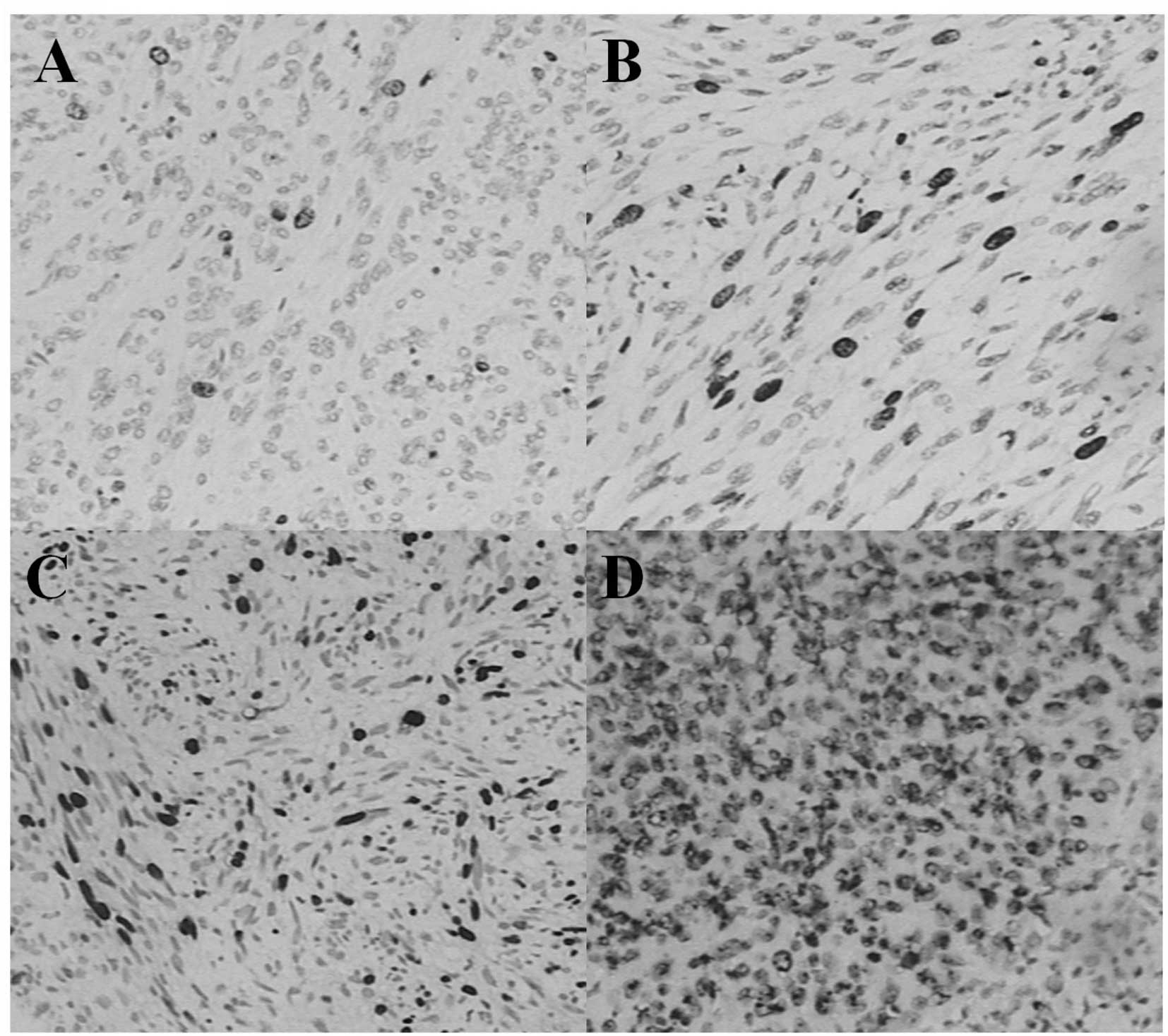
less than 5 per 50 high power field (HPF; 49%), 13 were 5–10 per 50
HPF (27%) and 12 were more than 10 per 50 HPF (24%; Fig. 2). Ki-67 LI was classified as ‘−’ in
21 cases (43%), ‘+’ in 15 cases (31%), ‘++’ in four cases (8%) and
‘+++’ in nine cases (18%) according to the previous evaluation
standards. According to the risk-grading system, one case was
classified as very low grade (2%), eight cases were classified as
low grade (16%), 22 as intermediate grade (45%) and 18 as high
grade (37%; Fig. 3). The
clinicopathological findings of the GISTs and major clinical
symptoms are summarized in Table
I.
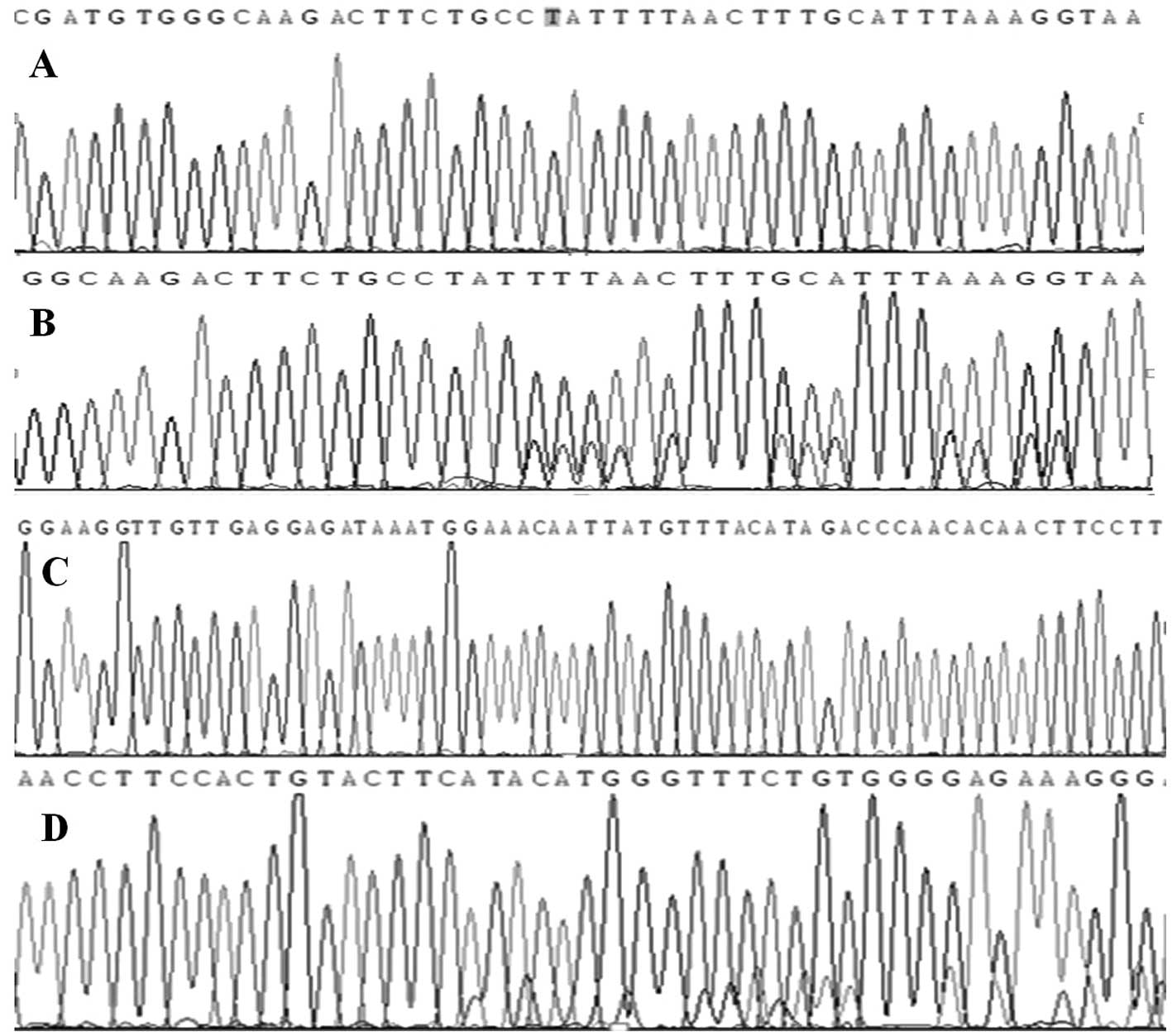
KIT mutations
Of the 48 GISTs analyzed, KIT exon 11 mutations were
detected in 29 cases (60%), KIT exon 9 mutations in 6 (13%) and
wild-type KIT in 13 (27%). The majority of the 29 tumors with exon
11 mutations were located between codons 550 and 570. The majority
of exon 9 mutations were insertions of six nucleotides, resulting
in duplications of amino-acid residues Ala 502-Tyr 503, which is
consistent with previous studies (Fig.
4).
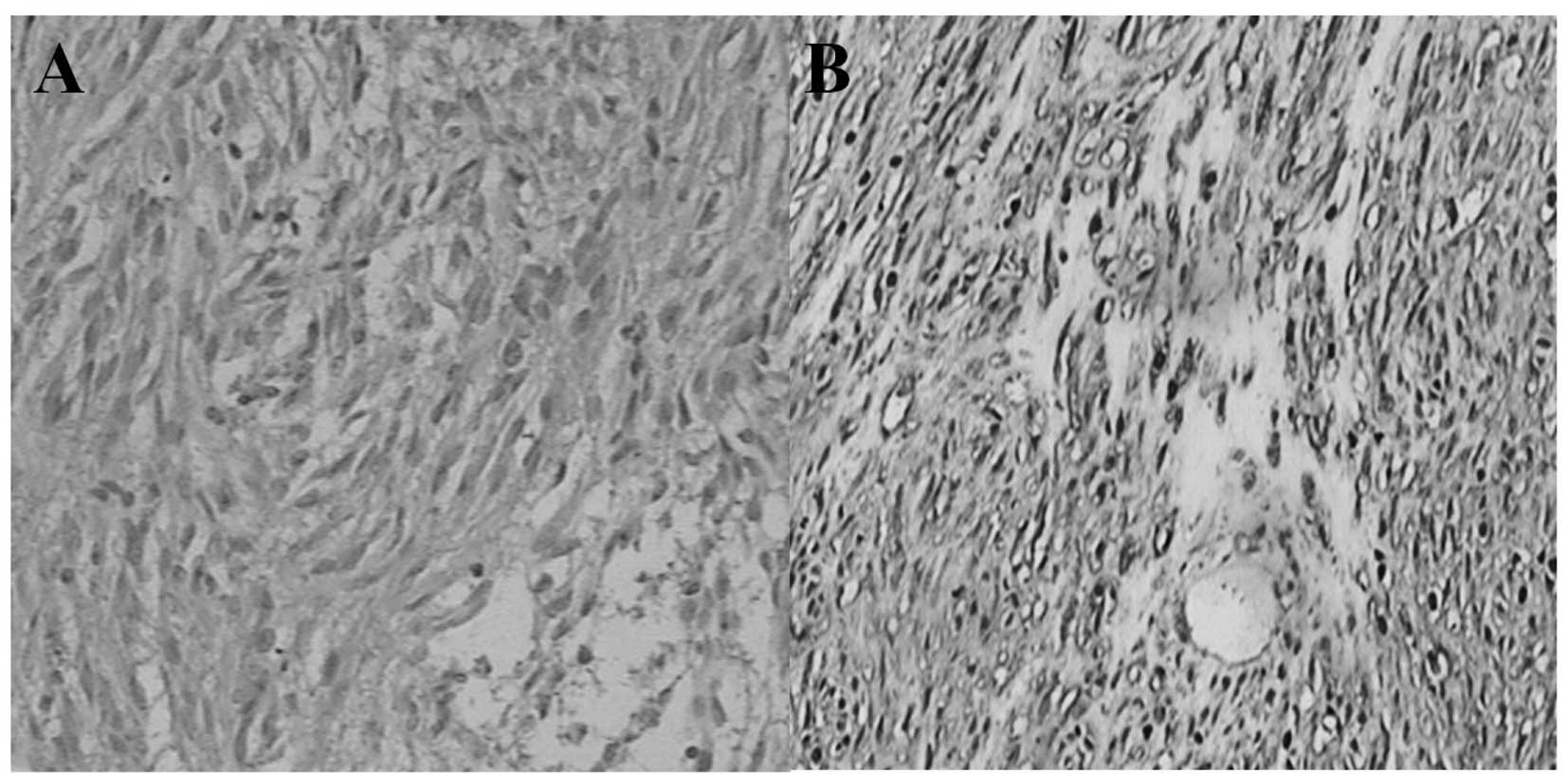
Expression of SCF in GIST tissue
The expression of SCF was detected by
immunohistochemical staining in all GIST samples. Positive SCF was
observed in 35 of the 49 (71%) GIST samples. The SCF-positive cases
presented moderate or strong staining in the membrane and plasma of
GISTs cells. Almost all of the GIST samples were KIT-positive (48
of 49, 98%). The KIT-positive cases presented diffusely moderate or
strong staining in the membrane and plasma of GIST cells (Fig. 5).
Expression of KIT-dimers in GIST
tissue
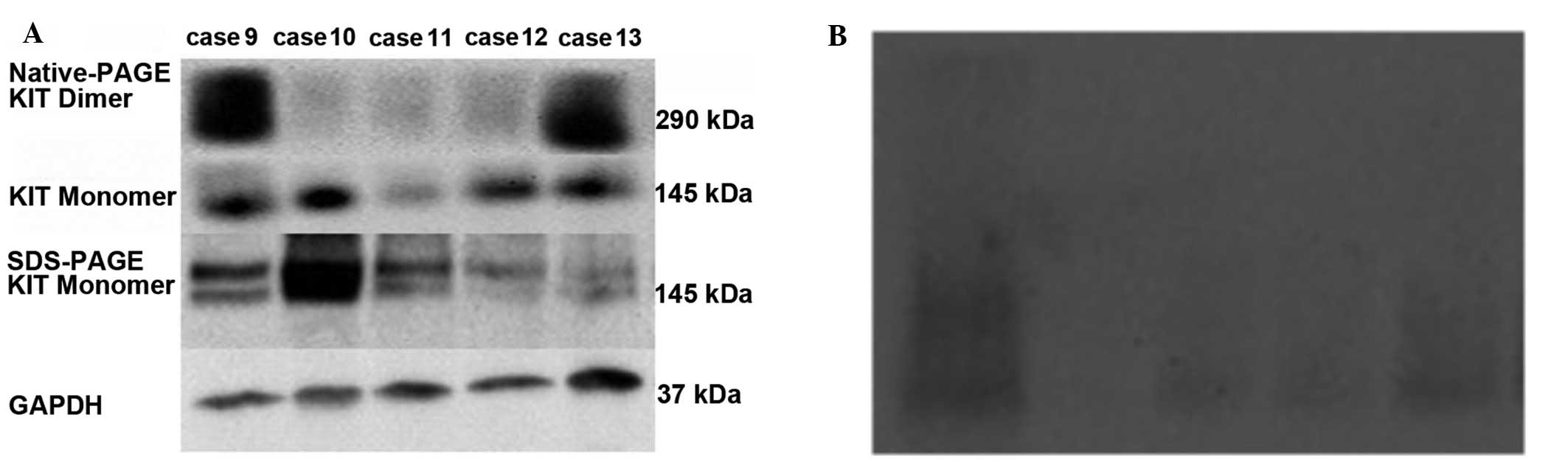
Signals for the KIT-dimer (molecular weight of
290/270 kDa) should be present above the band of 170 kDa on the
nitrocellulose membrane, while signals for KIT-monomers (molecular
weight of 145/125 kDa) should be present between the bands of 72
and 170 kDa. To detect the expression of KIT protein in GIST
tissues, all of the cases of the first portion were examined by
SDS-PAGE. As shown in Fig. 6A,
distinct bands were present at 72–170 kDa, confirming the presence
of KIT-monomers in tumor tissues (Fig.
6A). Two bands were obtained in certain cases, implying that
there were wild- and mutated-type KIT receptors in these GISTs,
whereas only one band was obtained in the other cases, implying
that there was one type of receptor present. However, no band was
present above 170 kDa, suggesting that the KIT-dimer was not
detected by SDS-PAGE, as the native crystal structure of
dimerization was destroyed by heating.
To further investigate the role of the KIT-dimer in
cellular events that occurred in GISTs, it is necessary to analyze
the non-denatured proteins. Bands were weakly present on the
nitrocellulose membrane by Native-PAGE (Fig. 6B) and thus it was difficult to
clarify the KIT-dimer expression, or identify the proteins of
various molecular weights since the marker was diffusive.
Finally, in order to determine the KIT-dimer in GIST
tissues without denaturing the crystal structure of the KIT-dimer,
certain methods were improved instead of using Native-PAGE in the
second portion. Notably, clear signals for the KIT-dimer (molecular
weight 290/270 kDa) were present above 170 kDa in 31% cases (15/49)
of the last portion (Table I),
while clear signals for the KIT-monomer (molecular weight of 145
kDa) were also observed in all tumor tissues (Fig. 6A).
Correlations between clinicopathological
factors and KIT-dimer expression
Since the dimerization of KIT receptors is essential
for the activation of specific intracellular signaling pathways,
including MAPK and PI3K/Akt, the clinicopathological and
immunohistochemical findings between the KIT-dimer-positive group
and the KIT-dimer-negative group were compared. It was revealed
that the tumor size of the KIT-dimer-positive patients was bigger
than that of the KIT-dimer-negative patients. KIT-dimer-positive
cases were more likely to belong to a higher risk classification
than KIT-dimer-negative cases (P<0.05, McNemar test; Table I). Statistical analysis demonstrated
that there was no significant difference in gender, age and tumor
location between the KIT-dimer-positive and KIT-dimer-negative
groups, indicating that there was no significant correlation
between gender, tumor location or age and the expression of
KIT-dimers (P>0.05; Table
I).
The association between KIT-dimer expression and
tumor proliferative activity was then examined. The Ki-67 index and
mitotic index were used to evaluate the potential of GIST cell
proliferation. The McNemar test indicated that the number of
mitotic cells in KIT-dimer-positive cases was significantly larger
than that in KIT-dimer-negative cases (P<0.05; Table I). There was also a significant
difference in the expression rate of Ki-67 between
KIT-dimer-positive and KIT-dimer-negative cases (P<0.05;
Table 1).
Correlations between c-kit mutations and
KIT-dimer expression
Since the autophosphorylation of KIT, one of the
mechanisms underlying KIT protein activation in GISTs, is due to
gain-of-function mutations of the c-kit gene, the KIT-dimer
expression between the c-kit exon 11 mutation group and the c-kit
exon 9 mutation group was compared, including the c-kit wild-type
gene. Correlations between c-kit mutations and KIT-dimer expression
were analyzed using the McNemar test. No correlation was revealed
between the presence of c-kit mutations and the expression of
KIT-dimers (P>0.05; Table
I).
Correlations between SCF expression and
KIT-dimer expression
Since the ligand-dependent mechanism is functionally
significant in the activation of the KIT protein, KIT-dimer
expression between the SCF-positive group and the SCF-negative
group was compared. The expression of the KIT-dimer was observed in
11 of 35 SCF-positive cases (31%) compared with 4 of 14
SCF-negative cases (29%). Notably, statistical analysis
demonstrated that there was no significant difference in the
expression rate of SCF between the KIT-dimer-positive cases and
KIT-dimer-negative cases (P>0.05, McNemar test; Table I).
Discussion
There are two mechanisms which underlie KIT protein
activation in malignant tumors. One is the autophosphorylation of
KIT due to gain-of-function mutations of the c-kit gene and the
second is ligand-dependent activation. Dimerization is essential to
the activation of KIT protein in all types of GISTs. In general,
KIT dimerization is thought to be mainly initiated by mutations in
the juxtamembrane domain of the c-kit gene in GISTs as previously
reported (8–10,21).
Previous clinical studies in vitro with imatinib resistance
demonstrated that SCF-dependent dimerization, without mutations in
the c-kit gene, is crucial to the activation of the GISTs signaling
pathway. In the current study, the expression of KIT-dimers and
monomers in GIST tissues was identified by western blot analysis.
Statistical analyses of the present study also indicated that there
was no significant correlation between KIT-dimer expression and
status of c-kit mutation. In addition, no significant correlation
was observed between KIT-dimer expression and SCF expression.
Finally, SCF expression and KIT-dimer expression were detected in
wild-type GISTs, while only KIT-dimer expression was detected in
mutated-type GISTs. The present study demonstrated that there was
no significant correlation between KIT-dimer expression, the status
of c-kit mutation and SCF expression, suggesting that
ligand-dependent SCF/KIT activation is an independent mechanism in
GISTs regardless of KIT autodimerization. With this unexpected
result, the expression of KIT-dimers was revealed to be
significantly associated with the tumor size, tumor grade mitotic
rate and Ki-67 labeling index, implying that KIT-dimer expression
is associated with high risk stratification and increases the risk
of GIST development.
Since KIT dimerization is not only the common
pathway of ligand-independent and ligand-dependent activation of
KIT, but relates to the action of RTKs in the survival and
proliferation of tumor cells, we infer that ligand-dependent
SCF/KIT dimerization is a considerable explanation for imatinib
resistance in wild-type GISTs, as it may only inhibit the
ligand-independent activation of KIT. Of note, previous clinical
studies (20,21) have confirmed that the expression of
SCF/KIT with no KIT mutations in uveal melanoma did not translate
into the clinical efficacy of imatinib. While receptor dimerization
is crucial for tumor proliferation, ligand-independent and
-dependent signaling are required to be switched off to achieve
complete inhibition of KIT activation. Previous studies have shown
dimercept and pertuzumab to be effective for the treatment of
breast carcinoma by intercepting the dimerization between the
epidermal growth factor receptor families (22,23).
Therefore, blocking a binding pocket necessary for receptor
dimerization might effectively inhibit tumor cell proliferation.
The previously mentioned results provide evidence that evaluation
of the KIT-dimer may be useful for predicting aggressive behavior
and assessment of tumor therapy.
In addition, SDS-PAGE is possibly the most commonly
used gel electrophoretic system for analyzing proteins. However, it
should be stressed that this method separates denatured proteins.
Occasionally, one is required to analyze native non-denatured
proteins, particularly when a protein in the gel is to be
identified with respect to its biological activity and binding
affinity. On such occasions it is necessary to use a non-denaturing
system. For example, the KIT-dimer may be present as a weaker (even
nonstaining) band in the gel and SDS-PAGE might destroy the
bivalency in the crystal structure of dimerization. The existence
of KIT-dimers may be proven once the major band is proven to be
active (24–28). Despite Native-PAGE conditions,
polypeptides retain their higher-order structures and interactions
with other polypeptides. We were unable to observe the presence of
the KIT receptor by native electrophoresis omitting the SDS. To the
best of our knowledge, although the function of KIT-dimers is
significant in tumorigenesis, few methods concerning how to detect
KIT-dimers have been reported. The present study introduced a
convenient method that may be used to directly detect the existence
of KIT-dimers in tissues. Since the SDS in electrophoresis buffer
has little effect on the native structure of a protein, the
standard electrophoresis buffer was used with SDS instead. It was
demonstrated that the native structure of a protein may be assayed
following electrophoresis. In order to obtain the previous result,
the gel must be prepared according to the standard Laemmli SDS
instructions, aside from excluding SDS or DTT from the sample
buffer and without heating the sample. It is advisable to run the
gel in a cold room, or circulate the buffer through a cooling coil
in ice. It is preferable to conduct the western-blotting transfer
at 200 mA for 140 min at 4°C. The pH of the TBST buffer should be
between 7.0 and 8.0.
In conclusion, the dimerization of KIT receptors
plays a crucial role in tumor cell proliferation, particularly in
increasing the risk of GIST development. Since there is no
significant association between KIT-dimer expression, the status of
c-kit mutation and SCF expression, blocking a binding pocket
necessary for receptor dimerization may be an effective treatment
for GISTs in the future. It appears viable to assess the inhibition
of KIT activation by detecting KIT-dimers.
References
|
1.
|
A UllrichJ SchlessingerSingnal
transduction by receptors with tyrosine kinase
activityCell61203212199010.1016/0092-8674(90)90801-K2158859
|
|
2.
|
Y YardenWJ KuangT Yang-FengL CoussensS
MunemitsuTJ DullHuman proto-oncogene c-kit: a new cell surface
receptor tyrosine kinase for an unidentified ligandEMBO
J63341335119872448137
|
|
3.
|
Y YardenJA EscobedoWJ KuangTJ Yang-FengTO
DanielPM TrembleStructure of the receptor for platelet-derived
growth factor helps define a family of closely related growth
factor receptorsNature323226232198610.1038/323226a03020426
|
|
4.
|
FH QiuHP RayY BrownPE BarkerS JhanwarFH
RuddleP BesmerPrimary structure of c-kit: relationship with the
CSF-1/PDGF receptor kinase family-oncogenic activation of v-kit
involves deletion of extracellular domain and C terminusEMBO
J71003111119882456920
|
|
5.
|
S BishayeeS MajunderJ KhireM DasLigand
induced dimerization of the platelet-derived growth factor
receptorJ Biol Chem264116991170519892545680
|
|
6.
|
P Blume-JensenL Claesson-WelshA SiegbahnKM
ZseboB WestermarkCH HeldinActivation of the human c-kit product by
ligand-induced dimerization mediates circular actin reorganization
and chemotaxisEMBO J10412141281991
|
|
7.
|
L CoussensC Van BeverenD SmithStructural
alteration of viral homologue of recap or proto-oncogene fms at
carboxyl terminusNature320277280198610.1038/320277a02421165
|
|
8.
|
AM TurnerLG BennettNL LinJ WypychTD
BartleyRW HuntHL AtkinsKE LangleyV ParkerF MartinVC
BroudyIdentification and characterization of a soluble form of the
c-kit receptorBlood83214521521995
|
|
9.
|
J WypychLG BennettMG SchwartzCL ClogstonHS
LuVC BroudyTD BartleyVP ParkerKE LangleySoluble kit receptor in
human serumBlood85667319957528574
|
|
10.
|
VC BroudyNL LinDF SabathThe fifth
immunoglobulin-like domain of the kit receptor is required for
proteolytic cleavage from the cell
surfaceCytokine15188195200110.1006/cyto.2001.090711563879
|
|
11.
|
LC CantleyThe phosphoinositide 3-kinase
pathwayScience29616551657200210.1126/science.296.5573.165512040186
|
|
12.
|
AD ReithC EllisSD LymanDM AndersonDE
WilliamsA BernsteinT PawsonSignal transduction by normal isoforms
and W mutant variants of the Kit receptor tyrosine kinaseEMBO
J102451245919911714377
|
|
13.
|
PS CohenJP ChanM LipkunskayaJL BiedlerRC
SeegerExpression of stem cell factor and c-kit in human
neuroblastoma. The Children’s Cancer GroupBlood84346534721994
|
|
14.
|
S HassanY KinoshitaC KawanamiK KishiY
MatsushimaA OhashiY FunasakaA OkadaT MaekawaW He-YaoT
ChibaExpression of proto-oncogene c-kit and its ligand stem cell
factor (SCF) in gastric carcinoma cell linesDig Dis
Sci43814199810.1023/A:10188514157049508539
|
|
15.
|
SJ HinesC OrganMJ KornsteinGW
KrystalCoexpression of the c-kit and stem cell factor genes in
breast carcinomasCell Growth Differ676977919957545433
|
|
16.
|
M InoueS KyoM FujitaT EnomotoG
KondohCoexpression of the c-kit receptor and the stem cell factor
in gynecological tumorsCancer Res543049305319947514496
|
|
17.
|
GW KrystalSJ HinesCP OrganAutocrine growth
of small cell lung cancer mediated by coexpression of c-kit and
stem cell factorCancer Res5637037619968542594
|
|
18.
|
T PietschU KyasU SteffensE YakisanMR
HadamWD LudwigK ZseboK WelteEffects of human stem cell factor
(c-kit ligand) on proliferation of myeloid leukemia cells:
heterogeneity in response and synergy with other hematopoietic
growth factorsBlood801199120619921381238
|
|
19.
|
M ToyotaY HinodaA TakaokaY MakiguchiT
TakahashiF ItohK ImaiA YachiExpression of c-kit and kit ligand in
human colon carcinoma cellsTumour
Biol14295302199310.1159/0002178427694350
|
|
20.
|
N Théou-AntonS TaboneD Brouty-BoyéCo
expression of SCF and KIT in gastrointestinal stromal tumours
(GISTs) suggests an autocrine/paracrine mechanismBr J
Cancer9411801185200616570044
|
|
21.
|
T NegriF BozziE ConcaOncogenic and
ligand-dependent activation of KIT/PDGFRA in surgical samples of
imatinib-treated gastrointestinal stromal tumours (GISTs)J
Pathol217103112200910.1002/path.245018973210
|
|
22.
|
T KoletsaI KostopoulosE CharalambousA
splice variant of HER2 corresponding to Herstatin is expressed in
the noncancerous breast and in breast
carcinomasNeoplasia10687696200818592003
|
|
23.
|
MC FranklinKD CareyFF VajdosInsights into
ErbB signaling from the structure of the ErbB2-pertuzumab
complexCancer
Cell4317328200410.1016/S1535-6108(04)00083-215093539
|
|
24.
|
CR ShawR PrasadGel electrophoresis of
enzymes, a compilation of recipesBiochem
Genet4297320197010.1007/BF004857804193186
|
|
25.
|
CR ShawAL KoenStarch gel zone
electrophoresis of enzymesChromatographic and Electrophoretic
TechniquesI Smith2HeinemannLondon, UK3323591968
|
|
26.
|
H HarrisDA HopkinsonHandbook of Enzyme
Electrophoresis in Human GeneticsNorth-HollandAmsterdam1976
|
|
27.
|
O GabrielLocating enzymes on gelsMethods
in EnzymologySP ColowickNO Kaplan22AcademicNew York,
NY578197110.1016/0076-6879(71)22042-5
|
|
28.
|
O GabrielDM GerstenStaining for enzymatic
activity after gel electrophoresisAnal
Biochem203121199210.1016/0003-2697(92)90036-71381872
|