Introduction
Tumor necrosis factor-related apoptosis-inducing
ligand (Apo2L/TRAIL), a type II transmembrane protein, is a member
of the TNF superfamily (1). In
humans, TRAIL binds to two death-inducing receptors, DR4/TRAIL-R1
and DR5/TRAIL-R2/KILLER. TRAIL has also been shown to bind three
decoy receptors, DcR1/TRID/TRAIL-R3, DcR2/TRAIL-R4 and a soluble
receptor osteoprotegerin (OPG) that was originally identified as a
receptor for OPGL/RANKL. The soluble form of TRAIL exhibits
apoptotic activity against various cancer cell lines with minimal
cytotoxicity toward normal tissues in vitro and in
vivo (2). Notable exceptions
are immature human and mouse dendritic cells (DCs) that are
sensitive to TRAIL-mediated apoptosis in vitro (3,4).
Ligands for the DRs, TNF and FasL, have been shown to induce
serious toxic effects following systemic administration (5,6). There
is also concern that certain rTRAIL variants may induce systemic
toxicity, highlighting the importance of preclinical assessment for
this ligand. Indeed, certain types of TRAIL show cytotoxicity to
normal cells. Polyhistidine-tagged recombinant human TRAIL has been
shown to induce apoptosis in normal human hepatocytes (5), recombinant human leucine zipper (LZ)-
and polyhistidine-tagged TRAIL have been shown to induce apoptosis
in normal keratinocytes (3,7), and recombinant LZ-TRAIL is cytotoxic
to human astrocytes in vitro (1). By contrast, other studies have
revealed that rTRAIL lacking exogenous sequences does not induce
apoptosis in normal human and cynomolgus monkey hepatocytes
(6), human mammary, renal or
prostatic epithelial cells, umbilical vein endothelial cells, lung
fibroblasts, colon smooth muscle cells, astrocytes or keratinocytes
(7–9). However, controversy remains concerning
which type of TRAIL is superior.
In this study, TRAIL-FT, which comprises TRAIL
(114–281aa) without any exogenous sequences, was expressed by a
prokaryotic expression system. Its identity was characterized and
its functions were analyzed in comparison with those of
TRAIL-HS, a tagged form of TRAIL (114–281aa) with a 45 aa exogenous
sequence including 6xHis-tag and S-tag.
This study was performed with the approval of the
ethical committee of Henan University, Henan, China.
Materials and methods
Construction and expression of TRAIL-HS
and TRAIL-FT
The primers were designed according to the cDNA
sequence of TRAIL provided in GenBank and synthesized by Invitrogen
Biotechnology Co., Ltd. (Shanghai, China). The primers were sense:
5′-CATGCCATGGTGAGAGAAAGAGGTCCTCAG-3′,
and anti-sense: 5′-TCCGCTCGAGCGGTTAGC CAACTAAC-3′. The
underlined sequences are NcoI and XhoI sites,
respectively. The total RNA of human PBMC was extracted using the
TRIzol reagent (Invitrogen Biotechnology Co., Ltd.). The cDNA was
synthesized using M-MLV reverse transcriptase (Takara Bio Inc.,
Shiga, Japan) with total RNA as the template. The TRAIL gene was
obtained by PCR amplification, cloned into pGEM-T-easy vector
(Promega, Madison, WI, USA) and sequenced. TRAIL extracellular gene
was digested from the sequencing vector and ligated to pET-30a and
pET-28a prokaryotic expression vectors with T4 ligase (New England
BioLabs, Inc., Ipswich, MA, USA). The TRAIL-HS and TRAIL-FT
proteins were expressed in E.coli BL21(DE3) induced by IPTG
(0.1 mM; Sigma, St. Louis, MO, USA) and purified by Ni-NTA and SP
column chromatography, respectively.
Western blotting
The two TRAIL proteins expressed in E.coli
BL21(DE3) were resolved by SDS-PAGE on 15% poly-acrylamide gels and
transferred to a nitrocellulose membrane using a horizontal
electrophoresis transfer system (Bio-Rad, Hercules, CA, USA). The
membrane was blocked with 5% non-fat milk for 1 h and then
incubated with poly-anti-TRAIL antibody (eBioscience, San Diego,
CA, USA) or anti-His-Tag antibody (Tiangen Biotech, Beijing, China)
at room temperature for 1 h. After washing twice with PBST, the
membrane was incubated with HRP-conjugated secondary antibody. The
blots were developed using enhanced chemiluminesence (ECL)
reagents. mDRA6 was used as a positive control.
Proliferation inhibition assay
Jurkat and Chang liver cells (American Type Culture
Collection, Manassas, VA, USA) were used to test the
antiproliferative activities of the two TRAIL proteins. Briefly,
100 μl Jurkat and Chang liver cells (8×105/ml)
were dispensed into the wells of 96-well cell culture plates. The
TRAIL-HS and TRAIL-FT proteins were diluted (final concentration:
10−5, 10−4, 10−3, 10−2,
10−1, 100, 101 or 102
pmol/ml) and added to the wells in triplicate. The plates were kept
in an incubator at 37°C with 5% CO2 for 12 h. After
adding 10 μl 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl
tetrazolium bromide (MTT; 10 mg/ml; Sigma), the plates were
incubated for an additional 4 h at 37°C. Following this incubation,
100 μl solubilization solution was added and incubation was
continued at 37°C for 12 h. The plate was then read at 570 nm using
a plate reader (Anthos Labtec Instruments GmbH, Salzburg, Austria).
The following formula was used to estimate the proliferation
inhibition rates of TRAIL-HS and TRAIL-FT: Proliferation inhibition
rate (%) = (1−A570(sample)) / (A570(control))
x 100.
Detection of cell apoptosis
Two experiments were carried out to detect the
apoptosis induced by the two recombinant TRAIL proteins. The Jurkat
cells were treated with TRAIL-HS or TRAIL-FT (20 ng/ml) for 100 min
and then washed once with PBS. PBS was used as the reagent control.
Each sample was then divided into two equal portions. One portion
was treated with 75% ethanol at −20°C for 24 h and then incubated
with 50 mg/ml RNaseA at 37°C for 30 min. PI (50 mg/ml) was used to
stain the cells for 30 min and the cells were analyzed by flow
cytometry (FACSCalibur; Becton-Dickinson, Franklin Lakes, NJ, USA)
using the software program CELLQUEST™. Ten thousand events were
counted. The other portion of the sample was distributed on a slide
with a cell centrifuge, fixed with methanol for 5 min and then
stained with Wright-Giemsa staining buffer. The slides were
observed under an immersion objective lens and images captured to
enable the identification of the apoptosis induced by the proteins
from the cell morphology.
Results
Expression and purification of TRAIL-FT
and TRAIL-HS proteins
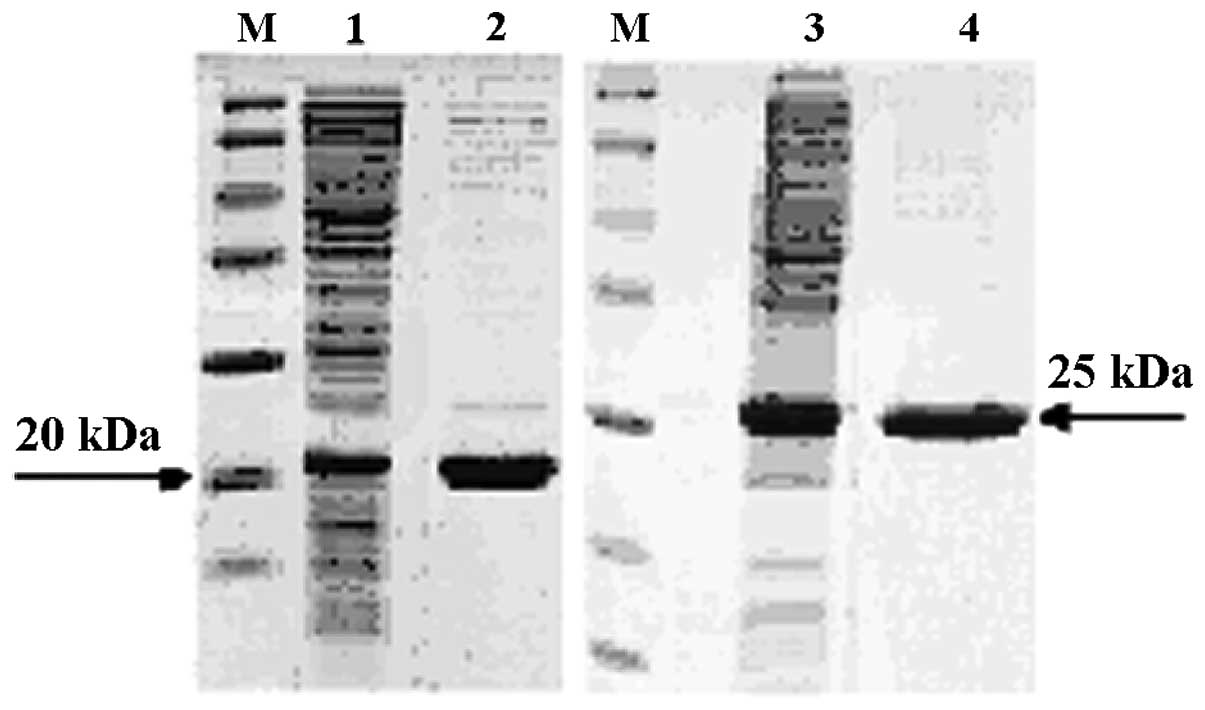
The two TRAIL proteins were purified using Ni-NTA
and SP chromatography columns. The results revealed that the target
proteins existed in the supernatants of the lysed E.coli
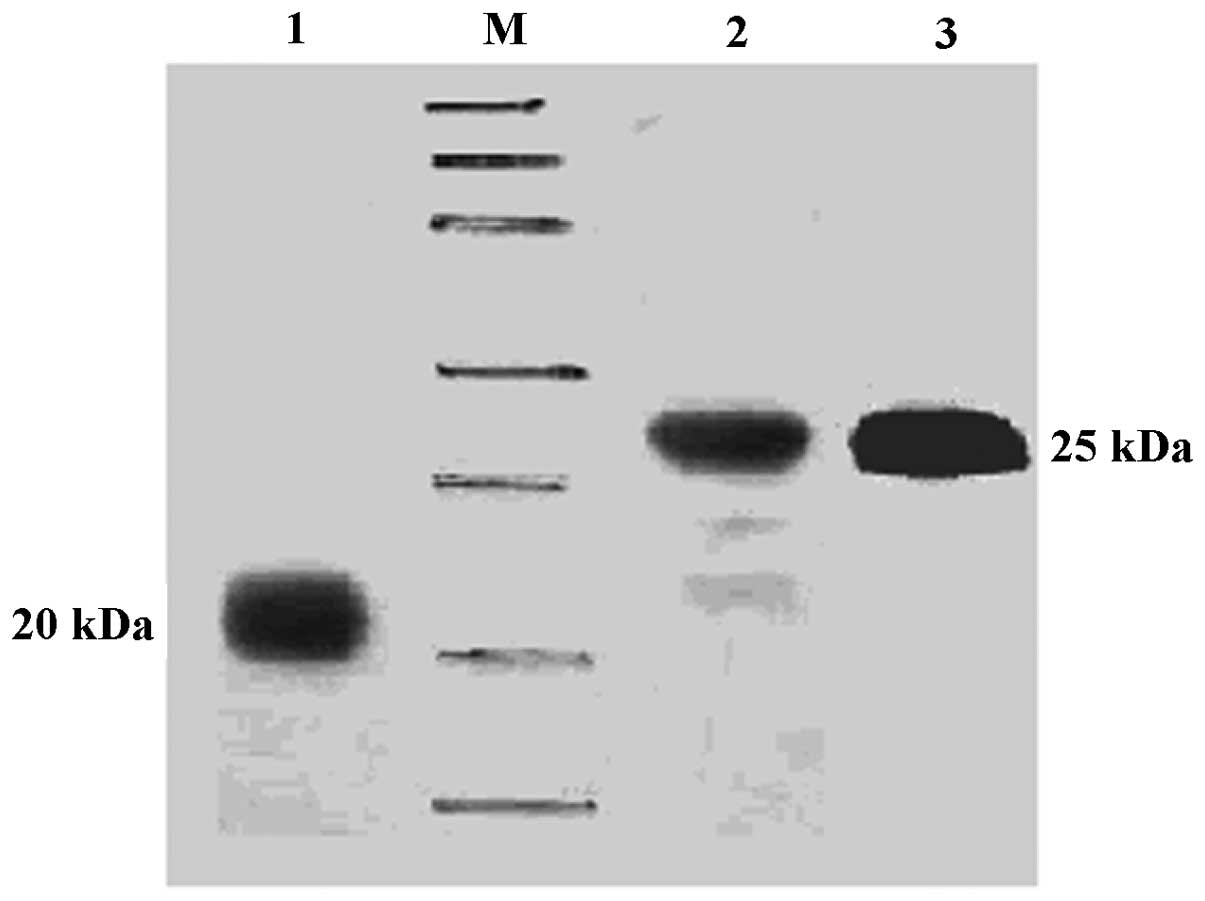
BL21(DE3). High purity proteins were obtained (Fig. 1). Western blot analysis indicated
positive reactions for TRAIL-FT and TRAIL-HS with poly-anti-TRAIL
and anti-His-Tag antibodies (Fig.
2).
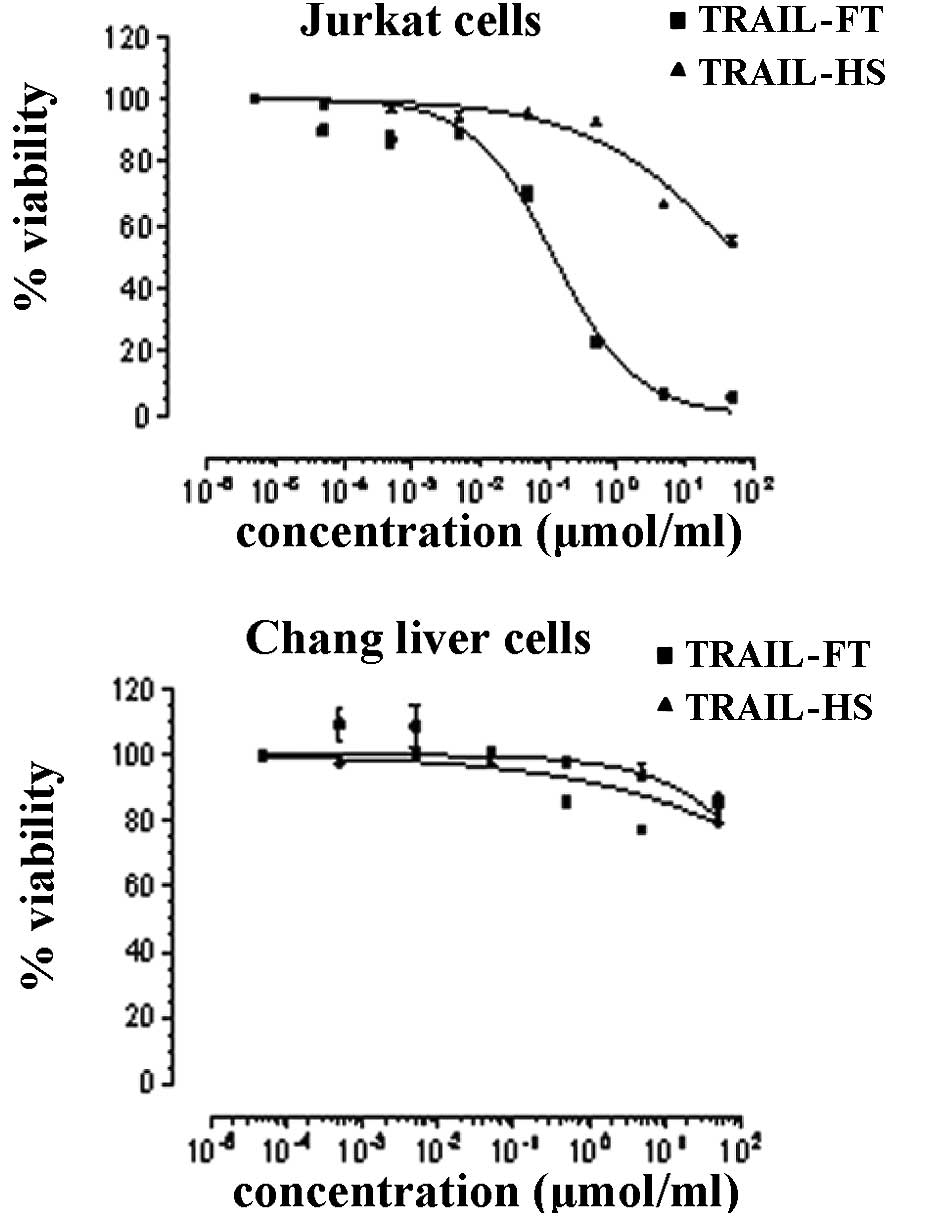
Inhibition of cell proliferation by
TRAIL-HS and TRAIL-FT proteins
The two proteins inhibited the proliferation of
Jurkat cells significantly at concentrations of
10−4–102 nmol/ml. The performance of the
TRAIL-FT protein is notable. Furthermore, when incubated with Chang
liver cells, the proteins revealed little or no cytotoxicity
(Fig. 3).
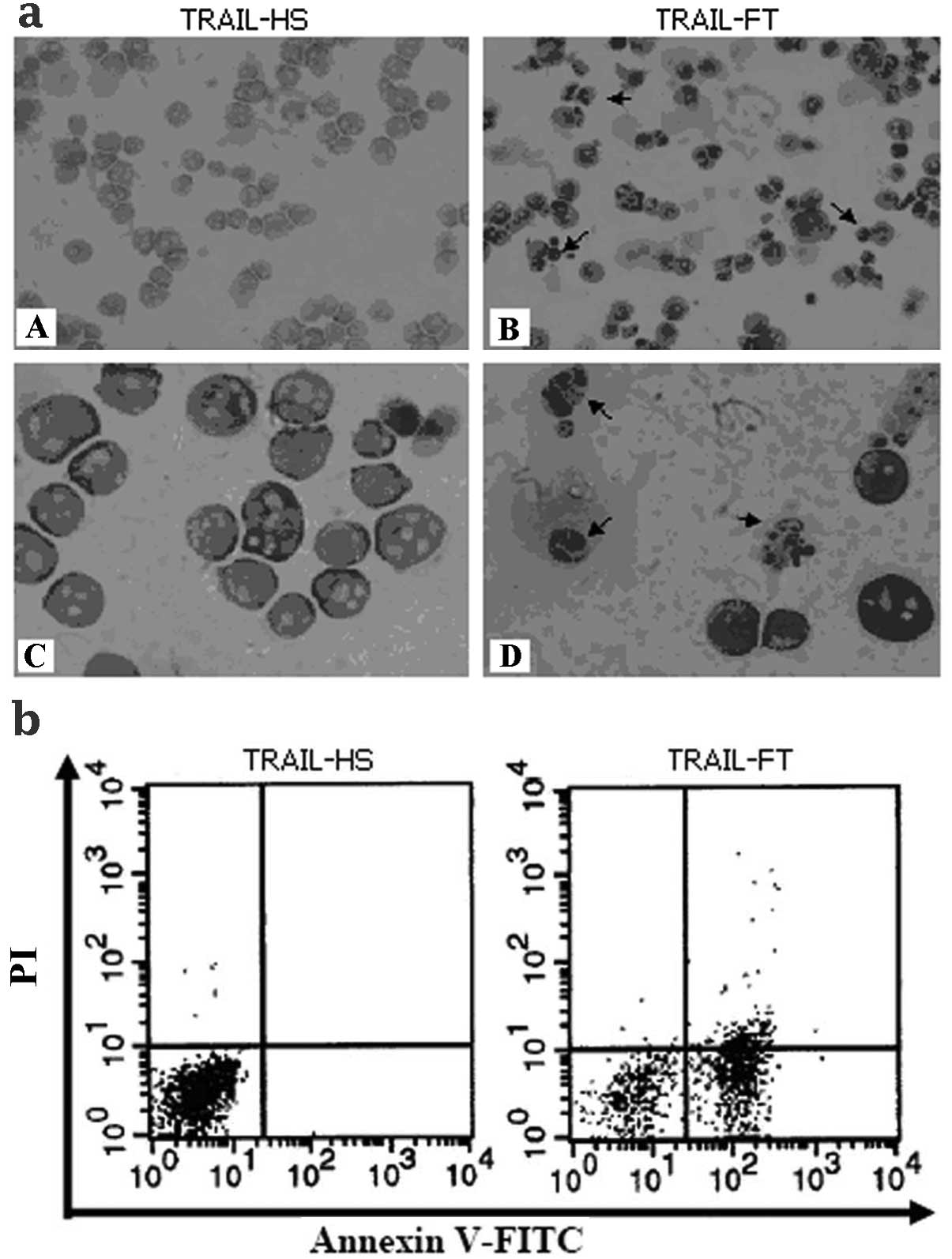
Apoptosis induced by TRAIL-FT and
TRAIL-HS
The results of Wright-Giemsa staining (Fig. 4a) revealed that the Jurkat cells
incubated with TRAIL-FT acquired the typical features of apoptosis,
including cell shrinkage, membrane blebbing and nuclear pyknosis,
unlike those incubated with TRAIL-HS. The results of FACS analysis
(Fig. 4b) indicate that TRAIL-FT is
more potent than TRAIL-HS in the induction of apoptosis.
Discussion
TRAIL has anticancer activity when used as a single
agent or in combination with chemotherapeutic agents. TRAIL induces
apoptosis by interacting with death receptors (DR4,DR5). When TRAIL
binds to DR4 and/or DR5, the receptors become trimerized to form
the death-inducing signaling complex (DISC) and recruit the adapter
protein FADD and caspase-8 or caspase-10. This leads to activation
of the executioner caspases, including caspase-3, and cleavage of
the death substrates to cause cell death (10). An earlier study indicated that a
polyhistidine-tagged version of rhTRAIL caused apoptosis in
cultured human and non-human primate hepatocytes, which raised
concerns about its safety (11).
Further studies suggested that hepatocyte toxicity caused by
polyhistidine-tagged TRAIL is attributable to low levels of zinc
(12,13). In our study, high-level expression
of soluble fusion proteins (TRAIL-FT and TRAIL-HS) were achieved in
E.coli BL21(DE3). Large amounts of pure and active rhTRAIL
proteins were purified by Ni-NTA and cation ion-exchange
chromatography. By structural analysis, we conclude that the tag
protein resulted in the formation of polymeric TRAIL and affected
its functions. Our study revealed that recombinant non-tagged TRAIL
protein is noteworthy as a potential anticancer drug. However,
further investigation of the mechanism of TRAIL-induced apoptosis
is required.
Acknowledgements
This study was supported by the
National Major Scientific and Technological Special Project for
‘Significant New Drugs Creation’ (2011ZX09506-004).
References
|
1.
|
SR WileyK SchooleyPJ SmolakIdentification
and characterization of a new member of the TNF family that induces
apoptosisImmunity3673682199510.1016/1074-7613(95)90057-88777713
|
|
2.
|
H WalczakRE MillerK AriailTumoricidal
activity of tumor necrosis factor-related apoptosis-inducing ligand
in vivoNat Med5157163199910.1038/55179930862
|
|
3.
|
M LeverkusH WalczakA McLellanMaturation of
dendritic cells leads to up-regulation of cellular FLICE-inhibitory
protein and concomitant down-regulation of death ligand-mediated
apoptosisBlood96262826312000
|
|
4.
|
H YagitaK TakedaY HayakawaTRAIL and its
receptors as targets for cancer therapyCancer
Sci95777783200410.1111/j.1349-7006.2004.tb02181.x15504243
|
|
5.
|
M JoTH KimDW SeolJE EsplenApoptosis
induced in normal human hepatocytes by tumor necrosis
factor-related apoptosis-inducing ligandNat
Med6564567200010.1038/7504510802713
|
|
6.
|
D LawrenceZ ShahrokhS MarstersDifferential
hepatocyte toxicity of recombinant Apo2L/TRAIL versionsNat
Med7383385200110.1038/8639711283636
|
|
7.
|
A AshkenaziRC PaiS FongSafety and
antitumor activity of recombinant soluble Apo2 ligandJ Clin
Invest104155162199910.1172/JCI692610411544
|
|
8.
|
E CretneyA ShankerH YagitaTNF-related
apoptosis-inducing ligand as a therapeutic agent in autoimmunity
and cancerImmunol Cell
Biol848798200610.1111/j.1440-1711.2005.01413.x16405656
|
|
9.
|
JZ QinP BaconV ChaturvediBJ NickoloffRole
of NF-κB activity in apoptotic response of keratinocytes mediated
by interferon-gamma, tumor necrosis factor-α, and tumor necrosis
factor-related apoptosis-inducing ligandJ Invest
Dermatol1178989072001
|
|
10.
|
GS WuTRAIL as a target in anti-cancer
therapyCancer Lett28515200910.1016/j.canlet.2009.02.02919299078
|
|
11.
|
M JoTH KimDW SeolApoptosis induced in
normal human hepatocytes by tumor necrosis factor-related
apoptosis-inducing ligandNat
Med6564567200010.1038/7504510802713
|
|
12.
|
D LawrenceZ ShahrokhS MarstersDifferential
hepatocyte toxicity of recombinant Apo2L/TRAIL versionsNat
Med7383385200110.1038/8639711283636
|
|
13.
|
AY SunYL ShenJC YinImprovement of
expression level and bioactivity of soluble tumor necrosis
factor-related apoptosis-inducing ligand (Apo2L/TRAIL) by a novel
zinc ion feeding strategyBiotechnol
Lett2812151219200610.1007/s10529-006-9073-z16799759
|