Introduction
The Notch signaling pathway is a highly conserved
evolutionarily signaling pathway which plays an important role in
regulating the process of development in species as diverse as
Drosophila to humans (1,2). The
Notch signaling pathway is composed of a Notch receptor, ligand and
CBF1/Su(H)/Lag-1 family (CSL) DNA binding protein. In mammals, four
homolog Notch receptors (Notch 1–4) and five Notch ligands (Jagged
1 and 2, and Delta-like 1, 3 and 4) have been identified. Both
Notch receptors and ligands are evolutionarily conserved
single-pass transmembrane proteins. After Notch ligands bind to the
receptors, the receptor undergoes at least two proteolytic cleavage
events, releasing the Notch intracellular domain (NICD), which is
activated into the cytoplasm, and then translocated to the nucleus,
where it binds to the CSL (CBF1 in humans, RBPJ in mice,
Suppressor of Hairless in Drosophila and Lag1 in C.
elegans) protein (3). After
binding to NICD, CSL turns from a transcription repressor to a
transcriptional activator (3). The
NICD/CSL complex then recruits the co-activator mastermind-like
(MAML) and p300 proteins to form a ternary complex that
subsequently activates the transcription of the Notch target genes
(4,5). Canonical target genes of Notch include
HES1, HES5 and Hey. The activated Notch signaling pathway
determines the cell fate, maintains the stem cell state and affects
cell proliferation, differentiation, apoptosis, organ formation and
morphogenesis (6). In recent years,
studies have found that abnormal Notch signaling is involved in
tumor formation, hereditary diseases, autoimmune diseases and
several other processes (7).
Several studies have demonstrated that Notch
signaling is involved in the development of lymphoid leukemias
(8), but only a few studies have
revealed abnormal Notch expression in myeoloid leukemias. Whether
abnormal Notch signaling can result in myelocytic leukemia remains
unclear. Certain studies, however, have shown that Notch can affect
myelopoiesis, with Notch ligands suppressing the differentiation of
progenitor cells to myeloid cells (9). Notch2 overexpression is implicated in
the development of chronic B-cell lymphoid leukemias, since B-cells
are able to survive significantly longer than normal cells. A study
by Ishiko et al found that Notch signaling did not affect
the proliferation of the chronic myeloid leukemia cell line K562,
but inhibited the development of erythroid/megakaryocytic cells by
suppresing GATA-1 activity and inducing apoptosis in cooperation
with 12-O-tetradecanoylphorbol-13-acetate (TPA) (10). In contrast, another study
demonstrated that activated Notch signaling inhibited the growth of
K562 cells, possibly by upregulating expression of Rb protein
(11), although the precise
mechanism is not clear.
In this study, K562 cells were used to observe the
effects of overexpression of the intracellular domain of Notch2
(ICN2). Overexpression of ICN2 inhibited the proliferation of K562
cells and caused G1 arrest. Notably, the expression level of NF-κB
and TGF-β1 genes were upregulated in K562 cells transfected with
ICN2, and Bcl-2 was downregulated, but Numb expression was
unchanged. These results may suggest that Notch signaling inhibits
the proliferation of K562 cells, possibly by regulating gene
expression.
Materials and methods
Cell culture and plasmid
transfection
The human CML cell line K562 was propagated in
RPMI-1640 medium (Hyclone, Logan, UT, USA) containing 10% fetal
bovine serum (FBS) (Hyclone). The cell lines were grown in a
humidified incubator at 37°C in the presence of 5% carbon dioxide.
K562 cells were propagated every two days. One day before
transfection, the cell medium was changed to ensure the cells were
viable. Log phase growth cells were washed, resuspended in
RPMI-1640, plated in 12-well plates at a density of
5×105/ml and cultured for 1 h. Plasmid pcDNA3.1-ICN2 was
transfected into the cells with Lipofectamine™ 2000 according to
the manufacturer’s instructions (K562-ICN2). Vector pcDNA3.1 was
used as control (K562-blank).
Cell proliferation assay
Cells were seeded in 96-well plates at a density of
1,500 cells per well and transfected with ICN. Cell proliferation
was assayed after 24 and 48 h of culture, by incubating in 20
μl methylthiazole tetrazolium (MTT) solution. Cells were
then incubated at 37°C for a further 6 h when 150 μl of
dimethyl sulfoxide (DMSO; Sigma, St. Louis, MO, USA) was added to
each well and subsequently mixed at room temperature for 10 min.
The spectrophotometric absorbance of the supernatant was measured
at 550 nm. Each assay was repeated at least three times and data
were analyzed with the LSD t-test.
Cell cycle analysis
K562 cells were plated at a density of
∼3×105/l in 12-well plates and transfected with plasmid
for 48 h. Cells were collected, fixed in 1% methanol-free
formaldehyde for 20 min and subsequently suspended in a 70% ethanol
solution. Cells were then suspended in 1 ml of 0.1% Triton X-100
solution and incubated in 500 μl propidium iodide solution
(50 μg/ml) containing 250 μg of DNase-free RNase A.
Cells were analyzed using fluorescence-assisted cell sorting
(FACS). Each assay was repeated in triplicate and data were
analyzed with the LSD-t test.
RNA extraction and semiquantitative
reverse transcription (RT)-polymerase chain reaction (PCR)
Total RNA was extracted from K562 cells with TRIzol
reagent (Roche Diagnostics, Mannheim, Germany) according to the
manufacturer’s instructions. cDNA was synthesized using a kit from
Takara with oligo-dT as a primer. PCR was run for 35 cycles with
cDNA from 0.1 μg of total RNA as a template. The PCR
products were resolved on 1.5% agarose gels and visualized by
GoldView staining. The RT-PCR primers used in this study were:
human Notch2 (5′ sense, TGGTGACCGAGATCCTG AAG; 3′ antisense,
TTGTTCACAGAGCCTTGTTG); Hes1 (5′ sense, TAGCTGATCAGTGGCGTGAC; 3′
antisense, ATCATCTGGCCTAGGAGACC); Hey1 (5′ sense, GAGAGG
TCCTCCATTGGAAT; 3′ antisense, ATGCACAACAAT GGCAACAG), Numb 5′
sense, TACCACGTCCTCACCTG TGG; 3′ antisense, TGAAGACTGCAGAACCATTG);
Bcl-2 (5′ sense, ACCTGACCACTAGCCTCCTG; 3′ antisense,
GCAGAGCACAGGATTCACAG); TGF-β1 (5′ sense, GCCTTGATGGAGAGCTTCAC; 3′
antisense, CTTGTG GTGGATGTGGACTG), NF-κB (5′ sense, AGTCTGTCC
AGGCTCGTCAT; 3′ antisense, GGACAGGAAGCTCCT GAATG); and human
β-actin I (5′ sense, ACTTGCGCA GAACAAGAGAT; 3′ antisense,
ACTGCCGCCTTCTCC TTAGA); human β-actin II (5′ sense, GATCTGGCACCA
CACCTTCT; 3′ antisense, AAGGAAGGCTGGAAGAGAGC).
Western blot analysis
To detect the expression of Notch2, whole cell
lysates were prepared using cell lysis buffer for western blot
analysis and immunoprecipitation [20 mM Tris (pH 7.5), 150 mM NaCl,
1% Triton X-100, sodium pyrophoshate, β-glycerophosphate, EDTA,
Na3VO4, leupeptin, 0.1 mM PMSF]. Cell
extracts were collected by centrifugation at 13,000 × g for 8 min
at 4°C. Protein concentrations of the cell extracts were determined
by BCA protein assay reagents (Beyotime Institute of Biotechnology,
Jiangsu, China), according to the manufacturer’s instructions.
Proteins were resolved by SDS-PAGE on 10% polyacrylmide gel. The
proteins were transferred to a PVDF membrane and detected using
immunoblotting. The Notch2 antibody was incubated with the membrane
for 12 h at 4°C, and immunoreactive proteins were visualized by
incubation with a goat anti-mouse immunoglobulin conjugated to
horseradish peroxidase (HRP), and the membrane was developed using
Pro-light HRP chemiluminescence reagents from Tiangen Biotech
(Beijing) Co., Ltd., Beijing, China. All antibodies were purchased
from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA).
Statistical analysis
To analyze the data, the mean ± SD was calculated.
One-way ANOVA was used to compare the mean values among multiple
groups, and the t-test was applied to compare the mean values
between pairs of groups. All data was processed by SSPS 10.0 (SPSS,
Inc., Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant result.
Results
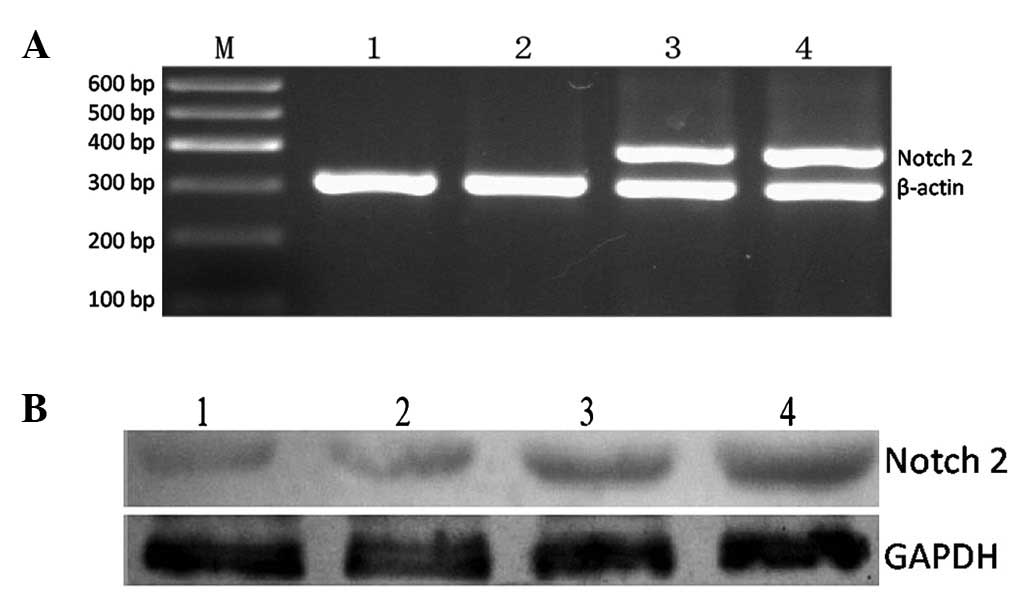
Notch2 gene is overexpressed successfully
in K562 cells
To confirm that the plasmid pcDNA3.1-ICN2 was
successfully introduced to the K562 cells, RT-PCR and western blot
analysis were employed to detect the expression of the gene and
protein, respectively. As shown in Fig.
1, the expression of Notch2 mRNA and protein was upregulated
significantly, suggesting that ICN2 was successfully introduced
into K562-ICN2 cells.
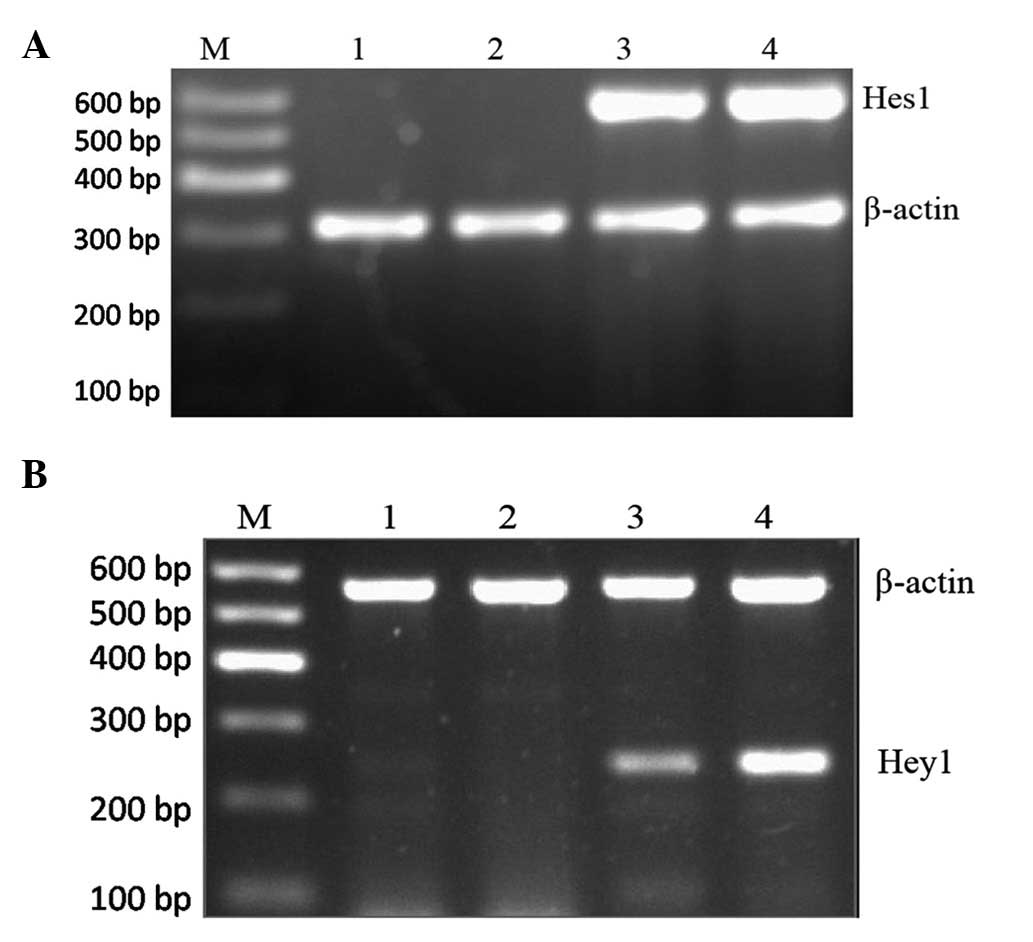
Notch2 signaling is activated in K562
cells
To further assess the activation of the Notch
signaling pathway, RT-PCR was used to detect the expression of
Notch target gene Hes1, and Hey1 of the transfected and control
group. As shown in Fig. 2, Hes1 and
Hey1 were not expressed in K562-blank cells, but they were
expressed strongly in the K562-ICN2 group, indicating that Notch
signaling was activated in K562-ICN2 cells.
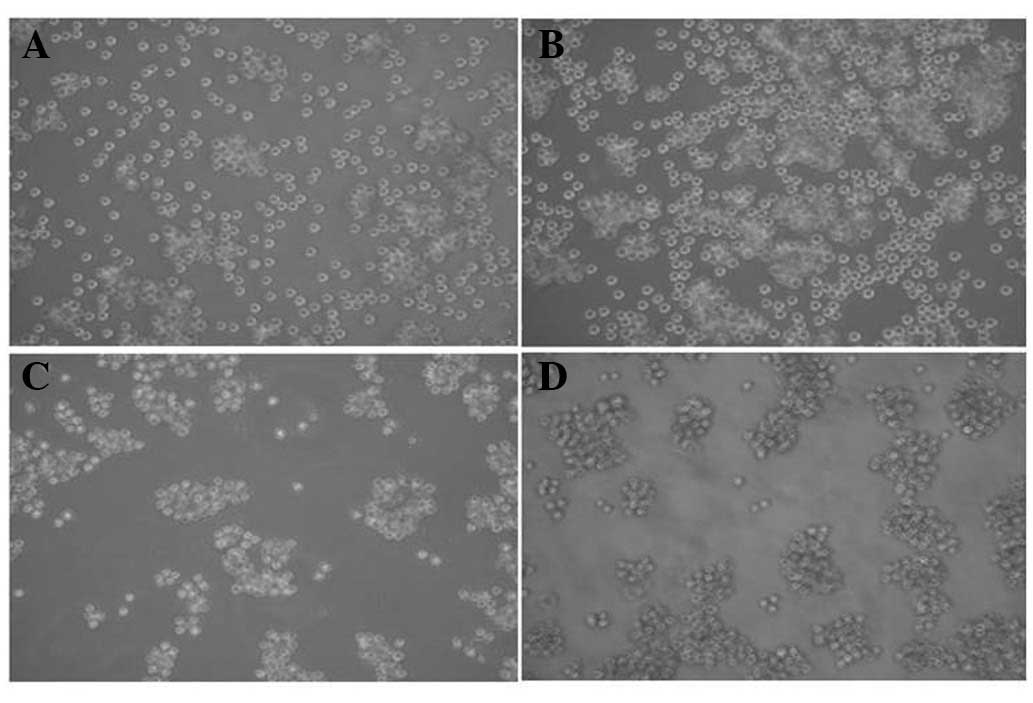
Overexpression of ICN2 changes the
morphology of K562 cells
As observed under an inverted phase contrast
microscope (Fig. 3), the
plasmalemma of K562-blank cells was transparent. The longer the
culture time, the larger the cell number. In contrast, the
K562-ICN2 cells were aggregated when cultured for 24 h with small
cells with a cloudy plasmalemma. The longer the culture time the
smaller the cell diameter and volume.
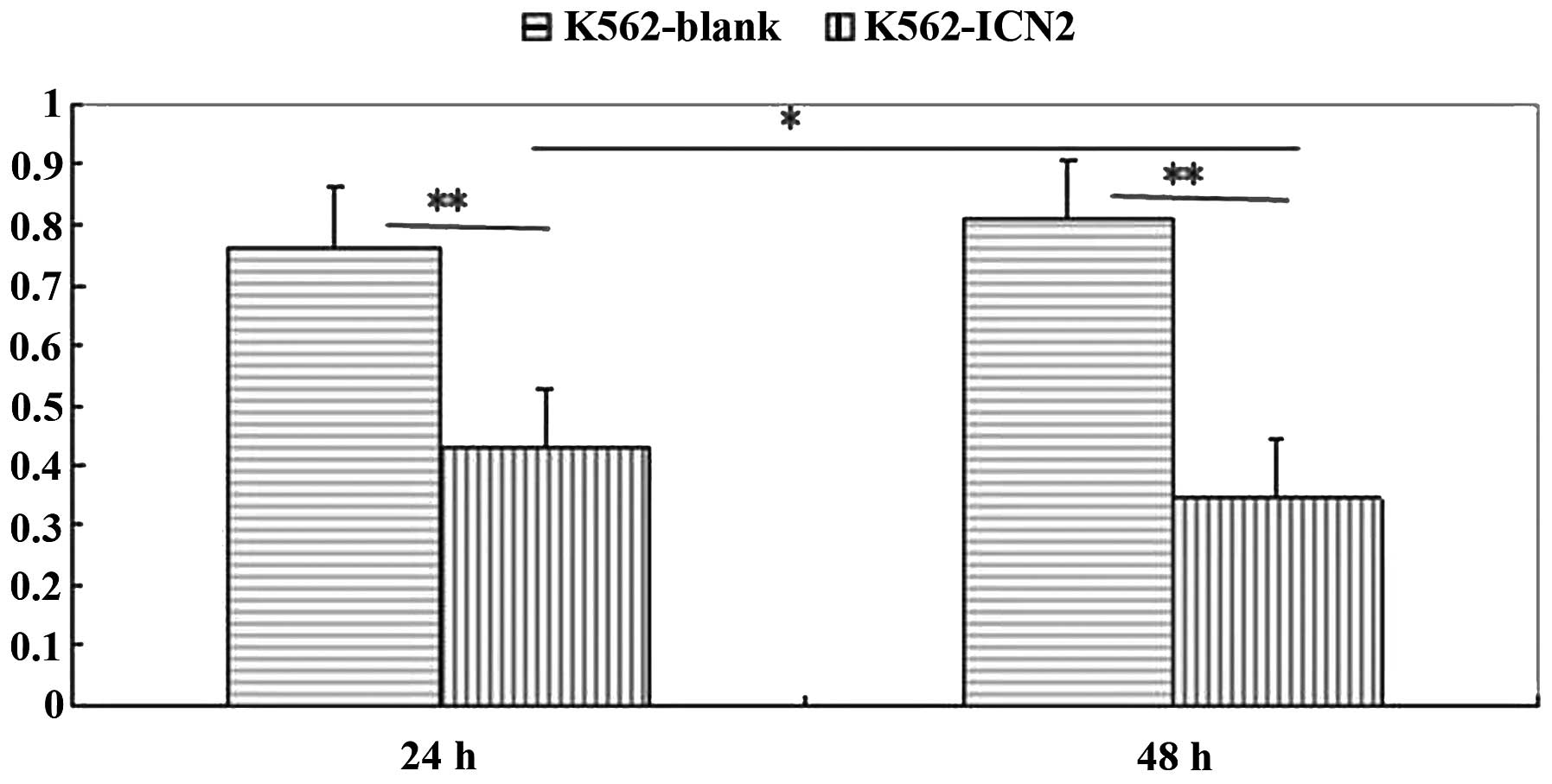
Activation of Notch by transfection with
ICN2 inhibits the proliferation of K562 cells
To determine whether Notch signaling affects the
proliferation of K562 cells, the growth of K562-ICN2 cells was
compared with the K562-blank cells using the MTT assay. As shown in
Fig. 4, the growth of K562-ICN2
cells was considerably slower than that of the K562-blank cells
(P<0.01). These data suggest that overexpression of the
constitutively active Notch signaling could inhibit the
proliferation of the human CML cell line K562 in vitro.
Notch activation rearranges the
distribution of the cell cycle of K562 cells
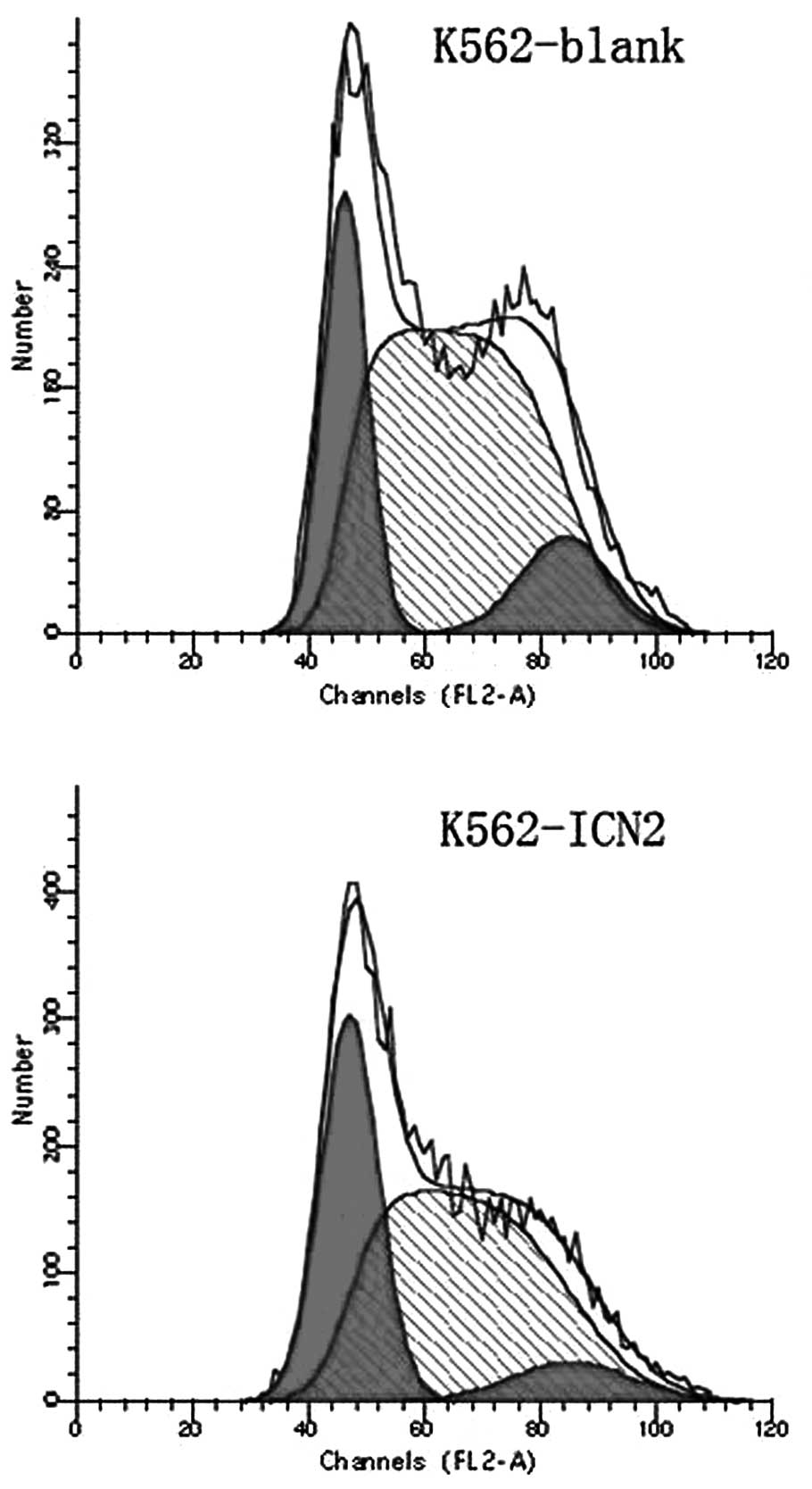
The effect of transfection with ICN2 for 48 h on the
course of the cell cycle using flow cytometry was investigated. As
shown in Fig. 5 and Table I, K562 cells showed a G1 arrest and
there were fewer S phase cells.
 | Table ICell cycle change of K562 cells after
transfection with ICN2 for 48 h (mean ± SD, %). |
Table I
Cell cycle change of K562 cells after
transfection with ICN2 for 48 h (mean ± SD, %).
| Group | G1 phase | S phase | G2+M phase | CV value |
|---|
| K562-blank | 21.52±2.51 | 63.37±3.44 | 10.82±0.96 | 4.96 |
| K562-ICN2 | 39.17±1.50a | 32.94±3.75a | 13.41±1.97 | 4.73 |
Activation of Notch signaling pathway
regulates the expression of certain genes in K562 cells
The molecular mechanism underlying the
growth-regulatory effect of Notch signaling on K562 was
investigated. The expression of the molecules related with cell
proliferation in K562 cells was measured. RT-PCR analysis showed
that the expression of NF-κB and TGF-β1 was upregulated, and that
the expression of Bcl-2 was downregulated. The expression of Numb,
however, was unchanged.
Discussion
Different Notch receptors have varying functions in
the development of tumors, depending on the cell context. Notch1
has been shown to inhibit the cell growth of hepatic carcinoma,
small cell lung cancer and prostate cancer in a previous study
(7). When all four Notch receptors
are activated they inhibit the growth of acute B-cell lymphoid
leukemia and induce apoptosis (12). In contrast, one study has proven
that overexpression of Notch3 promots the growth of human lung
cancer cells in vitro and inhibits the differentiation of
lung cancer cells in transgenic mice. Notch3 is expressed at a high
level in human pancreatic cancer cells and acute T-cell lymphoid
leukemia cells. The mechanism of the Notch receptor function as an
oncogene or tumor suppressor gene remains to be elucidated.
An important member of the Notch signaling family is
the Notch2 receptor, which plays a key role in regulating
developmental processes in the embryo (13). A study has shown that Notch2 was
upregulated in non-small cell lung carcinoma (NSCLC) and promoted
the proliferation of NSCLC cells (14). Expression of Notch2 is necessary for
marginal zone B-cell development and is related to CD23
overexpression in chronic B-cell lymphoid leukemia (15). Overexpression of ICN2 can induce
T-cell lymphoid leukemia and cells are more easily prone to
becoming CD8 cells when expression of Notch2 is inhibited.
Conversely, Notch2 is a new tumor suppressor; it inhibits tumor
progress in human breast cancer and small cell lung cancer
(16). In this study, activation of
the Notch2 signal pathway significantly inhibited the proliferation
of K562 cells and changed the cell morphology. These results
suggest that Notch2 gene is a potential tumor suppressor in chronic
myeloid leukemia.
Tumor cells cleave fast and have a high
proliferation ability. G1 phase is a DNA presynthetic phase of
cells which determines the cell cycle. A recent study found the p53
gene product is an important factor in the regulation of the cell
cycle and apoptosis, and p53 accumulated quickly when DNA was
damaged by extrinsic factors and G1 arrest occurred (17). Notch2 inhibited the proliferation of
K562 cells and may simulate the function of p53 to induce G1
arrest.
The crosstalk between Notch and other signaling
pathways is complicated, and the physiological correlation remains
unclear. Numb was considered to be a security device of WNT-Notch
signal transduction pathway. The silence, loss of function or
mutation of this gene can induce tumorigenesis (18). A study has found that there is
crosstalk between Notch and NF-κB when their roles in normal
development and cancer formation are considered (19). Transfected Notch1-ICD in the mouse
T-ALL cell line stimulates NF-κB expression through a canonical
pathway. A study on a human cervical cancer cell line found that
Notch1 activates NF-κB by interacting with the IκB kinase (IKK)
signal. This occurred in both the cytoplasm and the nucleus
(20). The Bcl-2 family of proteins
determined the mitochondrial events in cell death and mediated the
apoptosis induced by a numner of stimulants (21). Bcl-2 is expressed at a high
concentration in many tumor cells and is involved in drug
resistance. TGF-β1 is a pleiotropic anti-inflammatory factor, which
regulates T-cell differentiation. When γ-secretase inhibitor (GSI)
inhibits Notch signaling, TGF-β1 induces Foxp3 expression and naive
T-cell proliferation is blocked (22). TGF-β1 inhibits the growth of normal
cervical cells causing G1 arrest and apoptosis (23).
In this study, it was identified that overexpression
of ICN2 upregulated the expression of NF-κB and TGF-β1, and
downregulated the expression of Bcl-2, while the expression of Numb
was unaffected. These results suggest that enhanced expression of
Notch2 inhibits cell proliferation, suggesting a pathway which may
affect the cell cycle distribution by upregulating the NF-κB and
TGF-β1 genes and downregulating Bcl-2. Further research is required
to confirm our findings and to elucidate the mechanism of Notch in
chronic myeloid leukemia.
References
|
1
|
Artavanis-Tsakonas S, Rand MD and Lake RJ:
Notch signaling cell fate control and signal integration in
development. Science. 284:770–776. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Greenwald I: LIN-12/Notch signaling:
lessons from worms and flies. Genes Dev. 12:1751–1762. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mumm JS and Kopan R: Notch signaling: from
the outside in. Dev Biol. 228:151–165. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jarriault S, Le Bail O, Hirsinger E, et
al: Delta-1 activation of Notch-1 signaling results in HES-1
transactivation. Mol Cell Biol. 18:7423–7431. 1998.PubMed/NCBI
|
|
5
|
Kuroda K, Tani S, Tamura K, et al:
Delta-induced Notch signaling mediated by RBP-J inhibits MyoD
expression and myogenesis. J Biol Chem. 274:7238–7244. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ohishi K, Katayama N, Shiku H, et al:
Notch signaling in hematopoiesis. Cell Dev Bio1. 4:143–150.
2003.
|
|
7
|
Leong KG and Karsan A: Recent insights
into the role of Notch signaling in tumorigenesis. Blood.
107:2223–2233. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pancewicz J, Taylor JM, Datta A, et al:
Notch signaling contributes to proliferation and tumor formation of
human T-cell leukemia virus type 1-associated adult T- cell
leukemia. Proc Natl Acad Sci USA. 107:16619–16624. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
de Pooter RF, Schmitt TM, de la Pompa JL,
et al: Notch signaling requires GATA-2 to inhibit myelopoiesis from
embryonic stem cells and primary hemopoietic progenitors. J
Immunol. 176:5267–5275. 2006.PubMed/NCBI
|
|
10
|
Ishiko E, Matsumura I, Ezoe S, et al:
Notch signals inhibit the development of erythroid/megakaryocytic
cells by suppressing GATA-1 activity through the induction of HES1.
J Biol Chem. 280:4929–4939. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yin DD, Fan FY, Hu XB, et al: Notch
signaling inhibits the growth of the human chronic myeloid leukemia
cell line K562. Leuk Res. 33:109–114. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zweidler-McKay PA, He Y, Xu L, et al:
Notch signaling is a potent inducer of growth arrest and apoptosis
in a wide range of B-cell malignancies. Blood. 106:3898–3906. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen W, Jiang H, Wang M, et al: Effects of
chlorpyrifos exposure on kidney Notch2-Jagged1 pathway of early
prenatal embryo. Birth Defects Res B Dev Reprod Toxicol. 92:97–101.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bastide K, Uqolin N, Levalois C, et al:
Are adenosquamous lung carcinomas a simple mix of adenocarcinomas
and squamous cell carcinomas, or more complex at the molecular
level? Lung Cancer. 68:1–9. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gibb DR, EI Shikh M, Kang DJ, et al:
ADAM10 is essential for Notch2-dependent marginal zone B cell
development and CD23 cleavage in vivo. J Exp Med. 207:623–635.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mazur PK, Grüner BM, Nakhai H, et al:
Identification of epidermal Pdx1 expression discloses different
roles of Notch1 and Notch2 in murine Kras(G12D)-indenced skin
carcinogenesis in vivo. PloS one. 5:e135782010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang PM, Huang WC, Lin YC, et al: Loss of
IKKbeta activity increases p53 stability and p21 expression leading
to cell cycle arrest and apoptosis. J Cell Mol Med. 14:687–698.
2010.PubMed/NCBI
|
|
18
|
Cheng X, Huber TL, Chen VC, et al: Numb
mediates the interaction between Wnt and Notch to modulate
primitive erythropoietic specification from the hemangioblast.
Development. 135:3447–3458. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Z, Banerjee S, Ahmad A, et al:
Activated K-ras and INK4a/Arf deficiency cooperate during the
development of pancreatic cancer by activation of Notch and NF-κB
signaling pathways. PloS One. 6:e205372011.PubMed/NCBI
|
|
20
|
Song LL, Peng Y, Yun J, et al: Notch-1
associates with IKKalpha and regulates IKK activity in cervical
cancer cells. Oncogene. 27:5833–5844. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Frenzel A, Grespi F, Chmelewskij W, et al:
Bcl2 family proteins in carcinogenesis and the treatment of cancer.
Apoptosis. 14:584–596. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Morita Y, Ismail DM, Elkon KB, et al:
Dichotomous response to transforming growth factor β after T cell
receptor activation by naive CD4+ T cells from DBA/1 mice: enhanced
retinoic acid receptor-related orphan nuclear receptor γt
expression yet reduced FoxP3 expression. Arthritis Rheum.
63:118–126. 2011.
|
|
23
|
Rorke EA, Zhang D, Choo CK, et al:
TGF-beta-mediated cell cycle arrest of HPV16-immortalized human
ectocervical cells correlates with decreased E6/E7 mRNA and
increased p53 and p21(WAF-1) expression. Exp Cell Res. 259:149–157.
2000. View Article : Google Scholar : PubMed/NCBI
|