Introduction
Acute leukemia (AL) consists of a group of
heterogeneous malignancies in which immature and dysfunctional
hematopoietic progenitors proliferate and accumulate in the bone
marrow. DNA methylation plays a key role in the pathophysiology of
acute myeloid leukemia (AML) (1)
and acute lymphoblastic leukemia (ALL) (2,3). DNA
methyltransferase 3A (DNMT3A) is one of two human de novo
DNA methyltransferases essential for regulating gene expression
during cellular development and differentiation (4). DNMT3A mutations and deletions have
been analyzed in AML (5), chronic
myeloid leukemia (CML), chronic myelomonocytic leukemia (CMML),
myelodysplastic syndrome (MDS), lymphoma and myeloproliferative
neoplasms (MPNs) (6–11). The frequency of the mutations in
patients with different diseases varies between 0 (0/81, CML
patients in blast crisis) and 22.1% (62/281, AML patients). The
most common mutation has been identified at the site of amino acid
residue R882.
DNMT3A mutations have been found to be enriched in
the M4 (32.8%) and M5 subtypes (57.1%), according to the
French-American-British (FAB) classification system (12). The DNMT3A expression levels in
patients with DNMT3A mutations were observed to be marginally lower
than those without mutations. In addition, DNMT3A was also found to
be expressed in normal human CD34+ bone marrow cells and
its expression decreased with terminal myeloid differentiation
(5). However, to date, the
differences in the DNMT3A expression levels in various subtypes of
AML, according to FAB classification, have not been determined. In
a previous study, the results of a multivariate analysis indicated
that DNMT3A mutations represented independent predictive factors of
poor prognosis, including a reduced overall survival (OS) and
complete remission (CR) rate (7).
Qiao et al (13) reported
that the CR rate in acute leukemia (AL) patients that were
identified to positively express all the DNMT genes was
significantly higher than that of patients with partially positive
or negative expression, indicating that DNMT3A mutations and
expression may be associated with the pathogenesis and prognosis of
AL.
ALL is a heterogeneous malignancy caused by the
clonal proliferation of lymphocytes. The pathogenesis and causal
cancer genes associated with AML and ALL differ (14). Unlike AML, extremely little is known
about the mutation frequency of DNMT3A in ALL patients. Therefore,
in the present study, 99 Chinese AL patients were screened for
DNMT3A R882 mutations, with the aim of uncovering the frequency of
the R882 mutations in ALL and the relationship between ALL and AML.
In addition, DNMT3A expression levels were determined in these
samples and normal controls to determine whether expression levels
correlate with poor prognosis. The results demonstrate that the
DNMT3A mutation status in AML is an important factor to consider
for risk stratification of the disease.
Materials and methods
Patients, healthy subjects and bone
marrow mononuclear cell (BMMC) and peripheral blood mononuclear
cell (PBMC) collection
The study recruited 99 consecutive adult patients
with AL [AML, ALL and biphenotypic acute leukemia (BAL)]
newly-diagnosed at Qilu Hospital of Shandong University between
August 2011 and November 2012. The diagnosis was made according to
the FAB classification. For the clinical analysis, CR, partial
remission (PR) and non-remission (NR) were defined according to the
criteria of the International Working Group (15). Cytogenetic risk was determined in
the AML patients following a method described previously (16). The characteristics of the patients
at the time of sampling are presented in Tables I and II. The patients with AML were treated with
standard induction chemotherapy (anthracycline and cytarabine). The
patients with ALL were treated with standard induction chemotherapy
(vincristine, daunorubicin, L-asparaginase and prednisone). In
addition, a control group of 16 healthy donors was included. An
assessment of the patient history and a physical examination were
performed during the initial diagnosis. The corresponding
laboratory tests were performed. BMMCs and PBMCs were obtained from
41 ALL patients (bone marrow or whole blood), 57 AML patients, one
BAL patient and 16 control individuals (bone marrow) using
density-gradient centrifugation with the Ficoll-Hypaque technique
(Ficoll, Pharmacia LKB Biotechnology Inc., Piscataway, NY, USA).
The samples were then stored at −80°C.
 | Table I.Clinical characteristics of 57
patients with AML. |
Table I.
Clinical characteristics of 57
patients with AML.
| Characteristics | No DNMT3A
mutation | R882 mutation | Non-R882
mutation | Any DNMT3A
mutation | P-valuea |
|---|
| Patients, n | 47 | 9 | 1 | 10 | |
| Age at study entry,
years | 42.4±16.1b | 60.3±16.5b | 55 | 59.8±15.7b | 0.007c |
| Male gender (%) | 26 (55.3) | 3 (33.3) | 0 (0.0) | 3 (30.0) | 0.179d |
| Bone marrow blasts at
diagnosis, % | 77.75±20.84b | 84.9±19.7b | 87 | 84.9±19.7b | 0.073c |
| Normal karyotype,
n/total (%) | 11/13 (84.6) | 4/4 (100) | 1/1 (100.0) | 5/5 (100.0) | |
| White-cell count at
diagnosis, ×103 cells/mm3 | | | | | |
| Mean | 26.71±45.57b | 114.28±88.34b | 22.38 | 105.09±88.21b | <0.001b |
| Median | 10.93 | 103.00 | 22.38 | 94.47 | |
| Cytogenetic risk,
n/total (%) | | | | | 0.004d |
| Favorable | 18/45 (40.0) | 0/7 (0.0) | 0/1 (0.0) | 0/8 (0.0) | |
| Intermediate | 19/45 (42.2) | 7/7 (100.0) | 1/1 (100.0) | 8/8 (100.0) | |
| Adverse | 8/45 (17.8) | 0/7 (0.0) | 0/1 (0.0) | 0/8 (0.0) | |
| AML subtype, n
(%) | | | | | |
| M2 | 6 (12.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| M3 | 14 (29.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| M4 | 10 (21.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| M5 | 17 (36.2) | 9 (100.0) | 1 (100.0) | 10 (100.0) | |
 | Table II.Clinical characteristics of 41 ALL
patients and one BAL patient. |
Table II.
Clinical characteristics of 41 ALL
patients and one BAL patient.
|
Characteristics | T-cell
leukemia | B-cell
leukemia | ALL with unknown
phenotype | BAL |
|---|
| Patients, n | 6 | 31 | 4 | 1 |
| DNMT3A mutations, n
(%) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Age at study entry,
years | 28.3±24.7a | 37.4±16.7a | 43.0±24.6a | 60 |
| Male gender, n
(%) | 4 (66.7) | 16 (51.6) | 2 (50.0) | 1 (100.0) |
| Bone marrow blasts
at diagnosis, % | 95.0±0.0a | 86.1±13.8a | 83.5±12.0a | 97 |
| Normal karyotype,
n/total (%) | 4/4 (100.0) | 9/23 (39.1) | 0/2 (0.0) | 0/0 (0.0) |
| White cell count at
diagnosis, ×103 cells/mm3 | | | | |
| Mean | 52.1±67.8a | 63.0±98.4a | 2.54±1.94a | 5.08 |
| Median | 16.26 | 11.17 | 2.33 | 5.08 |
| Aberrant karyotype,
n/total (%) | | | | |
| (9,22)(q34;q11)
or BCR/ABL fusion gene | 0/4 (0.0) | 13/23 (41.9) | 0/2 (0.0) | 0/0 (0.0) |
The present study was approved by the ethics
committee of Qilu Hospital, Shandong University (Jinan, China).
Written informed patient consent was obtained from all participants
for the treatment and cryopreservation of BM and peripheral blood
according to the Declaration of Helsinki.
Genomic DNA isolation, PCR amplification
and sequencing
Genomic DNA samples from bone marrow or whole blood
of AML and ALL patients were extracted using the TIANamp genomic
DNA kit [Tiangen Biotech (Beijing) Co., Ltd., Beijing, China] or
the total DNA/RNA/protein extraction kit (Omega Bio-Tek, Inc.,
Norcross, GA, USA). A DNA fragment of 379 bp covering the R882 site
in exon 23 of the DNMT3A gene was amplified using the S1000 thermal
cycler (Bio-Rad, Hercules, CA, USA). Forward primer, 5′-TCC TGC TGT
GTG GTT AGA CG-3′; and reverse primer: 5′-TAT TTC CGC CTC TGT GGT
TT-3′. PCR was performed in a 25-μl volume containing 30 ng
DNA, 12.5 μl PCR mastermix, 1 μl forward primer, 1
μl reverse primer and ddH2O. The PCR conditions
were as follows: denaturation at 94°C for 5 min, followed by 35
cycles of denaturation at 94°C for 30 sec, annealing at 55°C for 30
sec, extension at 72°C for 30 sec and ending with an extension at
72°C for 10 min. The PCR products were sequenced bidirectionally
using the ABI 3730xl DNA analyzer (Applied Biosystems, Bedford, MA,
USA).
RNA preparation and real-time
quantitative PCR
The total RNA was extracted using TRIzol (Invitrogen
Life Technologies, Carlsbad, CA, USA), and the cDNA was prepared
using M-MLV reverse transcriptase (Promega Corporation, Madison,
WI, USA) according to the manufacturer’s instructions. Reverse
transcription was performed at 37°C for 15 min, followed by 85°C
for 5 sec. Real-time quantitative PCR (RQ-PCR) was performed using
the ABI Prism 7500 system (Applied Biosystems) according to the
manufacturer’s instructions. PCR was performed in a total volume of
10 μl, which included 5 μl 2X SYBR Green real-time
PCR master mix (Toyobo Co. Ltd., Osaka, Japan), PCR-grade water, 1
μl template cDNA and 0.5 μl forward and reverse
primers. The sequences of the target-specific primers were designed
from human cDNA sequences available in GenBank. DNMT3A forward,
5′-GCC ACC TCT TCG CTC CGC TG-3′ and reverse, 5′-GAT GAT GTC CAA
CCC TTT TCG CAA-3′; and β-actin forward, 5′-TGA CGT GGA CAT CCG CAA
AG-3′ and reverse, 5′-CTG GAA GGT GGA CAG CGA GG-3′. The thermal
cycling profile consisted of 95°C denaturation for 5 min, followed
by 40 cycles at 95°C for 15 sec, 65°C for 15 sec and 72°C for 45
sec. To exclude non-specific amplification and primer-dimer
formation, a dissociation curve analysis was performed and PCR
products were confirmed by agarose gel electrophoresis. PCR-grade
water was used instead of template cDNA for the negative control.
The fold-change in the gene expression was determined using the
2−ΔCT method with β-actin as an endogenous control. All
experiments were performed at least twice.
Statistical analysis
The Student’s t-test was used to compare the
differences in DNMT3A expression levels between the AML patients
with R882 mutations and the normal controls. The difference in the
DNMT3A expression levels between M5 subtype AML and the other AML
subtypes was also compared by Student’s t-test. Differences in the
DNMT3A expression levels were compared by an analysis of variance
in three groups. The clinical characteristics of the AML and ALL
patients, including gender, age, white cell count and other
factors, are presented in Tables I
and II. The AML and ALL patients
were categorized into high and low DNMT3A-expressing subgroups
using the median value as the cut-off. Fisher’s exact test was used
to compare the CR rate in patients with an intermediate-risk
profile. Pearson’s chi-square test was used to compare the CR rates
between other groups. P<0.05 was considered to indicate a
statistically significant difference. Statistical analysis was
performed using the SPSS 17.0 statistical software program (SPSS
Inc., Chicago, IL, USA).
Results
DNMT3A mutations
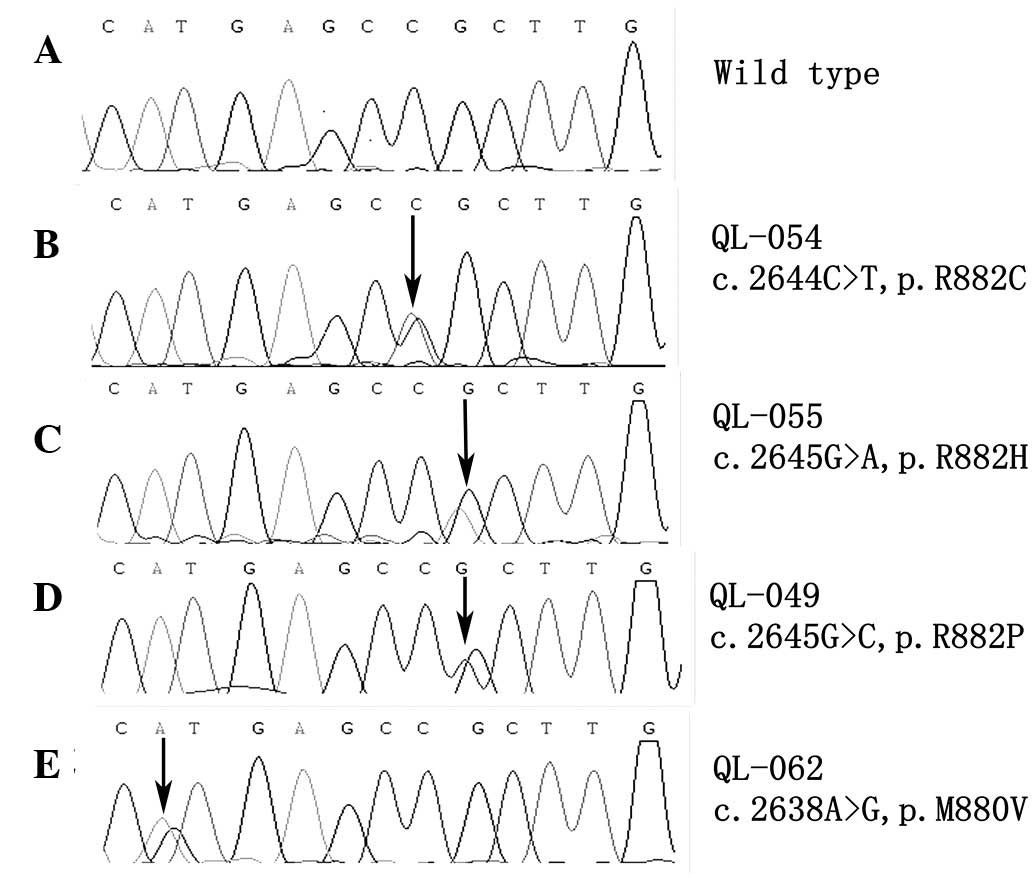
DNMT3A R882 mutational status was determined in a
cohort of 57 AML and 41 ALL patients and 1 BAL patient. Of the AML
patients, 6 were identified to exhibit the R882H variant, two the
R882C variant, one the R882P variant and one the M880V variant, a
novel single nucleotide polymorphism that leads to amino acid
substitution. The DNMT3A mutation frequency in AML was 17.5%
(10/57). Sequencing results of each type of mutation are presented
in Fig. 1. None of the DNMT3A
mutations were found in the ALL and BAL patients. The AML patients
with DNMT3A mutations revealed lower CR rates following induction
therapy compared with those with wild-type DNMT3A (0 vs. 62.8%;
P<0.001). The presence of a DNMT3A mutation was found to
correlate with a low CR rate in the AML patients with an
intermediate-risk profile (P=0.061).
Clinical features of patients with DNMT3A
mutations
The association between the status of DNMT3A
mutations and clinical features in AML was investigated. Patients
with DNMT3A mutations were classified with M5 subtype AML. The age
and white cell count of the AML patients with DNMT3A mutations were
higher than those without DNMT3A mutations. Gender and percentage
of bone marrow blasts at diagnosis were not found to be
significantly different between the two groups. DNMT3A mutations
were significantly enriched in 8/26 patients with a cytogenetic
profile associated with intermediate risk (30.8%; P=0.004; Table I). The clinical and genetic
characteristics of the 10 DNMT3A-mutated AML cases are presented in
Table III.
 | Table III.Clinical and genetic characteristics
of the ten DNMT3A-mutated AML cases. |
Table III.
Clinical and genetic characteristics
of the ten DNMT3A-mutated AML cases.
| UPN | Nucleotide
change | Consequence | Age, years | Gender | FAB | Karyotype | Aberrant expression
or mutation of other genes | Response or outcome
following induction chemotherapy |
|---|
| QL-049 | c.2645G>C | p.R882P | 37 | Female | M5 | 46, XX | None | PR |
| QL-053 | c.2645G>A | p.R882H | 79 | Male | M5 | 46, XY | None | Deceased |
| QL-054 | c.2644C>T | p.R882C | 44 | Male | M5 | 46, XY | None | NR |
| QL-055 | c.2645G>A | p.R882H | 41 | Female | M5 | Unknown | WT1 (+) | NR |
| QL-056 | c.2644C>T | p.R882C | 65 | Male | M5 | 46, XY | None | NR |
| QL-058 | c.2645G>A | p.R882H | 71 | Female | M5 | Unknown | None | NR |
| QL-062 | c.2638A>G | p.M880V | 55 | Female | M5 | 46, XX | WT1 (+) | NR |
| QL-067 | c.2645G>A | p.R882H | 60 | Female | M5 | 46, XX | WT1 (+) | NR |
| QL-084 | c.2645G>A | p.R882H | 63 | Female | M5 | 46, XX | NPM1 (+), CEBPA
(+) | NR |
| QL-092 | c.2645G>A | p.R882H | 83 | Female | M5 | 46, XX | FLT3 (+) | Deceased |
DNMT3A expression level
The DNMT3A expression levels were measured using
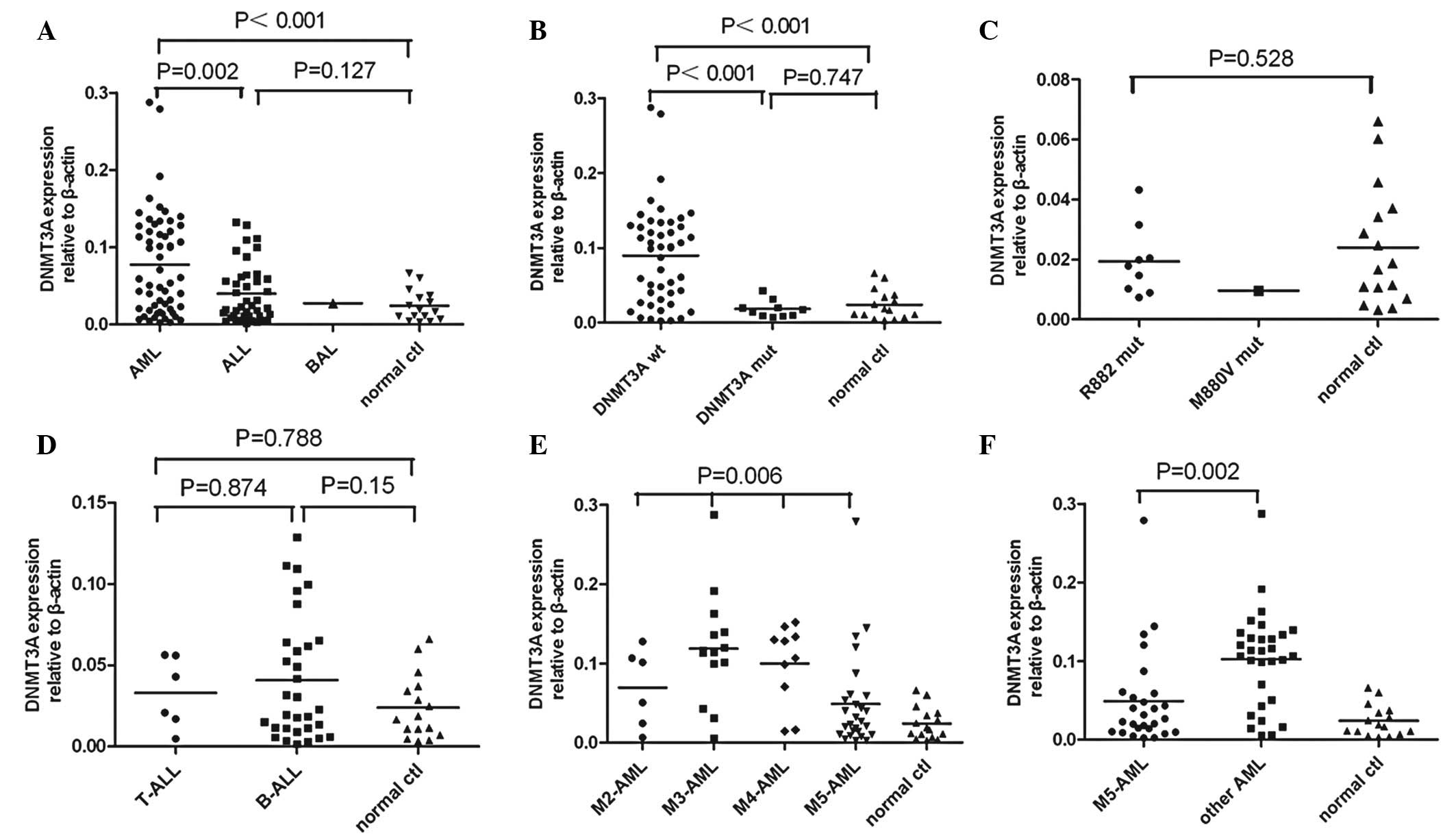
RQ-PCR. DNMT3A expression in the AML patients was found to be
significantly higher than that of the ALL patients or normal
controls (P=0.002 or P<0.001). DNMT3A expression was
significantly decreased in the AML patients with DNMT3A mutations,
including R882 and M880 mutations, compared with individuals
without mutations (P<0.001). The AML patients with wild-type
DNMT3A revealed significantly higher DNMT3A expression levels
compared with the normal controls (P<0.001). No statistical
difference was identified between the AML patients with DNMT3A
mutations and the normal controls (P=0.747). The ALL patients
demonstrated higher DNMT3A expression levels compared with the
normal controls, however, this difference was not statistically
significant (P=0.127). No difference was found in DNMT3A expression
between the T-cell ALL and B-cell ALL patients (P=0.874). DNMT3A
expression between the different AML subtypes (P= 0.006) was
significantly different. The M5 subtype AML patients were found to
exhibit significantly lower DNMT3A expression levels compared with
the patients with other subtypes of AML, including the M2, M3 and
M4 subtypes (P=0.002; Fig. 2).
To determine whether DNMT3A expression levels affect
the treatment response of AML patients, the patients were divided
into 2 groups; those with low or high DNMT3A expression (below or
above the median level, respectively). The CR rate was calculated
for each group according to the DNMT3A expression levels. The group
with low DNMT3A expression revealed a lower CR rate than that of
the high DNMT3A expression group (30.8 vs. 70.4%; P= 0.002). The
ALL patients were also divided into two groups using the same
method. No significant difference was observed in the CR rate (57.9
vs. 60%; P=0.894) between the two groups or between T-cell and
B-cell lymphoblastic leukemia (33.3 vs. 62.1%; P= 0.195).
Discussion
To date, DNMT3A mutations have been detected in AML,
CML, CMML, MDS, lymphoma and MPN. The frequency of DNMT3A mutations
in AML is the highest when compared with other heterogeneous
malignancies. Recently, Ribeiro et al reported that mutant
DNMT3A represents an independent prognostic marker in AML. When
patients with DNMT3A mutations at position R882 were analyzed, an
association with an inferior outcome was also observed (16). In the present study, 10 mutations
were identified in DNMT3A in 10/57 (17.5%) de novo AML
patients. This high frequency is consistent with results of
previous studies on DNMT3A mutations in AML patients (5,17). In
addition, the mutation of DNMT3A was found to correlate with a low
CR rate in AML patients with an intermediate-risk profile,
indicating that the mutation of DNMT3A represents a novel
prognostic index for intermediate-risk AML patients. However, none
of the DNMT3A R882 mutations were identified in this consecutive
series of ALL cases. Prior to the present study, Kim et al
reported that the frequency of DNMT3A mutations in adult ALL was
extremely low (0.8%, 1/124) (18).
Differences in the frequency of DNMT3A mutations between AML and
ALL may be associated with the different pathogenic mechanisms in
AML and ALL. This hypothesis must be studied further, using larger
cohorts to identify DNMT3A mutations in ALL patients and to
evaluate the prognostic impact of the mutations.
In the present study, DNMT3A R882 mutations were
observed to be recurrent in AML patients and associated with a poor
clinical outcome. DNMT3A is markedly over-expressed in the majority
of AML patients with wild-type DNMT3A when compared with normal
controls. However, no difference was identified in DNMT3A
expression between the AML patients with DNMT3A mutations and the
control individuals. These results indicate that DNMT3A mutations
may reduce the methyltransferase activity. Therefore, we
hypothesized that the reduced expression of DNMT3A is indicative of
a poor clinical outcome in AML patients due to decreased
methyltransferase activity. This hypothesis is consistent with the
observations that M880V and all R882 mutations are heterozygous and
that this mutation reduces methyltransferase activity in
vitro (19). Therefore, DNMT3A
expression may represent a potential biomarker for the prediction
of prognosis in AML.
In the present study, the DNMT3A expression levels
in AML patients were compared between various FAB subtypes, and a
significant difference was identified. DNMT3A expression in M5
subtype AML was lower than that of other AML subtypes. In addition,
the frequency of DNMT3A mutations in the AML patients with the M5
subtype was higher compared with the patients of other subtypes,
indicating that decreased DNMT3A expression caused by DNMT3A
mutations may be associated with the incidence and progression of
AML, particularly in the M5 subtype.
The difference in the CR rate between the two groups
of AML patients indicated that lower DNMT3A expression correlated
with an adverse treatment response. In the ALL patients, no
difference was found in the CR rate, which may be explained by the
marked difference in the DNMT3A mutation status and expression
levels between AML and ALL. These results indicate that the
function of DNMT3A in gene methylation in ALL may be distinct from
its role in AML.
A number of studies have confirmed that the DNA
methylation of specific genes is associated with the clinical
outcome (3,20,21)
and that the activity of DNA methyltransferases may contribute to
specific DNA methylation profiles. Previously, Challen et al
analyzed the effect of hematopoietic-specific conditional Dnmt3a
deletion on self-renewal in serial transplantation assays (14). Using conditional ablation, the study
reported that Dnmt3a loss progressively impaired mouse
hematopoietic stem cell (HSC) differentiation. Dnmt3a-null HSCs
were found to exhibit increased and decreased methylation at
distinct loci, including substantial CpG island hyper- and
hypomethylation. In the Dnmt3a-null HSCs, an extremely large number
of hypomethylated genes were found that are commonly overexpressed
in different types of leukemia, including AML and ALL. These
observations are indicative of a crucial role for Dnmt3a in the
pathogenesis of malignant neoplasms. However, the Dnmt3a-null
status in HSCs in mice is distinct from DNMT3A mutations in humans,
as all R882 mutations are heterozygous. Whether the same set of
genes is subjected to altered epigenetic patterning in
DNMT3A-mutant AML cells has not been investigated.
TET2 and NPM1 mutations are markedly associated with
DNMT3A mutations in T-cell lymphoma and adult AML, respectively
(5,10,17).
These studies indicate an oncogenic cooperation between DNMT3A and
other gene mutations, resulting in the deregulation of the cytosine
methylation and demethylation processes. In the present study, the
patients with DNMT3A mutations were older than the patients without
DNMT3A mutations in AML, which was consistent with the results
obtained by Ley et al (5).
Consistent with these observations, a low frequency of DNMT3A
mutations in pediatric AML was observed in studies by Ho et
al (0/180, 0%) (22) and
Hollink et al (3/140, 2.1%) (23). In pediatric AML, there is a 4–5-fold
lower frequency of NPM1 mutations compared with adult AML (24). These results may partially explain
why the frequency of DNMT3A mutations is low. Therefore, we
hypothesized that DNMT3A mutations alone are insufficient to
generate AML and other malignancies, and that second hits may be
required.
The results of the present study, in combination
with observations of previous studies, indicate that DNMT3A
mutations are associated with adverse outcomes in AML and that they
may represent a novel marker for the risk stratification of AML. By
contrast, DNMT3A mutations in ALL are rare. At present, the
mechanisms by which mutated DNMT3A regulates DNA methylation remain
unclear. Additional studies must be performed to identify and
understand the regulatory mechanisms of DNMT3A. Screening for
DNMT3A mutations may provide a novel tool for the prediction of
clinical outcome.
Acknowledgements
The present study was supported by
grants from the National Natural Science Foundation of China (nos.
81070422, 30871088, 81070407 and 81170515), the Specialized
Research Fund for the Doctoral Program of Higher Education of the
Ministry of Education (no. 20100131110060) and the Independent
Innovation Fund of Shandong University (IIFSDU21300072613160).
References
|
1.
|
Alvarez S, Suela J, Valencia A, et al: DNA
methylation profiles and their relationship with cytogenetic status
in adult acute myeloid leukemia. PLoS One. 5:e121972010. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Roman-Gomez J, Jimenez-Velasco A, Agirre
X, et al: Promoter hypermethylation and global hypomethylation are
independent epigenetic events in lymphoid leukemogenesis with
opposing effects on clinical outcome. Leukemia. 20:1445–1447. 2006.
View Article : Google Scholar
|
|
3.
|
Kraszewska MD, Dawidowska M, Larmonie NSD,
et al: DNA methylation pattern is altered in childhood T-cell acute
lymphoblastic leukemia patients as compared with normal thymic
subsets: insights into CpG island methylator phenotype in T-ALL.
Leukemia. 26:367–371. 2011. View Article : Google Scholar
|
|
4.
|
Holz-Schietinger C, Matje DM, Harrison MF
and Reich NO: Oligomerization of DNMT3A controls the mechanism of
de novo DNA methylation. J Biol Chem. 286:41479–41488. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Ley TJ, Ding L, Walter MJ, et al: DNMT3A
mutations in acute myeloid leukemia. N Engl J Med. 363:2424–2433.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Li X, Cen J, Wang Q, et al: Absence of
DNMT3A gene mutation in chronic myeloid leukemia patients in blast
crisis. Eur J Haematol. 88:455–457. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Shen Y, Zhu YM, Fan X, et al: Gene
mutation patterns and their prognostic impact in a cohort of 1185
patients with acute myeloid leukemia. Blood. 118:5593–5603. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Jankowska AM, Makishima H, Tiu RV, et al:
Mutational spectrum analysis of chronic myelomonocytic leukemia
includes genes associated with epigenetic regulation: UTX, EZH2 and
DNMT3A. Blood. 118:3932–3941. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Walter MJ, Ding L, Shen D, et al:
Recurrent DNMT3A mutations in patients with myelodysplastic
syndromes. Leukemia. 25:1153–1158. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Couronné L, Bastard C and Bernard OA: TET2
and DNMT3A mutations in human T-Cell lymphoma. N Engl J Med.
366:95–96. 2012.PubMed/NCBI
|
|
11.
|
Stegelmann F, Bullinger L, Schlenk RF, et
al: DNMT3A mutations in myeloproliferative neoplasms. Leukemia.
25:1217–1219. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Bennett JM, Catovsky D, Daniel MT, et al:
Proposals for the classification of the acute leukaemias.
French-American-British (FAB) co-operative group. Br J Haematol.
33:451–458. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Qiao SK, Xu SR, Guo XN and Wang Y:
Clinical significance of the expression of DNA methyltransferase
genes (DNMT) in acute leukemia patients. Zhongguo Shi Yan Xue Ye
Xue Za Zhi. 13:260–265. 2005.(In Chinese).
|
|
14.
|
Challen GA, Sun D, Jeong M, et al: Dnmt3a
is essential for hematopoietic stem cell differentiation. Nat
Genet. 44:23–31. 2011. View
Article : Google Scholar
|
|
15.
|
Cheson BD, Bennett JM, Kopecky KJ, et al:
Revised recommendations of the International Working Group for
diagnosis, standardization of response criteria, treatment
outcomes, and reporting standards for therapeutic trials in acute
myeloid leukemia. J Clin Oncol. 21:4642–4649. 2003. View Article : Google Scholar
|
|
16.
|
Ribeiro AF, Pratcorona M,
Erpelinck-Verschueren C, et al: Mutant DNMT3A: a marker of poor
prognosis in acute myeloid leukemia. Blood. 119:5824–5831. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Yan XJ, Xu J, Gu ZH, et al: Exome
sequencing identifies somatic mutations of DNA methyltransferase
gene DNMT3A in acute monocytic leukemia. Nat Genet. 43:309–315.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Kim MS, Kim YR, Yoo NJ and Lee SH:
Mutational analysis of DNMT3A gene in acute leukemias and common
solid cancers. APMIS. 121:85–94. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Yamashita Y, Yuan J, Suetake I, et al:
Array-based genomic resequencing of human leukemia. Oncogene.
29:3723–3731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Shen L, Kantarjian H, Guo Y, et al: DNA
methylation predicts survival and response to therapy in patients
with myelodysplastic syndromes. J Clin Oncol. 28:605–613. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Figueroa ME, Lugthart S, Li Y, et al: DNA
methylation signatures identify biologically distinct subtypes in
acute myeloid leukemia. Cancer Cell. 17:13–27. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Ho PA, Kutny MA, Alonzo TA, et al:
Leukemic mutations in the methylation-associated genes DNMT3A and
IDH2 are rare events in pediatric AML: a report from the Children’s
Oncology Group. Pediatr Blood Cancer. 57:204–209. 2011.PubMed/NCBI
|
|
23.
|
Hollink IH, Feng Q, Danen-van Oorschot AA,
et al: Low frequency of DNMT3A mutations in pediatric AML and the
identification of the OCI-AML3 cell line as an in vitro model.
Leukemia. 26:371–373. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Hollink IH, Zwaan CM, Zimmermann M, et al:
Favorable prognostic impact of NPM1 gene mutations in childhood
acute myeloid leukemia, with emphasis on cytogenetically normal
AML. Leukemia. 23:262–270. 2009. View Article : Google Scholar : PubMed/NCBI
|