Introduction
Advances in chemotherapeutic regimens for metastatic
colorectal cancer (MCRC) patients, including FOLFOX treatment
comprising of a combination of 5-fluorouracil (FU)/leucovorin (LV)
and oxaliplatin, have improved overall survival (OS) (1,2). It
has been reported that the efficacy rate of FOLFOX treatment varies
between 20 and 50% in MCRC patients (1,3–5).
Individuals who receive chemotherapy commonly suffer from side
effects, including myelosuppression, nausea, diarrhea and
peripheral neuropathy (1–3). Numerous patients undergoing FOLFOX
treatment have complained of oxaliplatin-induced peripheral
neuropathy. Therefore, several markers for predicting the efficacy
of FOLFOX treatment have been investigated to identify patients
with favorable treatment prognoses. Gene expression analysis,
associated with the metabolism of 5-FU and oxaliplatin, has been
intensively studied (6–9). It has been reported that thymidylate
synthase (TS) and thymidine phosphorylase (TP) mRNA
expression levels are useful markers for predicting the efficacy of
FOLFOX treatment in CRC patients with liver metastasis (10). In addition, advances in molecular
biology indicate that a number of drug metabolism genes have
polymorphisms that alter levels of expression. Among these,
polymorphisms have been identified in TS, excision repair
cross-complementing-1 (ERCC1) and ERCC2, glutathione
S-transferase π (GSTP1), GST θ1 (GSTT1), GST μ1
(GSTM1) and methylenetetrahydrofolate reductase
(MTHFR), which exert functions in drug metabolism and
antidotal effects on the 5-FU and oxaliplatin pathways. Studies
have demonstrated that specific polymorphisms of these genes are
associated with the efficacy of FOLFOX treatment in MCRC patients
(11–14).
TS and MTHFR are associated with the
metabolism of 5-FU, indicating that their altered expression
affects the response to 5-FU-based chemotherapy. The enzyme product
of TS is critical for catalyzing the methylation of
deoxyuridine-5′-monophosphate to deoxythymidine-5′-monophosphate in
de novo DNA synthesis. Fluorodeoxyuridine monophosphate
(FdUMP), the metabolic product of 5-FU, forms complexes with TS and
5,10-methylenetetrahydrofolate, resulting in the inhibition of DNA
synthesis. Two polymorphisms have been identified in TS, a
variable length tandem repeat polymorphism in the 5′-untranslated
region (UTR) that consists of two or three 28-bp repeated sequences
and a 6-bp insertion/deletion (6+/6−) in the 3′-UTR. A number of
studies have described correlations between genotype patterns of
polymorphisms in TS and the efficacy of FOLFOX treatment for
MCRC patients (11–14). However, current evidence is
insufficient to confirm a statistically significant correlation.
MTHFR is important for folate metabolism and catalyzing the
conversion of 5,10-methylenetetrahydrofolate to
5-methyltetrahydrofolate. Two important polymorphisms in
MTHFR, C677T and A1298C, have been studied (12–14)
and have been identified to affect the enzyme activity of MTHFR
(15,16), leading to the accumulation of
5,10-methylenetetrahydrofolate and the enhanced sensitivity of 5-FU
by forming complexes with TS and FdUMP. Studies have described the
correlation among these polymorphisms and the efficacy of FOLFOX
treatment for MCRC patients (12,14).
The expression levels of ERCC1, ERCC2,
GSTP1, GSTT1 and GSTM1 have been hypothesized
to be associated with the efficacy of platinum compounds, including
cisplatin and oxaliplatin. ERCC-1 and -2 are involved
in DNA repair and tolerance of DNA damage through the nucleotide
excision repair pathway. The enhanced expression of these proteins
may lead to the resistance to platinum drugs. A common C to T
transition at codon 118 of ERCC1 has been shown to increase
expression in patients with the T/T genotype compared with patients
with the C/T or C/C genotypes (17), despite the T/T polymorphism
producing the identical amino acid, asparagine. Studies have
revealed that patients with the T/T genotype have a poor outcome
compared with patients with the C/T or C/C genotypes (13). During oxaliplatin-based
chemotherapy, the prognosis for MCRC patients with the C/C genotype
is more encouraging than that of patients with other genotypes
(11,18,19).
The xeroderma pigmentosum group D (XPD) gene, ERCC2, has
three common polymorphisms in codons 156, 312 and 751. The
polymorphism at codon 751 (A>C: Lys>Gln) is associated with
the clinical outcome of MCRC patients receiving FOLFOX treatment
(13,20).
The GST family includes at least five subclasses
with major biological roles in the detoxification of genotoxic
compounds. GSTP1, GSTT1 and GSTM1 genotypes
have been extensively studied for drug response, including
oxaliplatin-based treatment (21–23). A
single nucleotide polymorphism at codon 105 (A>G: Ile>Val) of
GSTP1 affects enzyme activity (24). Several studies have demonstrated
that among MCRC patients receiving oxaliplatin-based treatment,
patients with GSTP1-105 A/G and G/G genotypes have a more
favorable outcome compared with patients with the GSTP1-105
A/A genotype (21–23).
In the present study, correlations were identified
between the polymorphism patterns of TS, MTHFR,
ERCC1, ERCC2, GSTP1, GSTT1 and GSTM1
and the clinical outcome, including the incidence of peripheral
neuropathy, in Japanese MCRC patients who were treated with
modified FOLFOX6 (mFOLFOX6).
Materials and methods
Patients and clinical procedures
The current study was performed in accordance with
the ethical guidelines for clinical studies with approval from the
institutional ethics committee. Informed consent was obtained from
all individuals.
The subjects included 63 CRC patients (22 females
and 41 males) who received mFOLFOX6 treatment as first-line
chemotherapy between 2005 and 2009. The mFOLFOX6 regimen was
comprised of intravenous infusions of oxaliplatin (85
mg/m2) and LV (200 mg/m2) for 2 h, followed
by a rapid intravenous bolus infusion of 5-FU (400
mg/m2) for 5 min and a continuous intravenous infusion
of 5-FU (2,400 mg/m2) for 46 h. This regimen was
repeated every 2 weeks. Table I
presents the patient characteristics. The median age of the
patients was 65 years old (range, 32–84 years old). The primary
site was the colon/rectosigmoid in 43 patients and the rectum in 20
patients. Performance status (PS), determined according to the
method of the Eastern Cooperative Oncology Group was 0 in 39
patients, 1 in 21 patients and 2 in 3 patients. The target lesions
were located in the liver of 43 patients, the lungs of 18 patients,
the peritoneum of 13 patients and the lymph nodes of 16 patients,
while in 7 patients the target lesions were detected in other
locations. The median number of oxaliplatin doses was 10 (range,
4–39) and the median relative dose intensity of oxaliplatin was 75%
(range, 28.1–100%). The response to mFOLFOX6 treatment was
evaluated during 4–6 courses of treatment according to the Response
Evaluation Criteria in Solid Tumors (version 1.1) (25). Complete response was observed in 3
patients, partial response in 23 patients, stable disease in 24
patients and progressive disease in 13 patients. Adverse events
were graded according to the Common Terminology Criteria for
Adverse Events (version 3.0). When an adverse event of >grade 3
severity occurred, mFOLFOX6 therapy was suspended until the
severity of the reaction improved to <grade 2. When mFOLFOX6
therapy was resumed, doses of oxaliplatin were reduced to 70–80% of
the previous dose.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Parameter | Value |
|---|
| Total patients,
n | 63 |
| Gender,
males:females | 41:22 |
| Age, yearsa | 65 (32–84) |
| Location, n |
|
Colon/rectosigmoid | 43 |
| Rectum | 20 |
| Performance status,
n |
| 0 | 39 |
| 1 | 21 |
| 2 | 3 |
| Number of target
organ(s) |
| 1 | 26 |
| >2 | 37 |
| Target organ,
n |
| Liver | 43 |
| Lung | 18 |
| Lymph node | 16 |
| Peritoneum | 13 |
| Others | 7 |
| Cycles of mFOLFOX6
therapy, na | 10.0 (4–39) |
| Relative dose
intensity, %a | 75.0
(28.1–100) |
| Response, n |
| CR | 3 |
| PR | 23 |
| SD | 24 |
| PD | 13 |
| Second line
chemotherapy, n |
| FOLFIRI | 29 |
| FOLFIRI +
bevacizumab | 15 |
| Other | 3 |
| Best supportive
care | 16 |
Of the 63 patients with CRC, 44 received folinic
acid/5-FU/irinotecan (FOLFIRI) either with (n=15) or without
bevacizumab (n=29) as a second-line chemotherapy treatment. The
FOLFIRI regimen comprised of intravenous infusions of irinotecan
(150 mg/m2) and LV (200 mg/m2) for 2 h,
followed by a rapid intravenous bolus infusion of 5-FU (400
mg/m2) for 5 min and a continuous intravenous infusion
of 5-FU (2,400 mg/m2) for 46 h, administered every 2
weeks. A total of 16 patients were observed without administration
of additional treatment.
DNA extraction and analysis of
polymorphisms
Genomic DNA was extracted from 23 blood samples and
40 normal colonic mucosae from each enrolled patient using the
QIAamp DNA Blood and QIAamp DNA Mini kits (Qiagen, Tokyo, Japan).
Polymorphisms were analyzed by polymerase chain reaction (PCR), a
PCR restriction fragment length polymorphism (PCR-RFLP) technique
and a PCR-invader method. Primer sequences and restriction enzymes
of all genes examined are presented in Table II.
 | Table IICharacteristics of polymorphisms with
primer sequences and restriction enzymes. |
Table II
Characteristics of polymorphisms with
primer sequences and restriction enzymes.
| Site | Polymorphism | Genotype | Restriction
enzymes | Primers | Detection
method |
|---|
| TS
5′-UTR | VNTR | 2R or 3R
alleles | |
5′-AGGCGCGCGGAAGGGGTCCT-3′
5′-TCCGAGCCGGCCACAGGCAT-3′ | PCR |
| TS
3′-UTR | 6 bp
insertion/deletion | 6+/6− | Dral |
5′-CAAATCTGAGGGAGCTGAGT-3′
5′-CAGATAAGTGGCAGTACAGA-3′ | PCR-RFLP |
| MTHFR (exon
4) | SNP | C/T, Ala677Val | Hinfi |
5′-TGAAGGAGATGTCTGCGGGA-3′
5′-AGGACGGTGCGGTGAGAGTG-3′ | Invader method |
| ERCC1 (exon
4) | SNP | C/T, Asn118Asn | Maell |
5′-GAGAGGGCTGAGCTGGAGACAG-3′
5′-CCAGCACATAGTCGGGAATTACGTC-3′ | PCR-RFLP |
| ERCC2 (exon
23) | SNP | A/C, Lys751Gln | Mboll |
5′-CAGGTGAGGGGGACATCTG-3′
5′-CTCTCCCTTTCCTCTGTTC-3′ | PCR-RFLP |
| GSTP1 (exon
5) | SNP | A/G, IIe105Val | Mspl |
5′-ACCCCAGGGCTCTATGGGAA-3′
5′-TGAGGGCACAAGCCCCT-3′ | PCR-RFLP |
| GSTT1 | Deletion | ±a | |
5′-TTCCTTACTGGTCCTCCTCACATCTC-3′
5′-TCACCGGATCATGGCCAGCA-3′ | PCR |
| GSTM1 | Deletion | ±a | |
5′-GAACTCCCTGAAAAGCTAAAGC-3′
5′-GTTGGGCTCAAATATACGGTGG-3′ | PCR |
Statistical analysis
Continuous data are presented as the median and
range. Mann-Whitney U, Fisher’s exact probability and χ2
tests were used where applicable. A survival analysis was conducted
using the Kaplan-Meier method. The log-rank test was used to
determine the significance of the survival curves. The OS period
was calculated between the time of surgery and the date of
mortality of any cause. OS was censored from the time of the
individuals last visit to the hospital or December 2010, depending
on which was the first event. Logistic regression was used to
determine independent predictors of adverse events. P<0.05 was
considered to indicate a statistically significant difference. All
statistical analyses were performed using a statistical software
package (StatFlex ver.6.0; Artech, Osaka, Japan).
Results
Correlation between polymorphisms in TS,
MTHFR, ERCC1, ERCC2, GSTP1, GSTT1 and GSTM1 and the response rate
to mFOLFOX6 treatment
Polymorphisms of GSTP1-105 were shown to
significantly correlate with the efficacy of mFOLFOX6 treatment
(Table III). The frequencies of
GSTP1-105 A/A, A/G and G/G genotypes were 70, 25 and 3%,
respectively. In the responder group, fewer patients expressed the
GSTP1-105 A/A genotype than the GSTP1-105 A/G or G/G
genotypes (P=0.01). No significant differences were identified
between the polymorphisms of other genes and the efficacy of
mFOLFOX6 treatment.
 | Table IIIFrequency of polymorphisms, response
rate and median PFS and OS. |
Table III
Frequency of polymorphisms, response
rate and median PFS and OS.
| Gene | Patients n=63, n
(%) | Responder, n
(%) | P-value | Median PFSb, months | P-value | Median OSb, months | P-value |
|---|
|
TS-5′UTR | | | 0.11 | | 0.56 | | 0.650 |
| 3R/3R | 46 (73) | 18 (39) | | 8.6 | | 27.0 | |
| 2R/3R | 13 (21) | 8 (62) | | 9.9 | | 25.4 | |
| 2R/2R | 3 (5) | 0 (0) | | 11.1 | | 31.8 | |
| Unknown | 1 (2) | | | | | | |
|
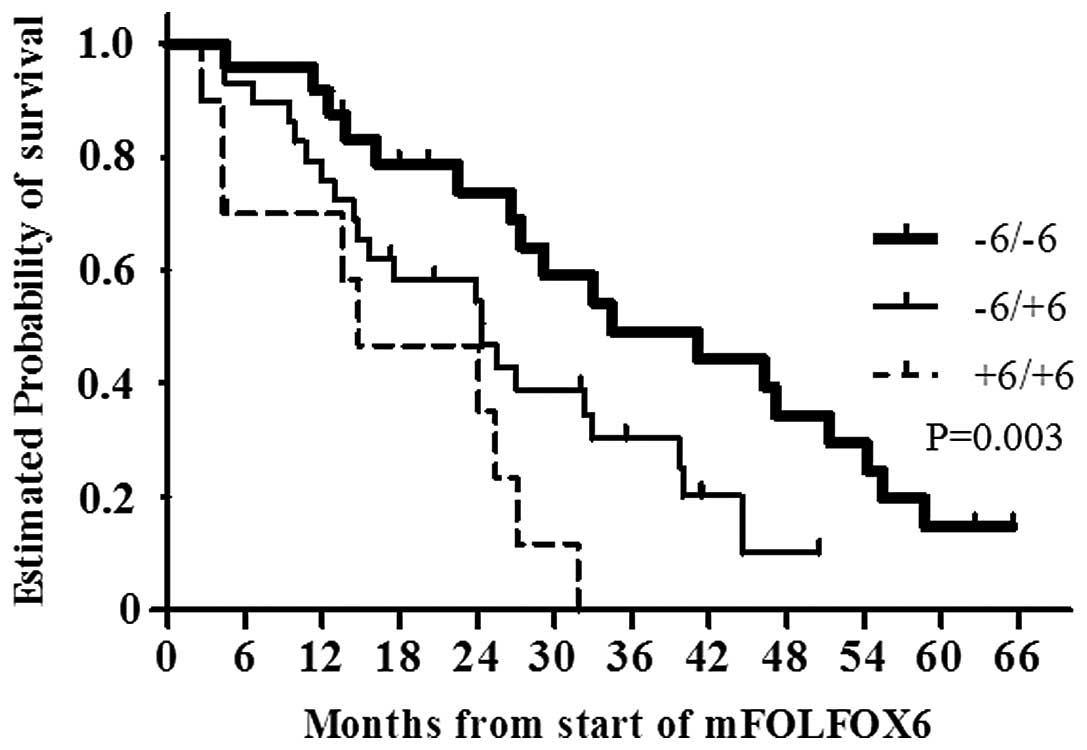
TS-3′UTR | | | 0.93 | | 0.48 | | 0.003 |
| −6/−6 | 24 (38) | 10 (42) | | 11.6 | | 34.4 | |
| −6/+6 | 29 (46) | 13 (45) | | 8.3 | | 24.4 | |
| +6/+6 | 10 (16) | 3 (30) | | 10.7 | | 14.8 | |
|
MTHFR-677 | | | 0.70 | | 0.80 | | 0.860 |
| C/C | 26 (41) | 12 (46) | | 9.9 | | 27.4 | |
| C/T | 30 (48) | 11 (37) | | 8.1 | | 27.0 | |
| T/T | 6 (10) | 3 (50) | | 8.3 | | 24.4 | |
| Unknown | 1 (2) | | | | | | |
|
ERCC1-118 | | | 0.71 | | 0.63 | | 0.380 |
| C/C | 30 (48) | 11 (37) | | 9.9 | | 27.4 | |
| C/T | 23 (37) | 11 (48) | | 8.1 | | 22.5 | |
| T/T | 10 (16) | 4 (40) | | 8.3 | | 32.9 | |
|
ERCC2-751 | | | 0.95 | | 0.05 | | 0.690 |
| A/A | 58 (92) | 24 (41) | | 10.3 | | 25.5 | |
| A/C | 5 (8) | 2 (40) | | 6.1 | | 29.2 | |
| C/C | 0 (0) | | | | | | |
|
GSTP1-105 | | | 0.05 | | 0.41 | | 0.260 |
| A/A | 44 (70) | 14 (32) | 0.01a | 8.6 | | 24.4 | |
| A/G | 16 (25) | 11 (69) | | 7.8 | | 31.1 | |
| G/G | 2 (3) | 1 (50) | | 11.8 | | 46.3 | |
| Unknown | 1 (2) | | | | | | |
| GSTT1 | | | 0.83 | | 0.47 | | 0.840 |
| Positive | 30 (48) | 13 (43) | | 8.1 | | 25.5 | |
| Negative | 32 (51) | 13 (41) | | 10.3 | | 27.1 | |
| Unknown | 1 (2) | | | | | | |
| GSTM1 | | | 0.73 | | 0.89 | | 0.480 |
| Positive | 23 (37) | 9 (39) | | 7.4 | | 22.5 | |
| Negative | 39 (62) | 17 (44) | | 10.7 | | 27.4 | |
| Unknown | 1 (2) | | | | | | |
Correlation between polymorphisms in TS,
MTHFR, ERCC1, ERCC2, GSTP1, GSTT1 and GSTM1 and PFS and OS in MCRC
patients treated with mFOLFOX6
Frequencies of the ERCC2-751 A/A, A/C and C/C
genotypes were 92, 8 and 0%, respectively (Table III). The median PFS of patients
with the ERCC2-751 A/A genotype was longer than that of
patients with the ERCC2-751 A/C genotype (10.3 and 6.1
months, respectively; P=0.05). There was no correlation between
polymorphisms of other genes and PFS. The median OS of the patients
with the TS-3′-UTR −6/−6 (n=24), −6/+6 (n=29) and +6/+6
(n=10) genotypes was 34.4, 24.4 and 14.8 months, respectively. The
OS of the patients with the TS-3′-UTR −6/−6 genotype was
significantly longer compared with that of the patients with other
genotypes (P=0.003; Fig. 1).
Correlation between polymorphisms in TS,
MTHFR, ERCC1, ERCC2, GSTP1, GSTT1 and GSTM1 and incidence of
peripheral neuropathy in patients treated with mFOLFOX6
The incidence of peripheral neuropathy of grades 2
(n=42) and 3 (n=2) was found to significantly correlate with the
GSTP1-105 (P=0.05) and GSTM1 (P=0.03) genotypes, as
identified by univariate regression analyses (Table IV). Peripheral neuropathy occurred
in the majority of patients with the GSTP1-105 A/G and G/G
genotypes compared with patients with the GSTP1-105 A/A
genotype. Individuals who were GSTM1-negative also had
peripheral neuropathy, whereas individuals who were
GSTM1-positive did not. A statistically significant
correlation between the incidence of peripheral neuropathy higher
than grade 2 and the GSTP1-105 (P=0.03) and GSTM1
(P=0.02) genotypes was determined using multivariate regression
analysis.
 | Table IVCorrelation between peripheral
neuropathy and polymorphisms. |
Table IV
Correlation between peripheral
neuropathy and polymorphisms.
| Gene | Patients, n | Patients with
>Grade 2, n (%) | Univariate
regression analysis | Multivariate
regression analysis |
|---|
|
|
|---|
| OR | 95% CI | P-value | OR | 95% CI | P-value |
|---|
|
TS-5′UTR |
| 3R/3R | 46 | 32 (70) | 1 | | | | | |
| 2R/2R, 2R/3R | 16 | 11 (69) | 0.962 | 0.281–3.289 | 0.95 | | | |
|
TS-3′UTR |
| −6/−6 | 24 | 19 (79) | 1 | | | | | |
| −6/+6, +6/+6 | 39 | 25 (64) | 0.470 | 0.144–1.533 | 0.21 | | | |
|
MTHFR-677 |
| C/C | 26 | 19 (73) | 1 | | | | | |
| C/T, T/T | 36 | 24 (67) | 0.737 | 0.243–2.235 | 0.59 | | | |
|
ERCC1-118 |
| C/C | 30 | 20 (67) | 1 | | | | | |
| C/T, T/T | 33 | 24 (73) | 1.333 | 0.453–3.921 | 0.60 | | | |
|
ERCC2-751 |
| A/A | 58 | 41 (71) | 1 | | | | | |
| A/C, C/C | 5 | 3 (60) | 0.622 | 0.095–4.062 | 0.62 | | | |
|
GSTP1-105 |
| A/A | 44 | 27 (61) | 1 | | | 1 | | |
| A/G, G/G | 18 | 16 (89) | 5.037 | 1.027–24.712 | 0.05 | 6.084 | 1.150–32.175 | 0.03 |
| GSTT1 |
| Positive | 30 | 21 (70) | 1 | | | | | |
| Negative | 32 | 22 (69) | 0.943 | 0.320–2.778 | 0.92 | | | |
| GSTM1 |
| Positive | 23 | 12 (52) | 1 | | | 1 | | |
| Negative | 39 | 31 (79) | 3.546 | 1.149–10.989 | 0.03 | 4.202 | 1.253–14.085 | 0.02 |
Discussion
In the present study, specific polymorphisms of
genes involved in 5-FU/oxaliplatin metabolism were demonstrated to
be significantly associated with the clinical outcome of Japanese
MCRC patients who received first-line chemotherapy with mFOLFOX6.
The response to mFOLFOX6 treatment in patients with the
GSTP1-105 A/G and G/G genotypes was significantly improved
compared with that of patients with the GSTP1-105 A/A
genotype. In addition, the ERCC2-751 and TS-3′-UTR
genotypes were shown to significantly correlate with PFS and OS,
respectively. The results indicated that polymorphisms in the
oxaliplatin-associated genes, GSTP1-105 and
ERCC2-751, were hypothesized to be important for the
prediction of primary clinical outcome, including drug responses
and PFS, for MCRC patients treated with mFOLFOX6. Second- and
third-line chemotherapy regimens also affected OS. Of the 63
patients, 44 were treated with FOLFIRI following the FOLFOX regimen
and 5-FU treatment continued throughout. Therefore, it is possible
that polymorphisms in the genes involved in 5-FU metabolism
contribute to OS in long-term observations.
Previous studies have revealed that GSTP1-105
genotypes are associated with the clinical outcome of MCRC patients
who receive 5-FU/oxaliplatin as first-line chemotherapy (11–13,21–23).
As GSTP1 expression is enhanced in CRC (26) it has been hypothesized to be
involved in the resistance to platinum compounds (27). The enzyme activity of the
GSTP1-105 A/G and G/G genotypes is lower than that of the
GSTP1-105 A/A genotype (24). In addition, clinical assessments of
the correlation between the GSTP1 genotype and the clinical
outcome in MCRC patients treated with 5-FU/oxaliplatin appears to
be consistent with basic studies. In the present study, patients
with the GSTP1-105 A/G and G/G genotypes were revealed to
have a significantly improved response to mFOLFOX6 treatment when
compared with patients with the GSTP1-105 A/A genotype.
Previous studies have indicated that the GSTP1-105 A/G and
G/G genotypes are significantly more common than the
GSTP1-105 A/A genotype among patients who respond to
5-FU/oxaliplatin treatment (22,23).
In addition to drug response, several studies have demonstrated
that MCRC patients with the GSTP1-105 A/G and G/G genotypes
have favorable outcomes following oxaliplatin-based treatment
compared with patients with the GSTP1-105 A/A genotype
(11,21). This tendency was also recognized in
the results of the current study. The frequencies of the
GSTP1-105 A/A, A/G and G/G polymorphisms were 70, 25 and 3%,
respectively, in the Japanese population, which is similar to
frequencies reported in other Asian populations, including Chinese
and Taiwanese (11,22,24).
In American and European populations, there is an almost equal
frequency of GSTP1-105 A/A and A/G carriers, which combine
to make a total of ~90% of all patients. By contrast, the frequency
of the GSTP1-105 G/G genotype is ~10% in these populations
(11–13,21,24).
Regardless of ethnic differences, the association of the
GSTP1-105 genotype with the clinical outcome is consistent
among all MCRC patients who receive 5-FU/oxaliplatin as first-line
chemotherapy.
In addition to GSTP1 and ERCC-1 and
-2, members of the nucleotide excision repair pathway are
involved in repair and tolerance of DNA damage and also encode key
enzymes for oxaliplatin metabolism. Several studies have
demonstrated that the ERCC1-118 and ERCC2-751
genotypes are associated with the clinical outcome of MCRC patients
receiving oxaliplatin-based treatment (11,13,18–20).
In the present study, the ERCC2-751 genotypes were
significantly associated with PFS, whereas no significant
difference was identified between the ERCC1-118 genotype and the
clinical outcome. The PFS of the patients with the ERCC2-751
A/A genotype was longer than that of patients with the
ERCC2-751 A/C genotype, and this was consistent with
previous studies (11,13,20).
The distribution of ERCC2-751 polymorphisms clearly differs
between Asian and Western individuals. Among Asians, the
frequencies of the ERCC2-751 A/A, A/C and C/C genotypes are
84–92, 8–16 and 0%, respectively (11,22),
whereas among Americans and Europeans the frequencies are 25–38,
50–61 and 11–15%, respectively (11–13,20,22,23).
There is a high possibility that the majority of Asians carry the
ERCC2-751 A/A genotype, leading to promising outcomes of
oxaliplatin-based chemotherapy.
A statistically significant association between
TS-3′-UTR genotypes and OS was identified in the current
study. Treatment with FOLFIRI was also administered as second-line
chemotherapy to ~70% of patients receiving mFOLFOX6 treatment, and
hence, the patients with increased survival rates were exposed to
5-FU for a long time. Numerous studies have indicated that MCRC
patients with lower TS expression have a favorable outcome
following 5-FU-based chemotherapy compared with patients with high
TS expression (10,28). A previous study revealed that
TS mRNA expression in rectal cancer patients with the
TS-3′-UTR −6/−6 and −6/+6 genotypes was significantly lower
compared with patients with the TS-3′-UTR +6/+6 genotype,
resulting in a favorable outcome following neoadjuvant 5-FU-based
chemoradiation (29). Among CRC
patients receiving 5-FU-based adjuvant treatment, the OS of
patients with the TS-3′-UTR −6/−6 genotype was significantly
longer compared with that of patients with other genotypes
(30). Although there are various
types of cancer, an encouraging association between clinical
outcome and the TS-3′-UTR −6/−6 genotype has been identified
in Asian gastric cancer patients receiving mFOLFOX6 treatment
(31). Several studies have
reported that there is no correlation between the TS-3′-UTR
genotype and the clinical outcome of MCRC patients receiving
5-FU/oxaliplatin treatment (12–14,23).
The frequency distribution of the TS-3′-UTR genotype may
lead to discrepancies in the clinical outcome. In the current
study, the frequencies of the TS-3′-UTR −6/−6, −6/+6 and
+6/+6 genotypes were 38, 46 and 16%, respectively, while in the USA
and Europe these genotypes are 10–16, 37-5 and 33–53%, respectively
(11–13,23).
Further studies may be required to clarify the association between
these differences in ethnicity and the efficacy of anti-cancer
drugs.
While mFOLFOX6 treatment improves the survival rate
of MCRC patients, adverse events, including myelosuppression,
nausea, diarrhea and peripheral neuropathy, are common. In
particular, peripheral neuropathy, caused by cumulative
administration of oxaliplatin, directly affects the quality of life
and is a major reason for the discontinuation of oxaliplatin
chemotherapy. Thus, predictive markers of peripheral neuropathy are
required for prospective evaluations. In agreement with previous
studies, the incidence of peripheral neuropathy higher than grade 2
was identified to significantly correlate with the GSTP1-105
and GSTM1 genotypes (13,22).
Notably, peripheral neuropathy in patients with the
GSTP1-105 A/G and G/G genotypes was of greater intensity
compared with that of patients with the GSTP1-105 A/A
genotype. A statistically significant correlation was identified
between the GSTP1-105 genotype and the clinical outcome.
Therefore, the GSTP1-105 polymorphism may serve as a
double-edged marker for predicting response to 5-FU/oxaliplatin
treatment and the intensity of oxaliplatin-associated peripheral
neuropathy.
In the present study, the association among gene
polymorphisms that affect the metabolism of 5-FU oxaliplatin and
the clinical outcome in Japanese patients with MCRC was identified.
Ethnic differences in the frequency distribution of polymorphisms,
which preclude the extrapolation of clinical studies between
Western and Asian populations, were also identified. Therefore, the
present study is likely to improve chemotherapy for individuals of
Asian descent. Consistent with studies in Western patients, the
polymorphisms of GSTP1-105, ERCC2-751 and the 3′-UTR
of TS were associated with the clinical outcome of FOLFOX
treatment in Japanese MCRC patients. Therefore, these polymorphisms
may be significant predictors of clinical outcome globally.
However, GSTP1-105 and GSTM1 genotypes may be more
useful as markers for severe oxaliplatin-induced peripheral
neuropathy in Japanese patients compared with Western patients.
References
|
1
|
de Gramont A, Figer A, Seymour M, et al:
Leucovorin and fluorouracil with or without oxaliplatin as
first-line treatment in advanced colorectal cancer. J Clin Oncol.
18:2938–2947. 2000.
|
|
2
|
Tournigand C, André T, Achille E, et al:
FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced
colorectal cancer: a randomized GERCOR study. J Clin Oncol.
22:229–237. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Goldberg RM, Sargent DJ, Morton RF, et al:
A randomized controlled trial of fluorouracil plus leucovorin,
irinotecan and oxaliplatin combinations in patients with previously
untreated metastatic colorectal cancer. J Clin Oncol. 22:23–30.
2004. View Article : Google Scholar
|
|
4
|
Reddy GK, Gibson AD and Price N: Evolution
of FOLFOX regimens in the treatment of advanced colorectal cancer.
Clin Colorectal Cancer. 4:296–299. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shimizu T, Satoh T, Tamura K, et al:
Oxaliplatin/fluorouracil/leucovorin (FOLFOX4 and modified FOLFOX6)
in patients with refractory or advanced colorectal cancer:
post-approval Japanese population experience. Int J Clin Oncol.
2:218–223. 2007. View Article : Google Scholar
|
|
6
|
Leichman CG, Lenz HJ, Leichman L, et al:
Quantitation of intratumoral thymidylate synthase expression
predicts for disseminated colorectal cancer response and resistance
to protracted-infusion fluorouracil and weekly leucovorin. J Clin
Oncol. 15:3223–3229. 1997.
|
|
7
|
Salonga D, Danenberg KD, Johnson M, et al:
Colorectal tumors responding to 5-fluorouracil have low gene
expression levels of dihydropyrimidine dehydrogenase, thymidylate
synthase and thymidine phosphorylase. Clin Cancer Res. 6:1322–1327.
2000.
|
|
8
|
Shirota Y, Stoehlmacher J, Brabender J, et
al: ERCC1 and thymidylate synthase mRNA levels predict survival for
colorectal cancer patients receiving combination oxaliplatin and
fluorouracil chemotherapy. J Clin Oncol. 19:4298–4304. 2001.
|
|
9
|
Kim SH, Kwon HC, Oh SY, et al: Prognostic
value of ERCC1, thymidylate synthase and glutathione S-transferase
pi for 5-FU/oxaliplatin chemotherapy in advanced colorectal cancer.
Am J Clin Oncol. 32:38–43. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kumamoto K, Kuwabara K, Tajima Y, et al:
Thymidylate synthase and thymidine phosphorylase mRNA expression in
primary lesions using laser capture microdissection is useful for
prediction of the efficacy of FOLFOX treatment in colorectal cancer
patients with liver metastasis. Oncol Lett. 3:983–989. 2012.
|
|
11
|
Stoehlmacher J, Park DJ, Zhang W, et al: A
multivariate analysis of genomic polymorphisms: prediction of
clinical outcome to 5-FU/oxaliplatin combination chemotherapy in
refractory colorectal cancer. Br J Cancer. 91:344–354. 2004.
|
|
12
|
Etienne-Grimaldi MC, Milano G,
Maindrault-Goebel F, et al: Methylenetetrahydrofolate reductase
(MTHFR) gene polymorphisms and FOLFOX response in colorectal cancer
patients. Br J Clin Pharmacol. 69:58–66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ruzzo A, Graziano F, Loupakis F, et al:
Pharmacogenetic profiling in patients with advanced colorectal
cancer treated with first-line FOLFOX-4 chemotherapy. J Clin Oncol.
25:1247–1254. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Boige V, Mendiboure J, Pignon JP, et al:
Pharmacogenetic assessment of toxicity and outcome in patients with
metastatic colorectal cancer treated with LV5FU2, FOLFOX and
FOLFIRI: FFCD 2000–05. J Clin Oncol. 28:2556–2564. 2010.PubMed/NCBI
|
|
15
|
Ma J, Stampfer MJ, Giovannucci E, et al:
Methylenetetrahydrofolate reductase polymorphism, dietary
interactions and risk of colorectal cancer. Cancer Res.
57:1098–1102. 1997.PubMed/NCBI
|
|
16
|
van der Put NM, Gabreëls F, Stevens EM, et
al: A second common mutation in the methylenetetrahydrofolate
reductase gene: an additional risk factor for neural-tube defects?
Am J Hum Genet. 62:1044–1051. 1998.PubMed/NCBI
|
|
17
|
Park DJ, Stoehlmacher J, Zhang W, et al:
ERCC1 gene polymorphism is associated with differential ERCC1 gene
expression. Proc AACR. 93:15912002.
|
|
18
|
Park DJ, Zhang W, Stoehlmacher J, et al:
ERCC1 gene polymorphism as a predictor for clinical outcome in
advanced colorectal cancer patients treated with platinum-based
chemotherapy. Clin Adv Hematol Oncol. 1:162–166. 2003.PubMed/NCBI
|
|
19
|
Chang PM, Tzeng CH, Chen PM, et al: ERCC1
codon 118 C>T polymorphism associated with ERCC1 expression and
outcome of FOLFOX-4 treatment in Asian patients with metastatic
colorectal carcinoma. Cancer Sci. 100:278–283. 2009.
|
|
20
|
Park DJ, Stoehlmacher J, Zhang W, et al: A
Xeroderma pigmentosum group D gene polymorphism predicts clinical
outcome to platinum-based chemotherapy in patients with advanced
colorectal cancer. Cancer Res. 61:8654–8658. 2001.
|
|
21
|
Stoehlmacher J, Park DJ, Zhang W, et al:
Association between glutathione S-transferase P1, T1 and M1 genetic
polymorphism and survival of patients with metastatic colorectal
cancer. J Natl Cancer Inst. 94:936–942. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen YC, Tzeng CH, Chen PM, et al:
Influence of GSTP1 I105V polymorphism on cumulative neuropathy and
outcome of FOLFOX-4 treatment in Asian patients with colorectal
carcinoma. Cancer Sci. 101:530–535. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zarate R, Rodríguez J, Bandres E, et al:
Oxaliplatin, irinotecan and capecitabine as first-line therapy in
metastatic colorectal cancer (mCRC): a dose-finding study and
pharmacogenomic analysis. Br J Cancer. 102:987–994. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Watson MA, Stewart RK, Smith GB, et al:
Human glutathione S-transferase P1 polymorphisms: relationship to
lung tissue enzyme activity and population frequency distribution.
Carcinogenesis. 19:275–280. 1998. View Article : Google Scholar
|
|
25
|
Eisenhauer EA, Therasse P, Bogaerts J, et
al: New response evaluation criteria in solid tumours: revised
RECIST guideline (version 1.1). Eur J Cancer. 45:228–247. 2009.
View Article : Google Scholar
|
|
26
|
Moscow JA, Fairchild CR, Madden MJ, et al:
Expression of anionic glutathione-S-transferase and P-glycoprotein
genes in human tissues and tumors. Cancer Res. 49:1422–1428.
1989.PubMed/NCBI
|
|
27
|
Ban N, Takahashi Y, Takayama T, et al:
Transfection of glutathione S-transferase (GST)-pi antisense
complementary DNA increases the sensitivity of a colon cancer cell
line to adriamycin, cisplatin, melphalan and etoposide. Cancer Res.
56:3577–3582. 1996.PubMed/NCBI
|
|
28
|
Popat S, Matakidou A and Houlston RS:
Thymidylate synthase expression and prognosis in colorectal cancer:
a systematic review and meta-analysis. J Clin Oncol. 22:529–536.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Stoehlmacher J, Goekkurt E, Mogck U, et
al: Thymidylate synthase genotypes and tumour regression in stage
II/III rectal cancer patients after neoadjuvant fluorouracil-based
chemoradiation. Cancer Lett. 272:221–225. 2008. View Article : Google Scholar
|
|
30
|
Dotor E, Cuatrecases M, Martínez-Iniesta
M, et al: Tumor thymidylate synthase 1494del6 genotype as a
prognostic factor in colorectal cancer patients receiving
fluorouracil-based adjuvant treatment. J Clin Oncol. 24:1603–1611.
2006. View Article : Google Scholar
|
|
31
|
Keam B, Im SA, Han SW, et al: Modified
FOLFOX-6 chemotherapy in advanced gastric cancer: Results of phase
II study and comprehensive analysis of polymorphisms as a
predictive and prognostic marker. BMC Cancer. 8:1482008. View Article : Google Scholar : PubMed/NCBI
|