Introduction
Acute lymphoblastic leukemia (ALL) comprises a
heterogeneous group of lymphoid malignancies, which affect 1–4.75
individuals per 100,000 worldwide (1–2).
Certain Western countries, including Italy, the United States,
Switzerland and Costa Rica account for the highest incidence of ALL
(3). Of all the leukemia cases in
children, ~80% are ALL. The incidence rate has increased
remarkably, with an annual percentage change of 0.8% between 1975
and 2006 (4,5). In children aged 1–4 years, the annual
percentage change reaches 1.2% (3).
The high mortality and morbidity rates considerably affect living
standards and survival quality. However, at present, the exact
etiology of ALL remains unknown, although the majority of studies
have considered it to be associated with infection, radiation,
chemical and genetic factors (3,4).
Over 80 common fragile sites have been identified in
the human genome database (6).
Common fragile sites are chromosomal regions that are observed in
metaphase chromosomes. The genes in these regions are more
susceptible to breakage, rearrangements and deletions compared with
other genes (7,8). Studies have demonstrated that the
genes associated with common fragile sites have important roles in
carcinogenesis (7,8).
Carcinogenesis is a complicated multipathway
processes involving genetic alterations, including the inactivation
of tumor suppressor genes. Fragile histidine triad (FHIT) and
WW-domain oxidoreductase (WWOX) are tumor suppressor genes that
encompass the two most active common fragile sites. Since FHIT and
WWOX were first identified by Ohta et al(9) and Bednarek et al(10), respectively, the loss or reduction
of FHIT and WWOX expression has been shown to be an important step
in the initiation of tumorigenesis in a variety of tumors,
including those in the breasts, lungs, esophagus, kidneys and
cervix (11–16). Despite the clear mechanism, evidence
has demonstrated that possible synergy may exist between FHIT and
WWOX in the pathogenesis of tumors.
p73 is another tumor suppressor gene, which was
identified in 1997 by Kaghad et al(17) and is considered to regulate cell
growth and induce a cell-cycle blockade and cell apoptosis. Given
the functional similarity between p53 and p73, it is not
unreasonable to suppose that p73 may also be important for the
inhibition of cancer development. It has been reported that WWOX
interacts with p73 via its first WW domain and promotes the
expression of p73 (18). The
tyrosine kinase, Src, phosphorylates WWOX at tyrosine 33 in the
first WW domain and enhances its binding to p73. Furthermore, the
high WWOX expression level boosts the expression of p73 and
triggers the redistribution of nuclear p73 to the cytoplasm,
resulting in the promotion of cell apoptosis (18). Therefore, the loss of WWOX
expression may decrease or suppress the functional role of p73 in
apoptosis, which may lead to the occurrence of tumors. All these
findings demonstrate that WWOX, FHIT and p73 have important roles
in the pathogenesis of tumors and that WWOX may cooperate with FHIT
and p73 in the development of tumors.
Epigenetic changes contribute greatly to leukemia
development (19). DNA methylation
is a well-studied mechanism in epigenetics. The hypermethylation of
numerous genes has been detected in various types of tumors and
hematological neoplasms (20).
Previous studies have shown that DNA methylation is the most
commonly detected alteration in ALL (21,22).
DNA methylation often results in the silencing of tumor suppressor
genes, although the gene sequence may not have been changed. This
mechanism has been identified as leading to the loss of function of
numerous tumor suppressor genes in various types of tumor cells
(23). To clarify how WWOX, FHIT
and p73 are involved in the development and progression of ALL, the
present study examined the methylation status and mRNA expression
of these three tumor suppressor genes in ALL.
Materials and methods
Patients and samples
Bone marrow (BM) samples from 112 ALL patients were
collected from the Hematology Department of The First Affiliated
Hospital of Guangxi Medical University (Nanning, Guangxi, China).
In addition to the 112 ALL patients (68 male and 44 female) of the
experimental group, 35 non-ALL patients (19 male and 16 female)
were included in the study as the control group. According to the
diagnosis status of the ALL patient group, there were 72 newly
diagnosed patients (NDPs), 26 complete remission patients (CRPs)
and 14 relapsed patients (RPs). The diagnosis of ALL was based on
morphology, histopathology, expression of leukocyte differentiation
antigens and the French American British (FAB) classification.
Prior consent from was obtained the patients for the use of these
clinical materials for research purposes and study approval was
granted from the Ethics Committee of The First Affiliated Hospital
of Guangxi Medical University.
WWOX, FHIT and p73 transcript
expression
Total RNA was extracted from the BM samples using
TRIzol (Tiangen Inc., Beijing, China), according to the
manufacturer's standard instructions. cDNA was synthesized from
each RNA using a random primer and Moloney murine leukemia virus
reverse transcriptase (Super-Script II; Gibco BRL, Gaithersburg,
ND, USA), according to the manufacturer's instructions. The PCR
primers used for the RT-PCR in the study are shown in Table I. β-actin expression was used as a
control. PCR was performed in a final volume of 25 μl, containing 1
μl cDNA, 2.5 μl 10X PCR buffer, 2 μl dNTPs, 1 μl forward primer, 1
μl reverse primer, 0.15 μl Taq DNA polymerase and 11.3 μl
sterilized DEPC water. RT-PCR was conducted in a PE480 DNA Thermal
Cycler. The amplification condition were as follows: 94°C for 45
sec, followed by 60°C (WWOX), 58°C (p73) or 60°C (FHIT) for 30 sec
and then 72°C for 45 sec, for 35 cycles. The amplification products
were analyzed with 2.0% agarose gel electrophoresis. The
electrophoresis results were observed with a gel imaging system and
photographs of the gels were taken.
 | Table IPrime sequence for RT-PCR and
MS-PCR. |
Table I
Prime sequence for RT-PCR and
MS-PCR.
| Gene primer | Primer sequence
(5′→3′) | PCR product size,
bp |
|---|
| β-actin-F |
AACAAGATGAGATTGGCA | 251 |
| β-actin-R |
AGTGGGGTGGCTTTTAGGAT | |
| rtpcr-WWOX-F |
AATCACACCGAGGAGAAGAC | 536 |
| rtpcr-WWOX-R |
AAAGTTGCTGCGTTGCACAC | |
| rtpcr-FHIT-F |
ATGTCGTTCAGATTTGGCCAAC | 340 |
| rtpcr-FHIT-R |
TCATAGATGCTGTCATTCCTGT | |
| rtpcr-p73-F |
CCCACCACTTTGAGGTCACT | 461 |
| rtpcr-p73-R |
CAGGGTGATGATGATGAGGA | |
| WWOX-MF |
AGGATTGGTTAGAATAACGC | 193 |
| WWOX-MR |
AAAATACCTAAAAAATCGCG | |
| WWOX-UF |
TGTAGGATTGGTTAGAATAATGT | 193 |
| WWOX-UR |
AAAAATACCTAAAAAATCACACT | |
| FHIT-MF |
TTTTCGTTTTTGTTTTTAGATAAGC | 157 |
| FHIT-MR |
AAAAATATACCCACTAAATAACCGC | |
| FHIT-UF |
TGGTTTTTGTTTTTGTTTTTAGATAAGT | 159 |
| FHIT-UR |
AAAATATACCCACTAAATAACCACC | |
| p73-MF |
GGACGTAGCGAAATCGGGGTTC | 68 |
| p73-MR |
ACCCCGAACATCGACGTCCG | |
| p73-UF |
AGGGGATGTAGTGAAATTGGGGTTT | 77 |
| p73-UR |
ATCACAACCCCAAACATCAACATCCA | |
Analysis of WWOX, FHIT and p73
methylation
The methylation status of the first exon region of
p73 and the promoter region of WWOX and FHIT was studied. High
molecular weight DNA was prepared from cell samples using standard
methods, and bisulphate modification of genomic DNA was performed
as previously reported (24). The
PCR primers used for the methylation-specific PCR (MS-PCR) in the
study are also shown in Table I.
The 20-μl reaction mixture contained 2.5 μl 10X PCR buffer, 2 μl
dNTPs, 1 μl forward primer, 1 μl reverse primer, 2 μl Taq DNA
polymerase, 2 μl modified DNA and 11.3 μl sterilized double
distilled water. Amplification was performed with a Thermo PCR
machine (Thermo Fisher Scientific, Waltham, MA, USA). The MS-PCR
reaction conditions for were as follows: Methylated WWOX, 94°C for
30 sec, 46°C for 30 sec and 72°C for 30 sec, for 40 cycles;
unmethylated WWOX, 94°C for 30 sec, 52°C for 30 sec and 72°C for 30
sec, for 40 cycles; methylated FHIT, 94°C for 30 sec, 51°C for 30
sec and 72°C for 30 sec, for 40 cycles; unmethylated FHIT, 94°C for
30 sec, 53°C for 30 sec and 72°C for 30 sec, for 40 cycles;
methylated p73, 94°C for 30 sec, 51°C for 30 sec and 72°C for 30
sec, for 40 cycles; and unmethylated p73, 94°C for 30 sec, 53°C for
30 sec and 72°C for 30 sec, for 40 cycles. DNA treated in
vitro with SSSI methyltransferase and untreated DNA was used as
the positive control for the methylated and unmethylated templates,
respectively. Negative control samples without DNA were included
for each set of PCR. The amplification products were analyzed with
2.0% agarose gel electrophoresis, then the electrophoresis results
were observed with a gel imaging system and images were captured.
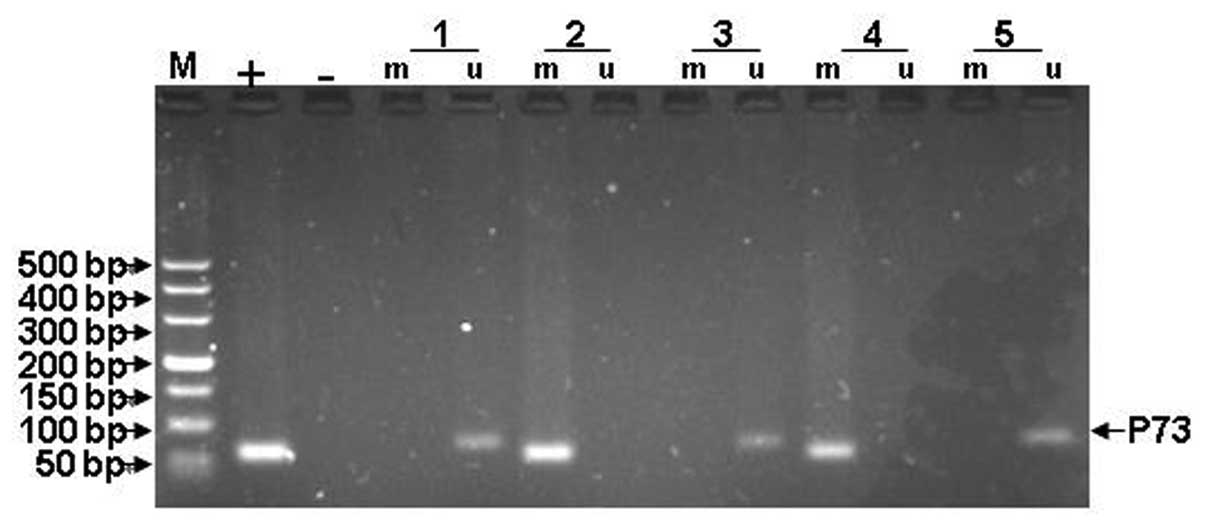
The presence of only the methylated amplification product was
considered to indicate complete methylation, while the presence of
the methylation and unmethylated amplification product indicated
partial methylation. All MSP was performed using the EZ DNA
Methylation-Gold™ kit (Zymo Research, Irvine, CA, USA).
Statistical analysis
All statistical analyses were performed using SPSS
13.0 software. The χ2 test was used to analyze the
associations in the mRNA expression and methylation frequency of
WWOX, FHIT and p73 between all the patients. Spearman's rank
correlation test was used to analyze possible linear correlations
between the expression of all three genes, the gene methylation
status and the possible association between gene expression and
methylation frequency. Fisher's exact test was used in data
numeration. P<0.05 (confidence >95%) was considered to
indicate a statistically significant difference.
Results
WWOX, FHIT and p73 transcript
expression
As the expression of WWOX, FHIT and p73 mRNA is too
low to detect routinely by RNA blot analysis of small samples,
RT-PCR amplification was performed to detect WWOX, FHIT and p73
expression, as previously described for leukemia and other tumors
(9,25,26).
As shown in Table II, the
expression of WWOX was not detected in 58 of 112 (51.8%) ALL
patients. However, only two out of 35 (5.7%) controls did not
exhibit the expression of WWOX. Similar results were observed for
FHIT and p73. Significant differences were observed in WWOX, FHIT
and p73 expression between the ALL patients and controls
(P<0.05). Considering that the results may differ in different
stages of ALL, the ALL patients were further classified by their
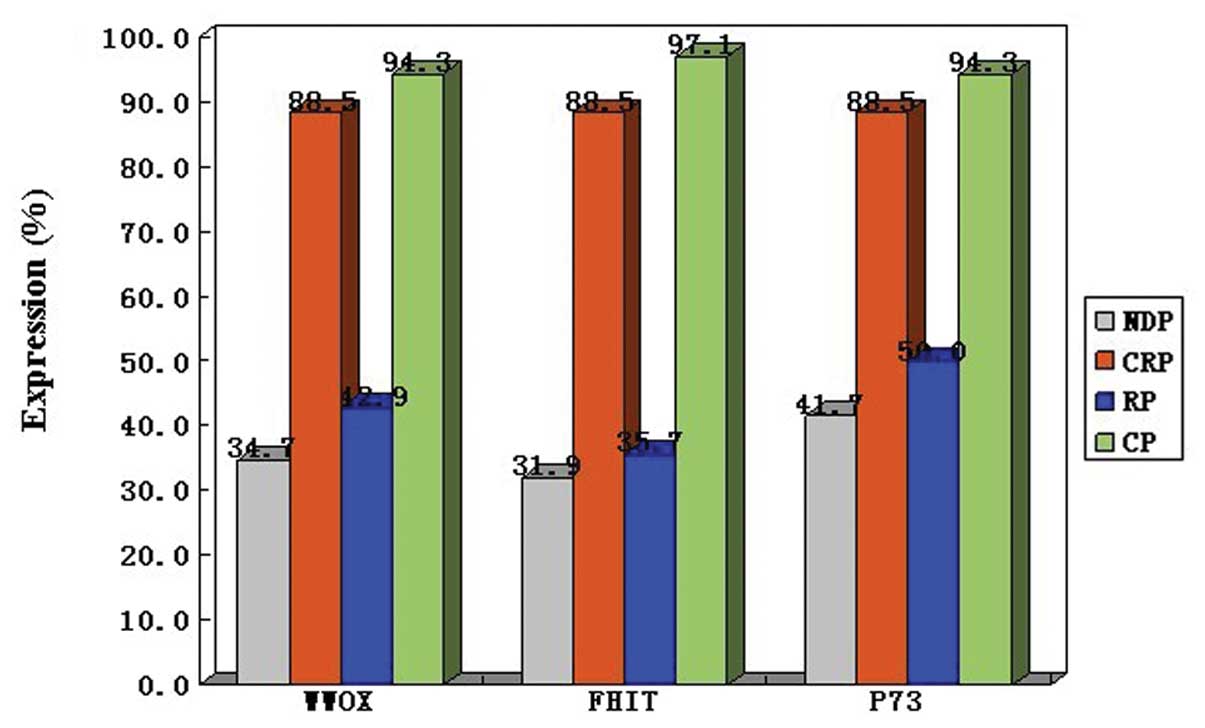
diagnosis status of NDP, CRP and RP. As a result, only 25 of 72
(34.7%) NDPs and 6 of 14 (42.9%) RPs exhibited WWOX expression.
However, 23 of 26 (88.5%) CRPs exhibited WWOX expression. Similar
results were detected for FHIT and p73. Significant differences
were observed in WWOX, FHIT and p73 expression levels between NDPs
and CRPs (P<0.05) and RPs and CRPs (P<0.05), but not between
NDPs and RPs (P>0.05). The electrophoresis gels of WWOX, FHIT
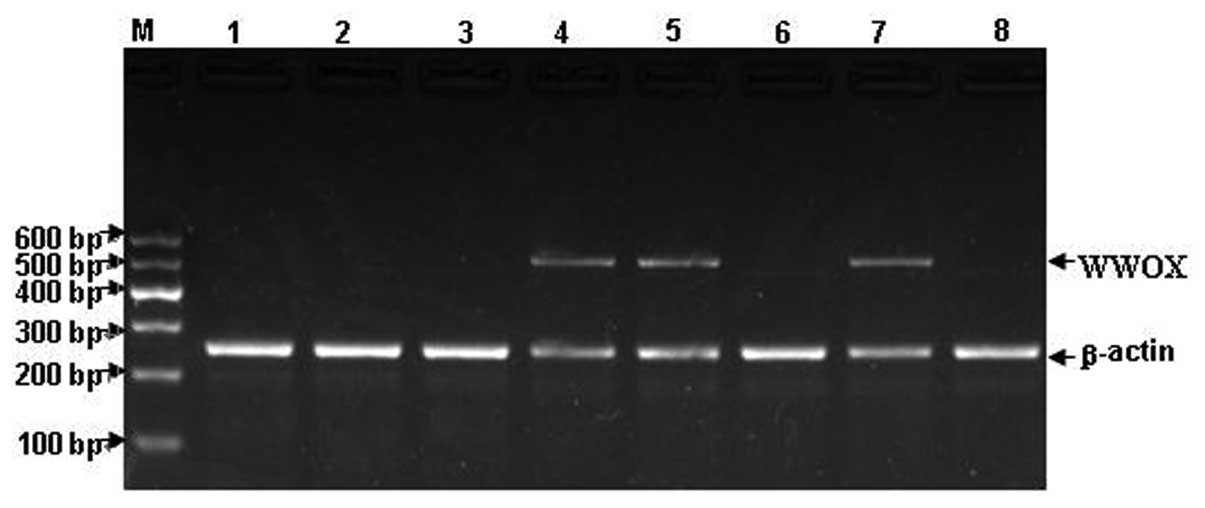
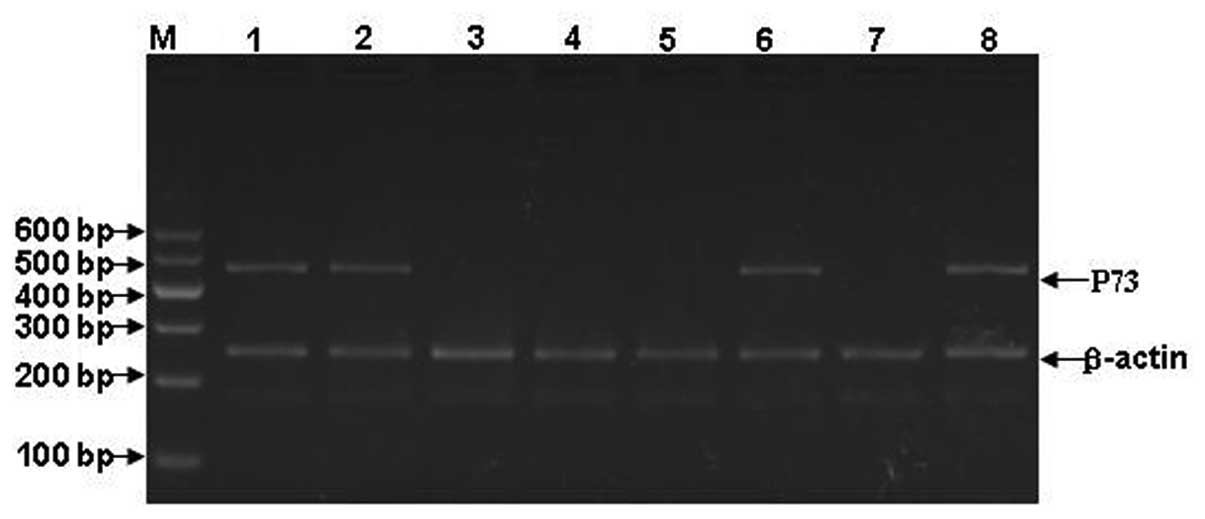
and p73 expression are shown in Figs.
1–3. The mRNA expression of
WWOX, FHIT and p73 in the patients with ALL is shown in Fig. 4.
 | Table IIWWOX, FHIT and p73 expression in ALL
and controls. |
Table II
WWOX, FHIT and p73 expression in ALL
and controls.
| | WWOX | FHIT | p73 |
|---|
| |
|
|
|
|---|
| Diagnosis | n | Detected | Not detected | Detected | Not detected | Detected | Not detected |
|---|
| ALL | 112 | 54 | 58 | 48 | 64 | 62 | 50 |
| NDP | 72 | 25 | 47 | 23 | 49 | 30 | 42 |
| CRP | 26 | 23 | 3 | 23 | 3 | 23 | 3 |
| RP | 14 | 6 | 8 | 5 | 9 | 7 | 7 |
| Controls | 35 | 33 | 2 | 34 | 1 | 33 | 2 |
Methylation status of WWOX, FHIT and
p73
As described in Table
III, although 50 of 112 (44.6%) patients exhibited methylation
in the WWOX promoter region, no methylation was detected in the
same areas of the control samples. Parallel results were observed
in the promoter region of FHIT and the first exon region of p73.
Significant differences were observed for the WWOX, FHIT and p73
methylation frequency between the ALL patients and controls
(P<0.05). Furthermore, the WWOX, FHIT and p73 methylation
frequency was examined in the ALL patients classified by their
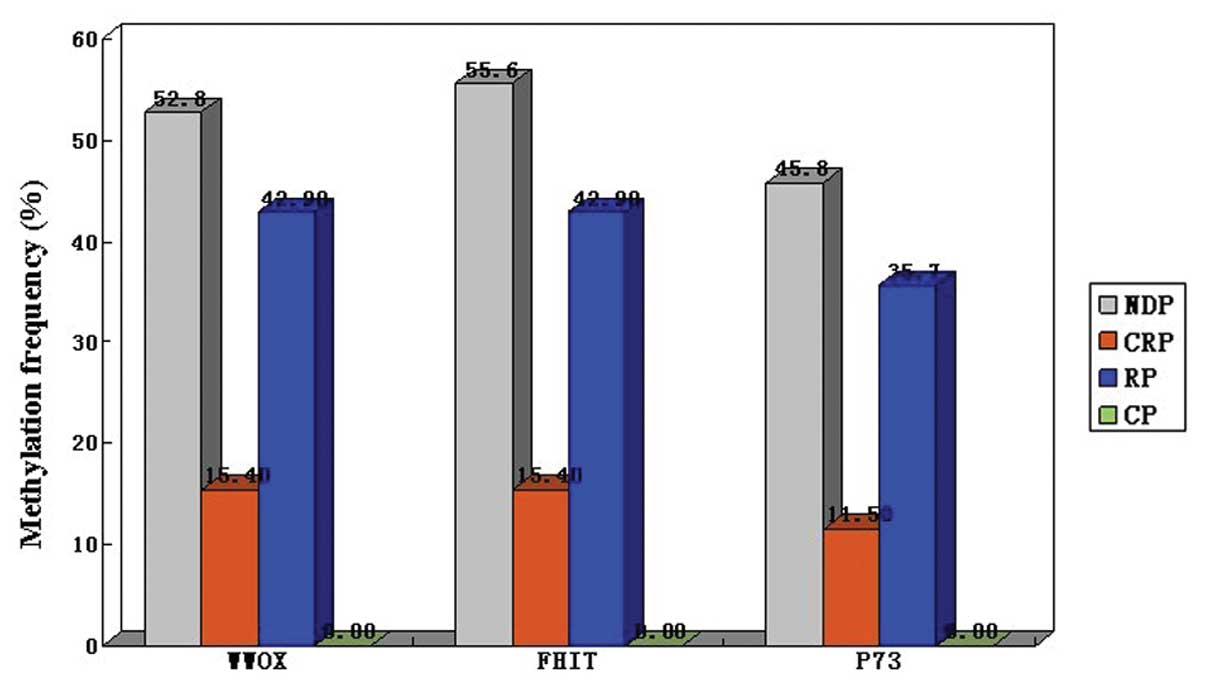
diagnosis status of NDP, CRP and RP. Consequently, 38 of 72 (52.8%)
NDPs, 4 of 26 (15.4%) CRPs and 6 of 14 (42.9%) RPs were observed to
have methylation in the WWOX promoter region. Significant
differences concerning the methylation frequency of the WWOX
promoter region were observed, but only for between NDP and CRP
(P<0.05) and not between NDP and RP (P>0.05) or CRP and RP
(P>0.05). Consistent results were also observed in the FHIT
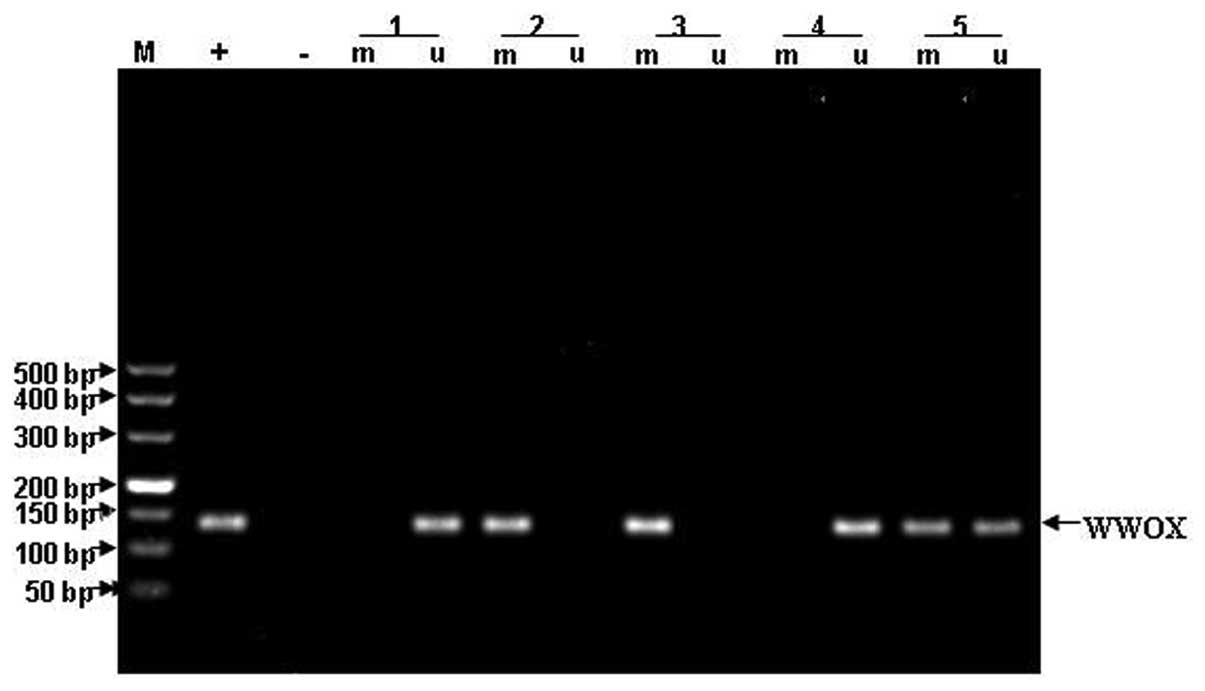
promoter region and the first exon region of p73. The gel images of
WWOX, FHIT and p73 methylation are shown in Figs. 5–7.
The methylation status of WWOX, FHIT and p73 in the ALL patients is
shown in Fig. 8.
 | Table IIIWWOX, FHIT and p73 methylation
frequency in ALL and controls. |
Table III
WWOX, FHIT and p73 methylation
frequency in ALL and controls.
| | WWOX | FHIT | p73 |
|---|
| |
|
|
|
|---|
| Diagnosis | n | Methylated | Unmethylated | Methylated | Unmethylated | Methylated | Unmethylated |
|---|
| ALL | 112 | 50 | 62 | 52 | 60 | 42 | 70 |
| NDP | 72 | 38 | 34 | 40 | 32 | 33 | 39 |
| CRP | 26 | 4 | 22 | 4 | 22 | 3 | 23 |
| RP | 14 | 6 | 8 | 6 | 8 | 5 | 9 |
| Controls | 35 | 0 | 35 | 0 | 35 | 0 | 35 |
Associations between methylation status
and mRNA expression
Spearman's rank correlation test showed that the
mRNA expression levels of WWOX, FHIT and p73 were inversely
correlated with the methylation frequency in the ALL patients. The
correlation coefficients were −0.661 (P<0.05) for WWOX, −0.685
(P<0.05) for FHIT and −0.536 (P<0.05) for p73.
Associations between WWOX, FHIT and
p73
Spearman's rank correlation test was also used to
analyze the associations between the three genes in ALL. The
correlation coefficient of mRNA expression between WWOX and FHIT
was 0.590 (P<0.05) and between WWOX and p73 was 0.520
(P<0.05). However, no significant correlation was observed in
the methylation frequency between WWOX and FHIT (P>0.05) or WWOX
and p73 (P>0.05).
Discussion
In previous studies, the loss or reduction of WWOX
and FHIT expression has been shown to be an important step in the
initiation of tumorigenesis in a variety of tumors, including those
in the breasts, lungs, esophagus, kidneys and cervix (11–16).
Furthermore, it has also been reported that altered or absent WWOX
expression may suppress the functional role of p73 in apoptosis,
leading to an increased possibility of tumor occurrence (18). The aim of the present study was to
determine whether there was a similar phenomenon in ALL cases. To
the best of our knowledge, this is the first study to focus on such
a subject.
In the present study, the mRNA expression of WWOX,
FHIT and p73 was analyzed in ALL. As in other tumors, a reduced
expression of WWOX, FHIT and p73 was observed in ALL. The present
study showed that the expression levels of WWOX and FHIT were
altered concordantly in ALL, and that the frequency of WWOX
expression, as determined by RT-PCR amplification of mRNA, was
almost the same as that of FHIT (48.2 vs. 42.9%). In previous
studies, the reported frequency of FHIT alterations in ALL has
varied; the expression of FHIT mRNA or protein has been reported to
be altered in 20–70% of cases (27–30). A
previous study investigating p73 expression in ALL revealed
relatively small differences, as p73 expression in ALL differed by
no more than 40% (31). It reported
that p73 expression is exhibited in 61–100% of ALL (31); p73 expression was detected in 55.4%
of ALL in the present study, which was slightly less than previous
results. The results of previous studies approximated to those of
the present study, which may to a certain extent provide evidence
for and support the conclusions of this study. In the present
study, the cases that showed WWOX alterations also showed
alterations to FHIT and exhibited almost the same mRNA expression.
These observations suggest that WWOX and FHIT may concordantly
affect the progression of ALL. Moreover, a number of authors have
shown that the suppressed transcription of WWOX and FHIT is
associated with the more advanced stages in various types of
cancer, including breast (32,35),
non-small cell lung (33) and
ovarian (34) cancers. The present
study demonstrated that a relatively low expression of WWOX and
FHIT correlates with NDP and RP diagnosis statuses in ALL patients,
supporting the hypothesis that the occurrence of ALL is a
progressive and multi-staged process, similar to that of other
tumors. The expression of p73 in ALL was not significantly higher
than that of WWOX (48.2 vs. 55.4%), although WWOX and p73
expression was significantly lower compared with the controls,
indicating that the loss of WWOX expression may inhibit the
apoptotic role of p73 and cause ALL to occur.
Despite the frequent suppression of WWOX, FHIT and
p73 expression in numerous cancers, complete gene inactivation by
the deletion of one allele and a second mutation or homozygous
deletion is extremely rare (36).
Based on these observations, it has been postulated that the
inactivation of WWOX, FHIT and p73 is driven by hemizygous
deletions or methylation status. However, loss of heterozygosity
(LOH) and somatic mutations are rare in hematological malignancies
(37). Epigenetic changes are of
great importance to leukemia development (19). Previous studies have shown that DNA
methylation is the most commonly detected alteration in AL
(21,22). Therefore, the methylation status was
investigated as the inactivating mechanism of WWOX, FHIT and p73 in
ALL in the present study. At present, FHIT and WWOX promoter
methylation associated with the loss of gene expression has been
reported for various types of cancers, including lung, breast,
esophageal and bladder cancer (38). Similarly, a higher methylation
frequency of WWOX, FHIT and p73 was shown in ALL in the present
study. Furthermore, the NDP and RP groups also showed the higher
methylation frequency of the three genes in ALL, suggesting that
the methylation of WWOX, FHIT and p73 in ALL was accumulated
through the progression of the disease. It has been reported that
30% of ALL cases have the methylated p73 status (39), which is close to the 37.5%
methylation frequency of p73 in the present study. Previous studies
concerning the frequency of FHIT methylation in ALL have differed,
ranging between 13.8 and 67.0% (40–43).
The present study demonstrated 46.4% FHIT methylation in the ALL
cases. The difference is possibly due to the diagnosis time for
ALL, which leads to the alteration of FHIT methylation frequency,
as numerous studies have demonstrated that FHIT methylation is an
accumulated process and different diagnosis times exhibit different
methylation frequencies (40–42).
Research on the methylation of the WWOX promoter region has mainly
focused on a few malignant tumors, such as stomach cancer (44), glioblastoma (45) and lung cancer (46). To date, no study has yet been
reported on the methylation of the WWOX promoter region in ALL.
Iliopoulos et al(38) studied the WWOX promoter methylation
status in pancreatic cancer and reported methylation in region −148
to −37, respective to the transcription initiation site (+1), in
pancreatic adenocarcinomas; the WWOX promoter methylation status
was associated with its expression and following treatment with
5-aza-deoxycytidine in Hs766T cells, WWOX was expressed again.
Maeda et al(44) reported
that 24 of 73 (32.9%) stomach cancer cases that showed absent WWOX
expression were highly methylated. These studies have suggested
that WWOX methylation causes the loss of or decreased expression of
WWOX. The present study showed that the mRNA expression of WWOX,
FHIT and p73 was reversely correlated with the methylation of WWOX,
FHIT and p73 in ALL, indicating that the highly methylated status
of WWOX, FHIT and p73 leads to silencing of the expression of these
genes, which may result in the loss of gene transcriptions and
promote the occurrence and development of ALL. Notably, significant
differences were detected in the mRNA expression of WWOX, FHIT and
p73 between the RP and CRP groups with ALL. However, no significant
difference was detected in the methylation frequency of the three
genes between the RP and CRP groups. We suggest that this confusing
phenomenon may be caused by the small sample size of the RP group,
as only 14 RPs were enrolled in this study.
The present study observed that the mRNA expression
of WWOX was positively correlated with not only FHIT, but also p73,
suggesting that to a certain extent, the decrease in the
transcription of p73 and FHIT may interact with the reduction of
WWOX transcription and further promote the occurrence and
development of ALL. However, the present study did not reveal any
associations in the methylation status among WWOX, FHIT and p73. It
may be inferred that WWOX methylation does not affect the
methylation of FHIT and p73.
In summary, the methylation status of WWOX, FHIT and
p73 may lead to the silencing of gene expression, promoting the
occurrence and development of ALL. Detecting the mRNA expression
and methylation status of WWOX, FHIT and p73 may aid in the
development of future treatment approaches for ALL.
Acknowledgements
This study was supported by grants from the Guangxi
Nature Science Fund ‘Research of WWOX in Nasopharyngeal Carcinoma’
(2010GXNSFA013184) and ‘Research of WWOX in Leukemia’ (Z2010372).
The present study was supported by students (Xiuli Huang and Zhe
Wang) of the First Affiliated Hospital of Guangxi Medical
University in the acquisition of data and identification of
background information relevant to the study. The authors would
like to thank them for their help, which has consequently improved
this manuscript.
References
|
1
|
Pui CH, Relling MV and Downing JR: Acute
lymphoblastic leukemia. N Engl J Med. 350:1535–1548. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Iughetti L, Bruzzi P, Predieri B and
Paolucci P: Obesity in patients with acute lymphoblastic leukemia
in childhood. Ital J Pediatr. 38:42012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dalmasso P, Pastore G, Zuccolo L, et al:
Temporal trends in the incidence of childhood leukemia, lymphomas
and solid tumors in north-west Italy, 1967–2001. A report of the
Childhood Cancer Registry of Piedmont. Haematologica. 90:1197–1204.
2005.PubMed/NCBI
|
|
4
|
Redaelli A, Laskin BL, Stephens JM,
Botteman MF and Pashos CL: A systematic literature review of the
clinical and epidemiological burden of acute lymphoblastic
leukaemia (ALL). Eur J Cancer Care (Engl). 14:53–62. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Smith MA, Seibel NL, Altekruse SF, et al:
Outcomes for children and adolescents with cancer: challenges for
the twenty-first century. J Clin Oncol. 28:2625–2634. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Turner BC, Ottey M, Zmonjic DB, et al: The
fragile histidine triad/common chromosome fragile site 3B locus and
repair-deficient cancer. Cancer Res. 62:4054–4060. 2002.PubMed/NCBI
|
|
7
|
Nunez MI, Ludes-Meyer J, Abba MC, et al:
Frequent loss of WWOX expression in breast cancer: correlation with
estrogen receptor status. Breast Cancer Res Treat. 89:99–105. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ilsley JL, Sudol M and Winder SJ: The WW
domain: linking cell signaling to the membrane cytoskeleton. Cell
Signal. 14:183–189. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ohta M, Inoue H, Cotticelli MG, et al: The
FHIT gene, spanning the chromosome 3p14.2 fragile site and renal
carcinoma-associated t (3;8) breakpoint, is abnormal in digestive
tract cancers. Cell. 84:587–597. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bednarek AK, Laflin KJ, Daniel RL, Liao Q,
Hawkins KA and Aldaz CM: WWOX, a novel WW domain-containing protein
mapping to human chromosome 16q2.33–2.41, a region frequently
affected in breast cancer. Cancer Res. 60:2140–2145. 2000.
|
|
11
|
Aqeilan RI and Croce CM: WWOX in
biological control and tumorigenesis. J Cell Physiol. 212:307–310.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sozzi G, Tornielli S, Tagliabue E, et al:
Absence of Fhit protein in primary lung tumor and cell lines with
FHIT gene abnormalities. Cancer Res. 57:5207–5212. 1997.PubMed/NCBI
|
|
13
|
Tseng JE, Kemp BL, Khuri FR, et al: Loss
of Fhit is frequent in stage I non-small cell lung cancer and in
the lungs of chronic smokers. Cancer Res. 59:4798–4803.
1999.PubMed/NCBI
|
|
14
|
Mori M, Mimori K, Shiraishi T, et al:
Altered expression of Fhit in carcinoma and precarcinomatous
lesions of the esophagus. Cancer Res. 60:1177–1182. 2000.PubMed/NCBI
|
|
15
|
Connolly DC, Greenspan DL, Wu R, et al:
Loss of Fhit expression in invasive cervical carcinomas and
intraepithelial lesions associated with invasive disease. Clin
Cancer Res. 6:3505–3510. 2000.PubMed/NCBI
|
|
16
|
Guler G, Uner A, Guler N, et al:
Concordant loss of fragile gene expression early in breast cancer
development. Pathol Int. 55:471–478. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kaghad M, Bonnet H, Yang A, et al:
Monoallelically expressed gene related to p73 at 1p36, a region
frequently deleted in neuroblastoma and other human cancers. Cell.
90:809–819. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Aqeilan RI, Pekarsky Y, Herrero JJ, et al:
Functional association between Wwox tumor suppressor protein and
p73, a p53 homolog. Proc Natl Acad Sci USA. 101:4401–4406. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Agrawal S, Unterberg M, Koschmieder S, et
al: DNA methylation of tumor suppressor genes in clinical remission
predicts the relapse risk in acute myeloid leukemia. Cancer Res.
67:1370–1377. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Esteller M: CpG island hypermethylation
and tumor suppressor genes: A booming present, a brighter future.
Oncogene. 21:5427–5440. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Garcia-Manero G, Daniel J, Smith TL, et
al: DNA methylation of multiple promoter-associated CpG islands in
adult acute lymphocytic leukemia. Clin Cancer Res. 8:2217–2224.
2002.PubMed/NCBI
|
|
22
|
Roman-Gomez J, Jimenez-Velasco A,
Castillejo JA, et al: Promoter hypermethylation of cancer-related
genes: A strong independent prognostic factor in acute
lymphoblastic leukemia. Blood. 104:2492–2498. 2004. View Article : Google Scholar
|
|
23
|
Nephew KP and Huang TH: Epigenetic gene
silencing in cancer initiation and progression. Cancer Lett.
190:125–133. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Herman JG, Graff JR, Myöhänen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: a novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kuroki T, Trapasso F, Shiraishi T, et al:
Genetic alterations of the tumor suppressor gene WWOX in esophageal
squamous cell carcinoma. Cancer Res. 62:2258–2260. 2002.PubMed/NCBI
|
|
26
|
Yendamuri S, Kuroki T, Trapasso F, et al:
WW domain containing oxidoreductase gene expression is altered in
non-small cell lung cancer. Cancer Res. 63:878–881. 2003.PubMed/NCBI
|
|
27
|
Lin PM, Liu TC, Chang JG, Chen TP and Lin
SF: Aberrant FHIT transcripts in acute myeloid leukemia. Br J
Haematol. 99:612–617. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Iwai T, Yokota S, Nakao M, et al: Frequent
aberration of FHIT gene expression in acute leukemias. Cancer Res.
58:5182–5187. 1998.PubMed/NCBI
|
|
29
|
Albitar M, Manshouri T, Gidel C, et al:
Clinical significance of fragile histidine triad gene expression in
adult acute lymphoblastic leukemia. Leuk Res. 25:859–864. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hallas C, Albitar M, Letofsky J, Keating
MJ, Huebner K and Croce CM: Loss of FHIT expression in acute
lymphoblastic leukemia. Clin Cancer Res. 5:2409–2414.
1999.PubMed/NCBI
|
|
31
|
Pluta A, Nyman U, Joseph B, Robak T,
Zhivotovsky B and Smolewski P: The role of p73 in hematological
malignancies. Leukemia. 20:757–766. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Campiglio M, Pekarsky Y, Menard S,
Tagliabue E, Pilotti S and Croce CM: FHIT loss of function in human
primary breast cancer correlates with advanced stage of the
disease. Cancer Res. 59:3866–3869. 1999.PubMed/NCBI
|
|
33
|
Donati V, Fontanini G, Dell'Omodarme M, et
al: WWOX expression in different histologic types and subtypes of
non-small cell lung cancer. Clin Cancer Res. 13:884–891. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nunez MI, Rosen DG, Ludes-Meyers JH, et
al: WWOX protein expression varies among ovarian carcinoma
histotypes and correlates with less favorable outcome. BMC Cancer.
5:642005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Płuciennik E, Kusińska R, Potemski P,
Kubiak R, Kordek R and Bednarek AK: WWOX - the FRA16D cancer gene:
expression correlation with breast cancer progression and
prognosis. Eur J Surg Oncol. 32:153–157. 2006.PubMed/NCBI
|
|
36
|
Alsop AE, Taylor K, Zhang J, Gabra H,
Paige AJ and Edwards PA: Homozygous deletions may be markers of
nearby heterozygous mutations: the complex deletion at FRA16D in
the HCT116 colon cancer cell line removes exons of WWOX. Genes
Chromosomes Cancer. 47:437–447. 2008. View Article : Google Scholar
|
|
37
|
Iwai M, Kiyoi H, Ozeki K, et al:
Expression and methylation status of the FHIT gene in acute myeloid
leukemia and myelodysplastic syndrome. Leukemia. 19:1367–1375.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Iliopoulos D, Guler G, Han SY, et al:
Roles of FHIT and WWOX fragile genes in cancer. Cancer Lett.
232:27–36. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Corn PG, Kuerbitz SJ, van Noesel MM, et
al: Transcriptional silencing of the p73 gene in acute
lymphoblastic leukemia and Burkitt's lymphoma is associated with 5′
CpG island methylation. Cancer Res. 59:3352–3356. 1999.PubMed/NCBI
|
|
40
|
Zheng S, Ma X, Zhang L, et al:
Hypermethylation of the 5′ CpG island of the FHIT gene is
associated with hyperdiploid and translocation-negative subtypes of
pediatric leukemia. Cancer Res. 64:2000–2006. 2004.
|
|
41
|
Iwai M, Kiyoi H, Ozeki K, et al:
Expression and methylation status of the FHIT gene in acute myeloid
leukemia and myelodysplastic syndrome. Leukemia. 19:1367–1375.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang Y, Takeuchi S, Hofmann WK, et al:
Aberrant methylation in promoter-associated CpG islands of multiple
genes in acute lymphoblastic leukemia. Leuk Res. 30:98–102. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Paulsson K, An Q, Moorman AV, et al:
Methylation of tumour suppressor gene promoters in the presence and
absence of transcriptional silencing in high hyperdiploid acute
lymphoblastic leukaemia. Br J Haematol. 144:838–847. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Maeda N, Semba S, Nakayama S, Yanagihara K
and Yokozaki H: Loss of WW domain-containing oxidoreductase
expression in the progression and development of gastric carcinoma:
clinical and histopathologic correlations. Virchows Arch.
457:423–432. 2010. View Article : Google Scholar
|
|
45
|
Kosla K, Pluciennik E, Kurzyk A, et al:
Molecular analysis of WWOX expression correlation with
proliferation and apoptosis in glioblastoma multiforme. J
Neurooncol. 101:207–213. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Baykara O, Demirkaya A, Kaynak K, Tanju S,
Toker A and Buyru N: WWOX gene may contribute to progression of
non-small-cell lung cancer (NSCLC). Tumour Biol. 31:315–320. 2010.
View Article : Google Scholar : PubMed/NCBI
|