Introduction
Glioblastoma multiforme (GBM) has been one of the
most lethal malignant brain tumors in adults for the past 30 years.
The best treatment to date is a combination of surgery, radiation
therapy and temozolomide administration; however, this does not
produce sufficient results. The overall survival time of patients
with GBM is between 26 and 52 weeks (1–3).
Previous studies have identified that a small population of tumor
cells called glioma stem cells (GSCs) is responsible for GBM
initiation, propagation, resistance and recurrence (4,5). Thus,
GSCs have become one of the hot topics of glioma research.
Solid tumors are unable to grow without the help of
vessels; necrosis appears in the center of a tumor when the volume
of the tumor reaches 8 mm3 without new vessels (6). Angiogenesis is one of the most
significant characteristics of malignant neoplasms, including
endothelial cell migration, proliferation and vessel remodeling.
Glioblastomas are rich in microvessels. Previous studies have
demonstrated that GSCs co-localize with microvessels (7), have a positive correlation with
microvessel density (MVD) (8) and
have multiple regulatory roles in endothelial cells (9).
The Hedgehog (HH) pathway is extremely important in
embryonic development, since it directs embryonic growth and cell
fate determination. The canonical HH pathway is activated by HH
ligands, mostly by Sonic Hedgehog (Shh) in the nervous system,
which binds to receptor complexes, such as the transmembrane
protein, Patched, and the G-protein-coupled protein, Smoothened
(Smo). Hedgehog-interacting protein (Hhip) is one of the inhibitory
ligands that binds to Shh. The binding of Shh to Patched releases
Smo. Smo then alters the activity of glioma-associated oncogene
homolog 1 (Gli1) protein and initiates the transcription of
downstream genes. The HH pathway is expressed in endothelial cells.
It has been demonstrated that the proliferation and migration of
endothelial cells are associated with the HH pathway (10).
The aim of the present study was to investigate
whether GSCs enhance proliferation and migration of endothelial
cells, and whether the HH pathway plays a role in this process.
Materials and methods
Cell culture
The mouse GL261 glioma and brain microvascular
endothelial cell lines (Cell Bank of Shanghai Institute of Cell
Biology, Chinese Academy of Sciences, Shanghai, China) were used in
this study. The two cell lines were cultured in Dulbecco's modified
Eagle's medium (DMEM), containing 10% fetal bovine serum (FBS), 100
U/ml penicillin and 100 mg/ml streptomycin. GSC spheres were
obtained from GL261 cells and cultured in serum-free DMEM/F12
medium with 10 ng/ml epidermal growth factor (EGF; PeproTech, Inc.,
Rocky Hill, NJ, USA), 10 ng/ml basic fibroblast growth factor
(bFGF; PeproTech) and 2 mg/ml B27 (Sigma-Aldrich, St. Louis, MO,
USA) (11,12).
For the co-culture system, Transwell cell culture
inserts were used (0.4 μm; Millipore, Billerica, MA, USA) to
explore the indirect effect of GSCs on endothelial cells. Briefly,
2×105 b.END3 cells were seeded in 6-well plates, while
1.5×105 GSCs were seeded in the cell culture chamber.
The insert was transfered into the well with endothelial cells
after 24 h of culture.
Transwell migration assay
Transwell cell culture inserts were selected (8.0
μm; Millipore). The cells were trypsinized and counted, then
1.5×105 cells were seeded in each cell culture insert.
Serum-free cell culture medium (400 μl) was added to each insert.
One milliliter medium with 5% FBS was added to the 6-well plate,
then the insert was transferred to the well and cultured at 37°C.
At the end of the migration assay, the insert was washed with
phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde.
The cells that were unable to migrate were wiped out and the rest
were stained by crystal violet solution (Beyotime Institute of
Biotechnology, Beijing, China). The cells were counted under a
microscope at each field.
Wound-healing assay
The b.END3 cells (2×104) were seeded in a
6-well plate cultured with DMEM in serum. Pipette tips (200 μl)
were used to scratch three parallel vertical lines in each well
subsequent to 12 h of culture. The wells were washed with PBS, then
the medium was changed to serum-free DMEM. Scratch lines were
observed under a microscope and scratch distances were measured,
with images captured at 6, 8, 12 and 24 h after scratching.
Gene knockdown assay
The murine Smo mRNA sequence (NM_176996.4) was
acquired from the NCBI database. Murine shRNA targeted to Smo was
designed, synthesized and packaged with lentivirus by SBO
Biomedical Technology (Shanghai, China). The endothelial cells were
seeded in a 6-well plate at 1×105 cells/well. Subsequent
to 24 h of culture, the lentivirus was diluted and added to culture
medium according to the manufacturer's instructions. The cells were
purified following proliferation.
Cell counting kit-8 (CCK-8) proliferation
assay
A CCK-8 proliferation assay kit (Beyotime Institute
of Biotechnology) was used in this experiment. A 48-h co-culture
medium and a 48-h control well culture medium were collected from
the empty control vector (NC b.END3; medium a and b) and siR-Smo
b.END3 cells (medium c and d). NC b.END3 and siR-Smo b.END3 cells
were seeded separately in a 96-well plate at 3×103
cells/well, using 40 wells each. Co-cultured medium or control
medium (both 150 μl) or 50 μl fresh medium was added to each well
subsequent to 12 h of culture. The specific grouping was as
follows: 40 wells with NC b.END3 plus medium a, 40 wells with NC
b.END3 plus medium b, 40 wells with siR-Smo b.END3 plus medium c
and 40 wells with siR-Smo b.END3 plus medium d. At each time-point,
the medium in each well was changed to 200 μl fresh medium mixed
with 10 μl CCK-8 solution for 10 wells from each group. The
absorbance at 450 nm was detected following 2 h of incubation.
Quantitative polymerase chain reaction
(qPCR)
The total RNA of the b.END3 cells was extracted with
RNAiso reagent (Takara Bio Inc., Shiga, Japan). Reverse
transcription and PCR were performed using a Takara RNA PCR (AMV)
kit with primers designed for murine genes. The primers were
synthetized by Invitrogen (Carlsbad, CA, USA). The sequences of
each primer pair and the product size are presented in Table I. The results were normalized
against the level of glyceraldehyde 3-phosphate dehydrogenase
(GAPDH) as the internal control. qPCR was performed in triplicate
for each experiment, including for the non-template controls.
 | Table IPrimers used in this study. |
Table I
Primers used in this study.
| Gene | Primer sequence
(5′→3′) | Product size, bp |
|---|
| GAPDH | F:
CCTGCACCACCACATGCTTA | 85 |
| R:
TCATGAGCCCTTCCACAATG | |
| Gli1 | F:
GAGAATGGGGCATCGTCGTCA | 168 |
| R:
CGGGTACTCGGTTCGGCT | |
| Hes1 | F:
TCAACACGACACCGGACAAAC | 155 |
| R:
ATGCCGGGAGCTATCTTTCTT | |
| β-catenin | F:
GCTTCTATGAAGACCCCAGTTC | 311 |
| R:
CAGTGGGCTAGGTGTCAGGA | |
| Shh | F:
AGGGGGTTTGGAAAGAGG | 168 |
| R:
GTCGGGGTTGTAATTGGG | |
| Hhip | F:
GAGAAGGGACAGGCGGGTGA | 212 |
| R:
GGGAATGCGGGGAGCAGGGA | |
Western blotting
The proteins of the b.END3 cells were extracted by
RIPA lysis buffer (Beyotime Institute of Biotechnology) with the
protease inhibitor phenylmethanesulfonyl fluoride (Beyotime
Institute of Biotechnology). The proteins were separated with 10%
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF)
membranes (Millipore). The membranes were blocked with 5% milk,
then incubated with primary rat anti-mouse Gli1 (1:1,000; R&D
Systems, Minneapolis, MN, USA), rat anti-mouse Shh (1:1,000;
R&D Systems), rabbit anti-mouse Hhip (1:1,000; Abcam,
Cambridge, MA, USA) and rabbit anti-mouse GAPDH (1:1,500; Goodhere,
Hangzhou, China) antibodies overnight at 4°C. The membranes were
then washed and incubated with goat anti-rat and goat anti-rabbit
horseradish peroxidase (HRP)-conjugated secondary antibodies
(1:100; ZSGB-BIO, Beijing, China) for 2 h at room temperature. The
proteins were detected by enhanced chemiluminescence detection
reagent. GADPH was used as the loading control.
Enzyme-linked immunosorbent assay
(ELISA)
Cell culture supernatant was acquired from the wells
of the GSCs, b.END3 cells and the co-culture. Medium was removed
and centrifuged at 3,000 × g after 12, 24 and 48 h of culture, then
stored at −80°C immediately. An ELISA kit for mouse Shh N-Terminus
was purchased from R&D Systems and an ELISA kit for mouse Hhip
was purchased from USCN Life Science Inc. (Wuhan, China). ELISA was
performed strictly according to the manufacturer's
instructions.
Statistical analysis
All experiments were conducted at least three times
and the results were from representative experiments. Data are
expressed as the mean ± standard deviation and the statistical
significance between the experimental and control groups was
analyzed with SPSS 16.0 statistical software (SPSS Inc., Chicago,
IL, USA). When two groups were compared, the unpaired Student's
t-test was used. P<0.05 was considered to indicate a
statistically significant difference.
Results
Migration of endothelial cells is
enhanced when cultured with GSCs
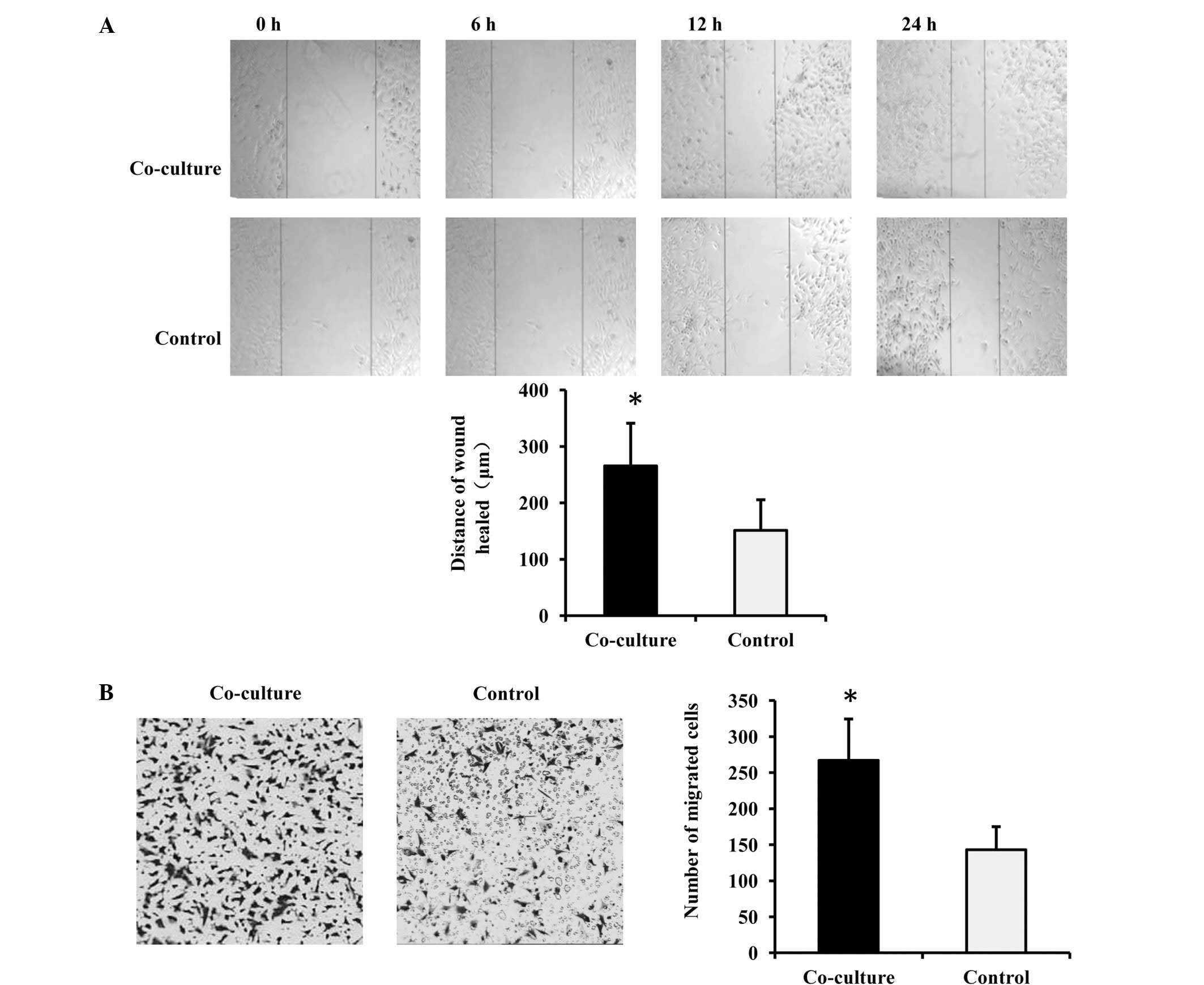
To explore the effect of GSCs on endothelial cells,
a Transwell co-culture system was selected for the in vitro
model in order to rebuild an approximate in vivo niche,
which we consider to be better than just using a GSC-conditioned
medium. In this model, the two types of cells interact via soluble
factors, but do not have direct connections. The b.END3 cells were
seeded in the lower chambers and GSCs were seeded in the upper
chambers. When the wound-healing assay was processed in the
co-culture wells, it was clear that the co-cultured b.END3 cells
exhibited enhanced migration, since the scratches in the
co-cultured wells were narrower than in the control (Fig. 1A). The endothelial cells in tumor
angiogenesis were guided by chemokines, so a Transwell migration
assay was generated to confirm the observation with the help of
serum. Subsequent to co-culture for 48 h, more b.END3 cells
migrated through the membrane and appeared on the other surface
(Fig. 1B). This result indicated
that GSCs enhanced the migration of the endothelial cells.
Proliferation of endothelial cells is
enhanced by GSCs
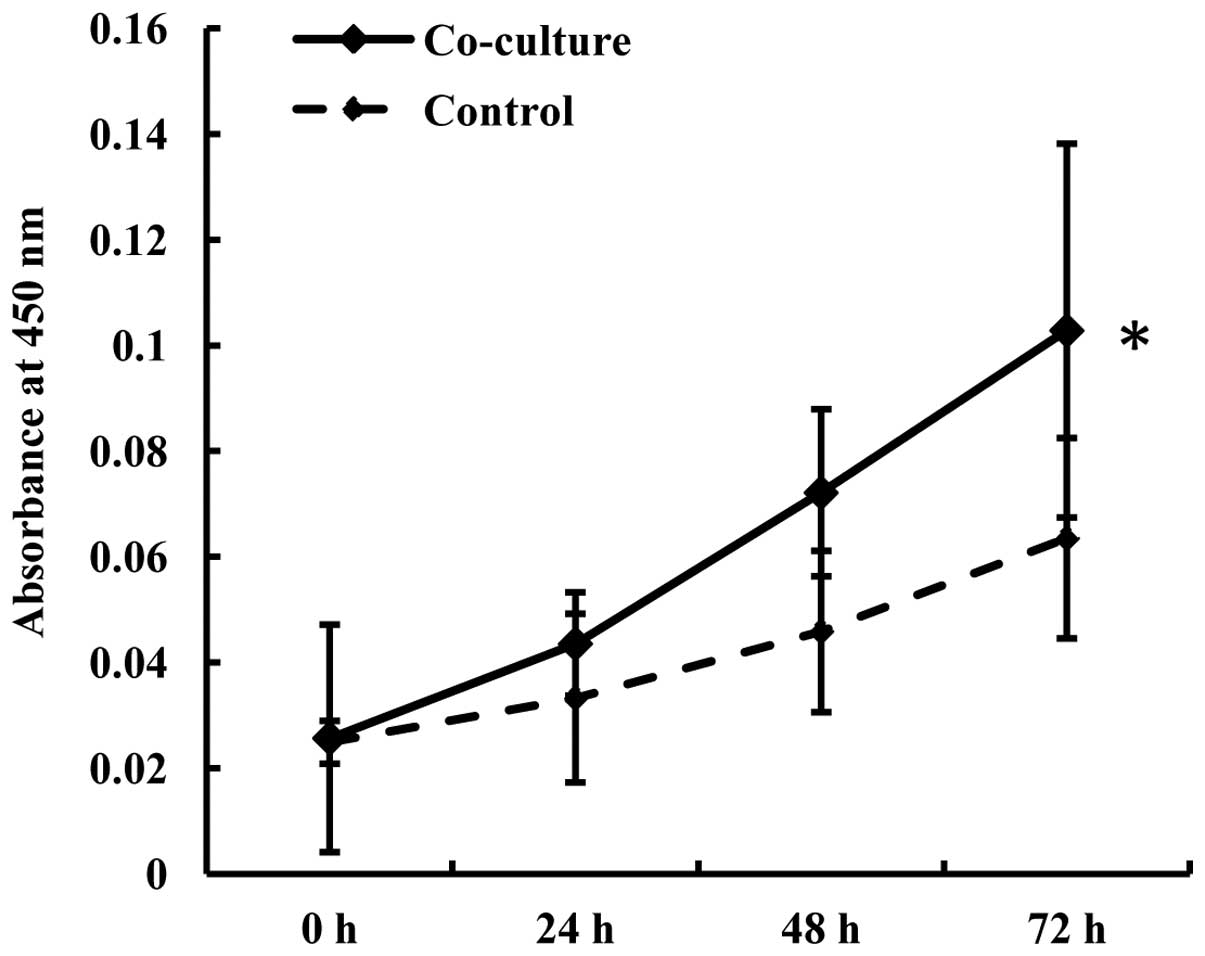
A proliferation assay was performed to determine
whether GSCs would affect the proliferation ability of the
endothelial cells. The endothelial cells were cultured with or
without GSCs for 48 h, then seeded in a 96-well plate. The medium
in each well consisted of 50 μl fresh medium mixed with 150 μl 48-h
co-cultured medium or 150 μl control medium, respectively. The
proliferation of the b.END3 cells was shown to be accelerated after
co-culture and was positively related to the culture time (Fig. 2).
HH pathway in endothelial cells is
activated by GSCs
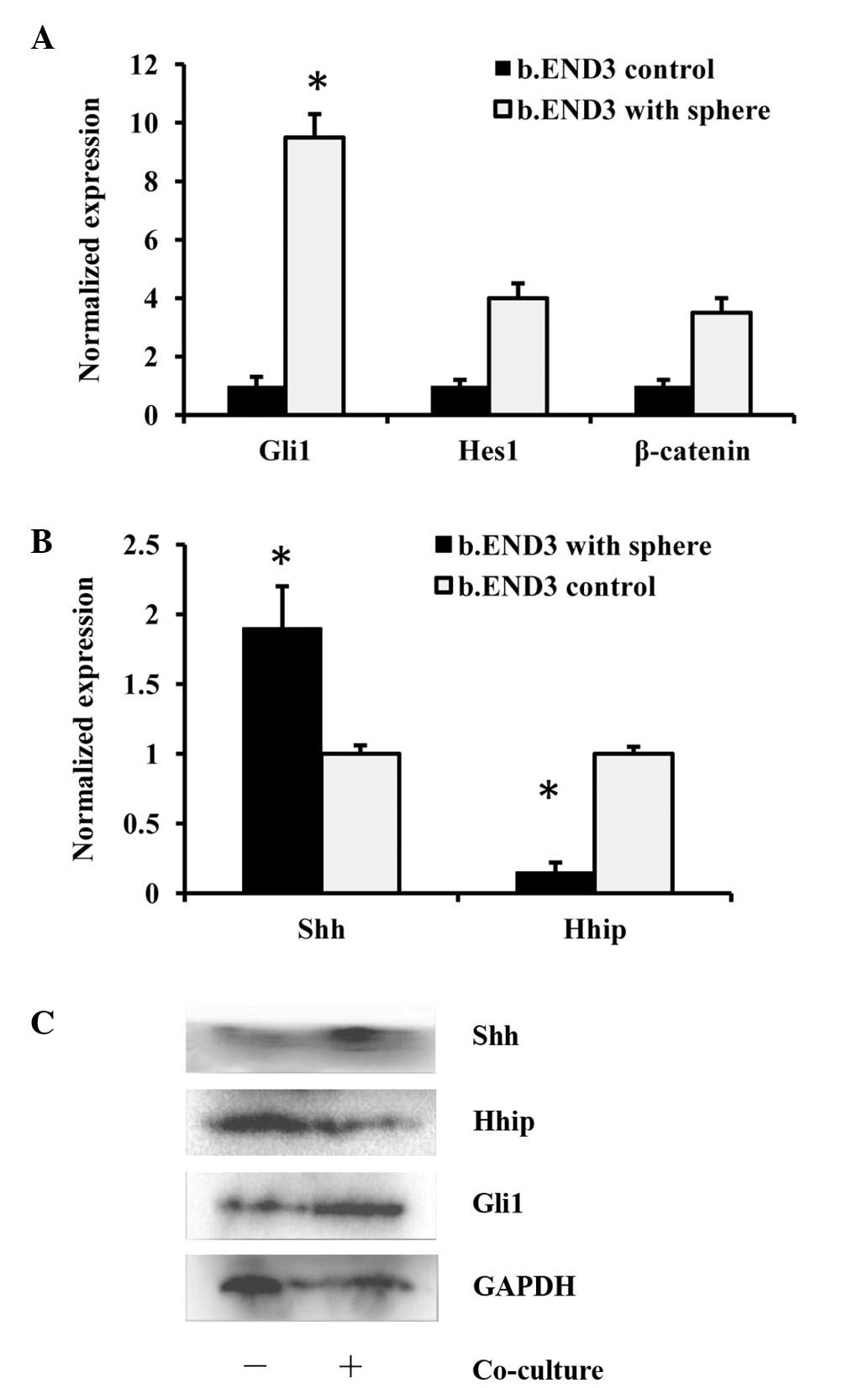
To determine the mechanism behind the migration and
proliferation of the endothelial cells caused by GSCs, three
possible pathways were selected that may have been involved. The
HH, Notch and β-catenin pathways all participate in endothelial
cell proliferation, migration, angiogenesis and the functioning of
endothelial cells. Although the three pathways were all affected in
the b.END3 cells following the 48-h co-culture with GSCs, the Gli1
gene, which is the key component of the HH pathway, was induced to
the highest extent at the mRNA level (Fig. 3A). It was also demonstrated that
ligands of the HH pathway, Shh and Hhip, had altered expression
(Fig. 3B), which was confirmed at
the protein level (Fig. 3C). These
results indicated that the HH pathway may be the main mediator of
the effect of GSCs on the b.END3 cells.
Migration ability of endothelial cells is
inhibited following Smo gene knockdown
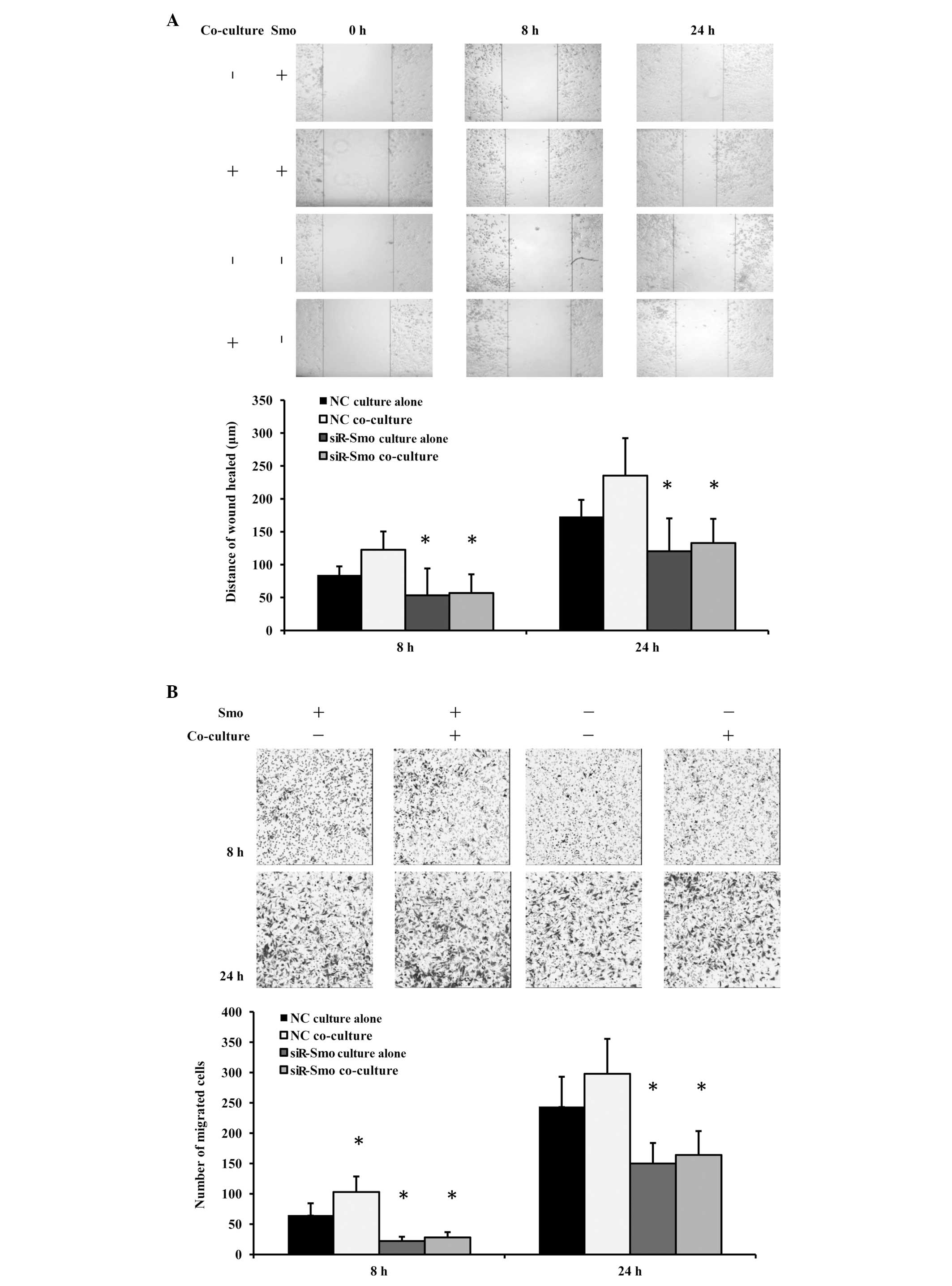
To further confirm the interaction of the HH pathway
in GSC-enhanced b.END3 cell mobility, Smo gene expression was
knocked down in the b.END3 cells, then the HH pathway was partially
blocked. Migration assays were repeated using siR-Smo-b.END3 cells
and control cells. Early and late time-points were selected in
order to observe the effect pattern. As expected, the migration
ability of the b.END3 cells was inhibited when the HH pathway was
knocked down. Furthermore, the siR-Smo-b.END3 cells co-cultured
with GSCs did not retrieve the normal level of migration ability in
the wound-healing assay (Fig. 4A)
and Transwell migration assay (Fig.
4B). These results indicated that the HH pathway was the
molecular mechanism behind the effect of GSCs on the migration
ability of the b.END3 cells. Additionally, it was observed that the
phenomenon was more significant at an early stage than a late
stage. We suspect that this was due to the unstable overactivation
of the HH pathway.
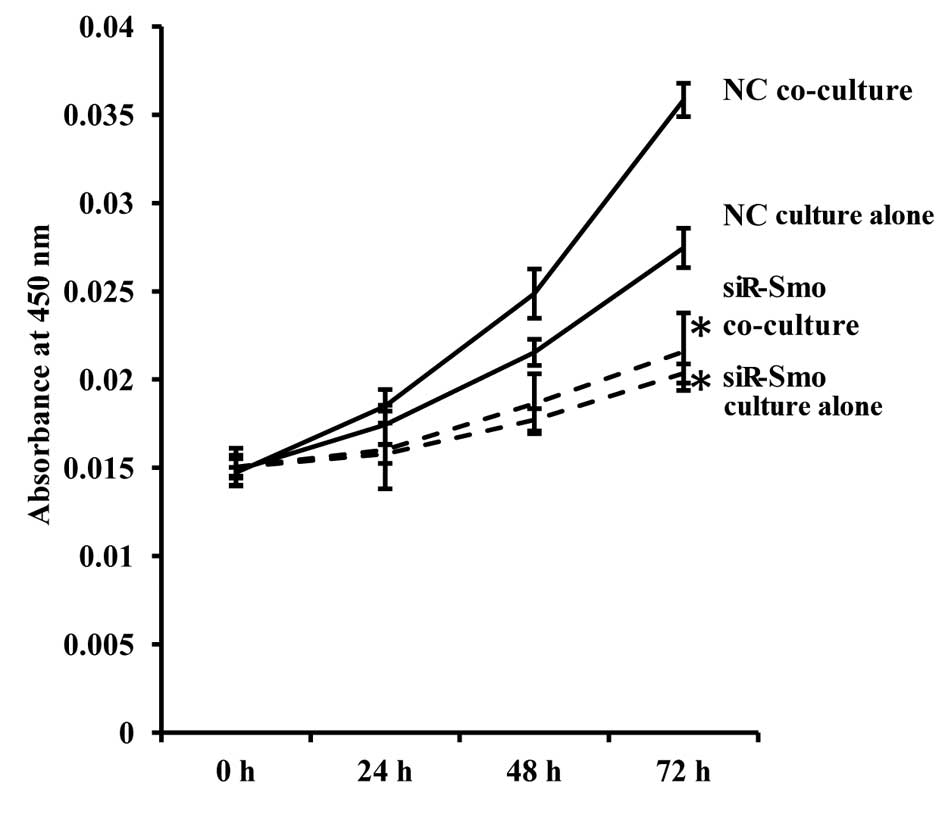
Proliferation of endothelial cells is
reduced following Smo gene knockdown
The proliferation assay was repeated to explore the
importance of the HH pathway in GSC-enhanced b.END3 cell
proliferation. Consistent with our expectations, the proliferation
of the endothelial cells was significantly decreased after Smo was
inhibited, and this was not restored by culture with GSCs (Fig. 5).
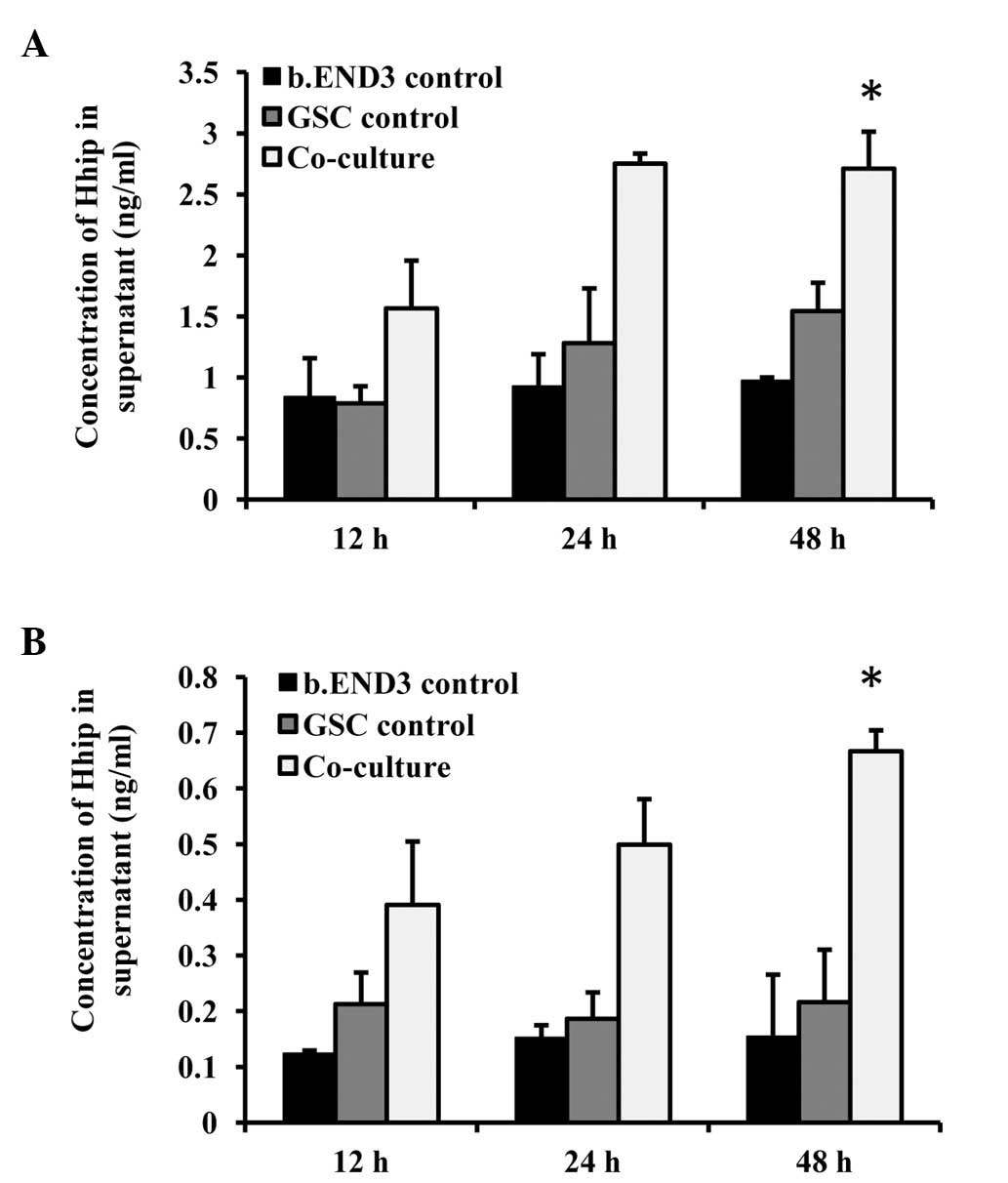
Shh and Hhip secretion is altered in the
co-culture system
To further explore the upstream factor of the HH
pathway in the co-culture, Shh and Hhip, the activator and
inhibitor of the HH pathway, were detected. As we considered that
the regulation of the endothelial cells by the GSCs in vivo
was mediated indirectly due to the low percentage of GSCs, ELISA
was used to detect the amount of Shh and Hhip secreted into the
supernatant. The Shh concentration was increased in the co-cultured
wells dependent on the culture time (Fig. 6A). A higher concentration of Hhip
protein was also detected in the co-culture supernatant (Fig. 6B). Since Hhip was inhibitory, we
hypothesize that the GSCs induced Shh expression, increasing the
local concentration of Shh, and that Hhip expression was the
feedback effect.
Discussion
Glioma is the most common malignant tumor in the
brain, accounting for 33.3–58.9% of all brain tumors, and with an
incidence that is still increasing (13). Modern surgery and other treatments
are not effective enough, due to the unique biological behavior,
high invasive growth and recurrence of glioblastomas. GSCs have
been isolated and identified. Researchers hope this small group of
cells, which self-renew and undergo multipotential differentiation,
are responsible for glioblastoma initiation, propagation and
recurrence, and may provide an indication towards a cure (4,14).
Angiogenesis is closely related to tumor growth. GBM
is rich in microvessels, making it a perfect model to study
angiogenesis and cancer stem cells (CSCs). It has been discovered
that the location of GSCs is close to the microvessels (10). Tumor microvessel endothelial cells
have been shown to be morphologically different from normal
endothelial cells, with elevated migration and resistance to
necrosis (15). The mechanism under
tumor angiogenesis is complex. Angiogenesis begins with the
gemmation and migration of endothelial cells. Cell cords then form
by endothelial cell proliferation and circulating endothelial cell
recruitment. Microvessels mature with a series of vascular
remodeling.
The mechanism of GSC-regulated tumor angiogenesis is
a hot topic of research (16–18).
Vascular endothelial growth factor is considered to be one of the
most important factors (19). Bone
morphogenic protein (20), formyl
peptide receptor (21), stromal
cell derived factor 1 and its receptor CXCR4 (22) and hypoxia-inducible factor (23) have all been reported to participate
in the regulation of angiogenesis.
The present study indicates a new mechanism of tumor
angiogenesis. The results demonstrated that GSCs regulate the gene
expression of nearby endothelial cells, specifically through the HH
pathway, thereby affecting their biological behavior. The HH
pathway is a classical pathway of cell survival, proliferation and
migration, and Shh is one of its classical activators, which
functions in autocrine and paracrine ways (24). The HH pathway is activated in
endothelial cells and their progenitors. Overexpression of the HH
pathway affects the proliferation, migration and remodeling of
vasculature (25). In the present
study, it was observed that the Gli1 gene, which is the key gene in
the HH pathway, was overexpressed in the endothelial cells when
indirectly co-cultured with GSCs, indicating HH pathway activation.
The proliferation and migration of the endothelial cells were
induced. Once the HH pathway had been knocked down, the phenomenon
disappeared. Next, the cause of this regulation was investigated
and Shh was shown to be overexpressed and secreted in the
endothelial cells when cultured with GSCs. Therefore, GSCs may
regulate Shh expression in nearby endothelial cells via soluble
factors, then activate their HH pathway. The present results also
provide further evidence of a CSC niche where GSCs interact with
endothelial cells.
Progression has been made in the study and treatment
of cancer using anti-angiogenesis drugs. However, adverse effects
have been observed. Microvasculature fracture causes local anoxia,
which is another niche for CSCs, causing local appearance of the
cells. In future studies, we aim to interfere with the interaction
between GSCs and microvessels, for example the HH pathway, to
inhibit tumor invasion and propagation and to provide opportunities
for tumor resection.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (no. 30870965) and the National
Basic Research Program of China (973 program, no.
2010CB529403).
References
|
1
|
Berger MS: Glioma surgery: a century of
challenge. Clin Neurosurg. 58:7–9. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Quick A, Patel D, Hadziahmetovic M, et al:
Current therapeutic paradigms in glioblastoma. Rev Recent Clin
Trials. 5:14–27. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chaichana KL, McGirt MJ, Laterra J, et al:
Recurrence and malignant degeneration after resection of adult
hemispheric low grade gliomas. J Neurosurg. 112:10–17. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dietrich J, Diamond EL and Kesari S:
Glioma stem cell signaling: therapeutic opportunities and
challenges. Expert Rev Anticancer Ther. 10:709–722. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
O'Brien CA, Kreso A and Jamieson CH:
Cancer stem cells and self-renewal. Clin Cancer Res. 16:3113–3120.
2010. View Article : Google Scholar
|
|
6
|
Liu Y, Carson-Walter EB, Cooper A, et al:
Vascular gene expression patterns are conserved in primary and
metastatic brain tumors. J Neurooncol. 99:13–24. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Calabrese C, Poppleton H, Kocak M, et al:
A perivascular niche for brain tumor stem cells. Cancer Cell.
11:69–82. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li M, Niu C, Gao G, et al: The study of
the relation between proliferating brain tumor stem cells with
micro-vascular system. Chin J Neurooncol. 8:82–87. 2010.(In
Chinese).
|
|
9
|
Barami K: Relationship of neural stem
cells with their vascular niche: implications in the malignant
progression of gliomas. J Clin Neurosci. 15:1193–1197. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fu JR, Liu WL, Zhou JF, et al: Sonic
hedgehog protein promotes bone marrow-derived endothelial
progenitor cell proliferation, migration and VEGF production via PI
3-kinase/Akt signaling pathways. Acta Pharmacol Sin. 27:685–693.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang Q, Dong J, Zhu Y, et al: Isolation
and culture of tumor stem cells from human brain glioma tissues.
Zhonghua Zhong Liu Za Zhi. 28:331–333. 2006.(In Chinese).
|
|
12
|
Zhou K, Yang H, Zhou W, Zhang C, Shu H and
Wang B: Effect of drug blockade of type-3 metabotropic glutamate
receptor on proliferation and differentiation in glioma stem cells.
Di 3 Jun Yi Da Xue Xue Bao. 34:1701–1706. 2012.(In Chinese).
|
|
13
|
Hess KR, Broglio KR and Bondy ML: Adult
glioma incidence trends in the United States, 1977–2000. Cancer.
101:2293–2299. 2004.PubMed/NCBI
|
|
14
|
Singh SK, Clarke ID, Terasaki M, et al:
Identification of a cancer stem cell in human brain tumors. Cancer
Res. 63:5821–5828. 2003.PubMed/NCBI
|
|
15
|
Charalambous C, Chen TC and Hofman FM:
Characteristics of tumor-associated endothelial cells derived from
glioblastoma multiforme. Neurosurg Focus. 20:E222006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bao S, Wu Q, Sathornsumetee S, et al: Stem
cell-like glioma cells promote tumor angiogenesis through vascular
endothelial growth factor. Cancer Res. 66:7843–7848. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Borovski T, De Sousa E, Melo F, Vermeulen
L and Medema JP: Cancer stem cell niche: the place to be. Cancer
Res. 71:634–639. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang R, Chadalavada K, Wilshire J, et al:
Glioblastoma stem-like cells give rise to tumour endothelium.
Nature. 468:829–833. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Folkins C, Shaked Y, Man S, et al: Glioma
tumor stem-like cells promote tumor angiogenesis and vasculogenesis
via vascular endothelial growth factor and stromal-derived factor
1. Cancer Res. 69:7243–7251. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Scharpfenecker M, van Dinther M, Liu Z, et
al: BMP-9 signals via ALK1 and inhibits bFGF-induced endothelial
cell proliferation and VEGF-stimulated angiogenesis. J Cell Sci.
120:964–972. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yao XH, Ping YF, Chen JH, et al:
Glioblastoma stem cells produce vascular endothelial growth factor
by activation of a G-protein coupled formylpeptide receptor FPR. J
Pathol. 215:369–376. 2008. View Article : Google Scholar
|
|
22
|
Ping YF, Yao XH, Jiang JY, et al: The
chemokine CXCL12 and its receptor CXCR4 promote glioma stem
cell-mediated VEGF production and tumor angiogenesis via PI3K/AKT
signalling. J Pathol. 224:344–354. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Medici D and Olsen BR: Rapamycin inhibits
proliferation of hemangioma endothelial cells by reducing
HIF-1-dependent expression of VEGF. PLoS One. 7:e429132012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma G, Xiao Y and He L: Recent progress in
the study of Hedgehog signaling. J Genet Genomics. 35:129–137.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Asai J, Takenaka H, Kusano KF, et al:
Topical sonic hedgehog gene therapy accelerates wound healing in
diabetes by enhancing endothelial progenitor cell-mediated
microvascular remodeling. Circulation. 113:2413–2424. 2006.
View Article : Google Scholar
|