Introduction
Laryngeal squamous cell carcinoma (LSCC) represents
the second most common malignant neoplasm of the respiratory tract
after lung cancer (1). LSCC has a
strong propensity to metastasize to regional lymph nodes, which
decreases the cure and survival rates (2,3).
Multiple steps and factors are involved in the process of malignant
cancer progression (4,5). This process is under the control of
many metastasis-associated genes. Among these,
metastasis-associated gene 1 (MTA1) is positively correlated
with cancer metastasis in many cancer types. Toh et
al(6) first cloned MTA1
from the highly metastatic mammary adenocarcinoma cell line by
differential cDNA library screening. MTA1 is a subunit of
the nucleosome remodeling and deacetylase (NURD) complex, which is
involved in chromatin remodeling and histone deacetylation in gene
expression regulation (7).
MTA1 functions as a transcriptional coregulator, regulating
the downstream target genes that encode effector proteins
controlling cancerous processes (8). MTA1 overexpression is
positively correlated with in vitro migration and invasion
ability in KYSE150 and B16F10 melanoma cell lines, and inhibition
of MTA1 protein expression results in growth inhibition of
cancer cell lines (9,10). Sasaki et al(11,12)
reported that MTA1 mRNA was overexpressed in thymoma and
advanced lung cancer. It has also been reported that MTA1
may be involved in initiating carcinogenesis (13,14).
Miyatani et al(15) compared
the expression of MTA1 in normal esophageal epithelium,
normal gastric epithelium and gastro-esophageal junction cancer,
and found that MTA1 levels were significantly higher in
cancer samples than in their normal counterparts. Moreover, in
tonsil cancer, MTA1 is positively correlated with lymphatic
metastasis (16). It has also been
indicated that MTA1 is correlated with tumor angiogenesis
and poor outcome in patients with early-stage non-small cell lung
cancer (NSCLC) (17). This line of
evidence indicates that MTA1 may become a new marker for
predicting cancer metastasis, or even cancer outcome.
Concerning the molecular mechanism of MTA1 in
cancer cell metastasis, MTA1 has been reported to be
involved in cancer development in several ways.
MTA1-interacting coactivator has been identified as a
molecule that interacts with MTA1 to regulate estrogen
receptor-α transactivation (18).
Yoo et al(19) reported that
MTA1 stabilizes hypoxia-inducible factor-1α protein by
recruiting histone deacetylase 1, and is correlated with
angiogenesis in cancer development (20). Since MTA1 is a histone
deacetylase (HDAC)-interacting protein that modulates the
epigenetic status of its target genes, it is expected to widely
influence the expression pattern of the cancer-related gene
spectrum. Ghanta et al(21)
revealed, using a profiling assay, that MTA1 regulation was
partially under the control of p53. When p53 is functional,
MTA1 mainly focuses on inflammatory and antimicrobial
responses; when p53 is absent, MTA1 predominantly targets
genes in cancer signaling. MTA1 is correlated with cigarette
smoking in NSCLC, indicating its importance in the smoking-related
progression of this type of cancer (22). MTA1 has also been reported to
regulate the anoikis of human prostate cancer cells (23), which reveals a new subfield of
MTA1 mechanisms.
MTA1 is a corepressor responsible for
estrogen receptor repression at the transcriptional level (24). A naturally occurring MTA1
variant, MTA1s, can sequester estrogen receptor-α in the
cytoplasm (25). Estrogen receptor
involvement is the first insight into the p53-independent function
of MTA1 in the DNA damage response involving the
p21/WAF1-proliferating cell nuclear antigen pathway (26). MTA1 is required for the
ATR-mediated DNA damage checkpoint function (27). UV radiation stabilizes MTA1
and increases MTA1 binding to ATR. Other molecules found to
be associated with MTA1 expression include RECK (28), HDAC1 (15) and MMP-9 (29). Silencing MTA1 by RNA
interference (RNAi) reverses the malignant phenotypes, including
adhesion, migration and invasiveness of cervical cancer cells
(SiHa) via altered expression of p53 and the E-cadherin/β-catenin
complex (30).
No systematic biological studies have been performed
on LSCC to date. This study aimed to determine the biological role
of MTA1 in LSCC using gain-of-function and RNAi
techniques.
Materials and methods
Cell lines
The human LSCC cell line HEP-2 and the human
keratinocyte HaCaT cell line (State Key Laboratory of Molecular
Oncology, Beijing, China) were cultured in RPMI-1640 and DMEM
medium (Gibco-BRL, Grand Island, NY, USA), respectively,
supplemented with 10% (v/v) fetal calf serum (FCS) (Hyclone
Laboratories, Inc., Logan, UT, USA), 2 mM L-glutamine and
antibiotics (penicillin-streptomycin at 100 U/ml) in a humidified
atmosphere of 5% CO2 at 37°C. The keratinocyte HaCaT
cell line is an immortalized normal epithelial cell line.
Reagents
Lipofectamine 2000 was purchased from Invitrogen
Life Technologies (Carlsbad, CA, USA). siRNA sequence was
chemically synthesized by Jikai Co. (Shanghai, China). The MTA1
primary antibody was from Santa Cruz Biotechnology, Inc. (sc-9446;
Santa Cruz, CA, USA) and the horseradish peroxidase-conjugated
secondary antibody was from Zhongshan Biotech, Co., Ltd (Zhongshan,
China). The ECL detection system was purchased from Amersham
Biosciences (Piscataway, NJ, USA). The Boyden chamber system and
polycarbonate membrane (8 μm pore size) were obtained from Neuro
Probe, Inc. (Canada). Matrigel was purchased from BD Biosciences
(San José, CA, USA).
siRNA and plasmid transfection
The 21-nt siRNA sequence was chemically synthesized
(Jikai Co.). The target sequences of the siRNA for the MTA1
gene (MTA1-siRNA) were as follows: Sense,
5′-GAACAUCUACGACAUCUCCdTdT-3′ and antisense,
5′-GGAGAUGUCGUAGAUGUUCdTdT-3′ (9).
The MTA1-siRNA was dissolved in sterilized and RNase-free
water and annealed. The final concentration was 20 μM.
Lipofectamine 2000 (20 μl/ml, Invitrogen Life Technologies) was
used to transfect the HEP-2 cell line according to the
manufacturer’s instructions. A sequence non-specific to any known
gene was used as a negative control (Jikai Co.). The
pcDNA3-MTA1 plasmid was provided by Dr Mahoney (Jefferson
Institute of Molecular Medicine, Thomas Jefferson University,
Philadelphia, PA, USA), and the transfection was performed
according to the manufacturer’s instructions. The cells transfected
with pcDNA3-MTA1 were selected by G418 prior to use.
Western blotting analysis
Cells were grown to 80% confluence and rinsed twice
with 1X PBS prior to harvesting. Total cell protein was extracted
using PBS buffer containing aprotinin (2 μg/ml), PMSF (100 μg/ml),
leupeptin (2 μg/ml) and 1% Nonidet P-40. Protein concentration was
determined using the Gene Quant Pro-91738 protein assay system
(Bio-Rad Laboratories, Inc, Hercules, CA, USA). Samples were
briefly electrophoresed in 10% SDS-PAGE and transferred to
nitrocellulose membranes using a semidry transfer system.
Nonspecific binding was blocked for 2 h in 5% fat-free milk in PBS
buffer, pH 7.6. Blots were first incubated with MTA1 primary
antibody (1:200) (Santa Cruz Biotechnology, Inc.) for 2 h at 37°C,
and then with corresponding horseradish peroxidase-conjugated
secondary antibody (1:2,000, Zhongshan Biotech) for 1 h at room
temperature. Signals were visualized using the ECL detection system
according to the manufacturer’s instructions (Amersham
Biosciences). The detection was repeated three times.
Migration and invasion assay
Migration assays were performed using a Boyden
chamber system (Neuro Probe, Inc., Gaithersburg, MD USA) with a
fibronectin-precoated (0.5 mg/ml) polycarbonate membrane (8 μm pore
size) (AP48; Neuro Probe, Inc.) as described previously, with minor
modifications (9). The invasion
chamber was identical to the migration chamber, but with the 250
μg/ml Matrigel (BD Biosciences) precoated polycarbonate membrane.
For both assays, the bottom chambers were filled with medium
containing 10% FCS, RPMI-1640 and 2% BSA as chemoattractant, and
medium containing serum-free RPMI-1640, and 0.2% BSA was added into
the top chambers. Treated or control cells (2×104 per
well) were added to the top chambers, followed by a 10-h incubation
at 37°C and 5% CO2. Three independent experiments were
performed for each set. The cells migrated through and adhered to
the bottom of the membrane were then fixed and stained with Giemsa
dye. The cells that migrated to the lower side of the membrane were
mounted under a microscope and averaged. This experiment was
repeated three times.
Adhesion assay
Adhesion assay was performed by the MTT assay. The
same numbers of MTA1-siRNA-, control-siRNA- and
pcDNA3-MTA1-transfected HEP-2 cells (1×105) were
plated into the Matrigel-precoated (50 μg/ml) 96-well plate in
triplicate. The groups of cells were washed for 30, 60 and 90 min,
respectively, to remove the non-adherent cells. After washing, the
adherent cells were measured with MTT assay at 490 nm wavelength.
The OD values reflect the proportion of cells that adhered to the
Matrigel-coated 96-well plate. This experiment was repeated in
triplicate.
Wound healing assay
To perform the wound healing assay,
pcDNA3-MTA1-, MTA1-siRNA- and
control-siRNA-transfected HEP-2 cells were implanted into the
Matrigel (50 μg/ml)-coated 35-mm culture dishes, as described by
Qian et al(9). When the
cells grew to 80% confluence, a sterilized tip was used to draw a
line with the same width on the bottom of the dishes. Images were
captured at 8, 16 and 24 h after the wounding. Data shown in the
text are representative of three independent repeats.
RT-PCR analysis
Total RNA was isolated from HEP-2 and HaCaT cells,
and RT-PCR was performed according to the manufacturer’s
instructions to detect gene expression (K0011 RT-PCR kit, Vigorous
Biotechnology, Beijing, China). The primers used for amplification
were as follows: MTA1 (forward primer:
5′-CCGGGCCTGCGAGAGCTGTTACAC-3′, reverse primer:
5′-CACGGCTTCCAGCGGCTTGCGTAC-3′); β-actin (forward primer:
5′-ACCACAGTCCATGCCATCAC-3′, reverse primer:
5′-TCCACCACCCTGTTGCTGTA-3′). The cycling conditions were as
follows: Initial denaturation at 94°C for 5 min, followed by 28
cycles at 94°C for 30 sec, 55°C for 40 sec and 72°C for 30 sec.
Statistical analyses
The data were analyzed by ANOVA. The statistical
analysis was performed using SPSS 11.0 software (SPSS, Inc,
Chicago, IL, USA), and P<0.05 was considered to indicate a
statistically significant difference.
Results
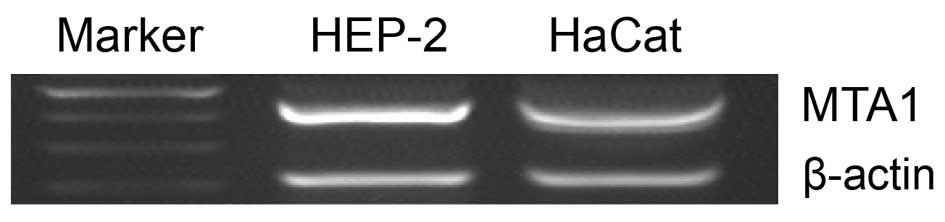
MTA1 mRNA expression in HEP-2 and HaCaT
cell lines
Expression of MTA1 mRNA in Hep-2 and HaCaT
cell lines was evaluated by RT-PCR. The level of MTA1 mRNA
was significantly higher in the LSCC cell line, HEP-2, than in the
immortalized keratinocyte HaCaT cell line (Fig. 1). This result indicates that LSCC
cells have higher levels of MTA1 expression than normal cell
lines, as indicated by the active role of MTA1 in LSCC.
Thus, for further experiments in this study, the HEP-2 cell line
was selected to study MTA1 biological functions.
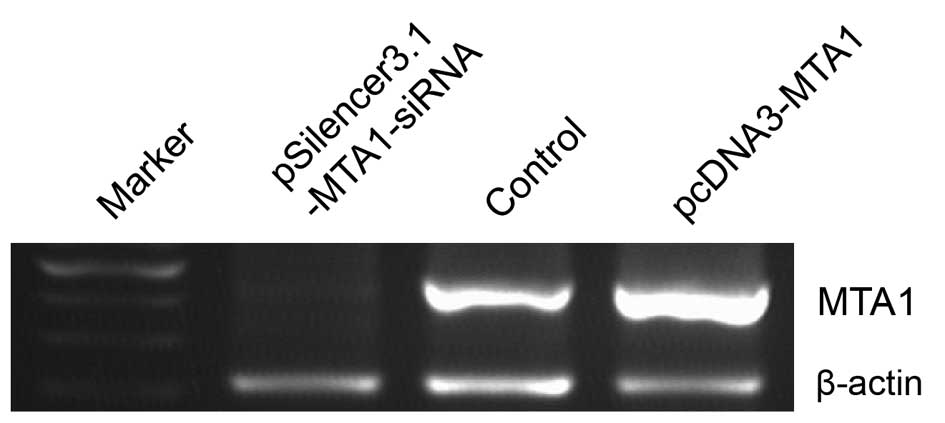
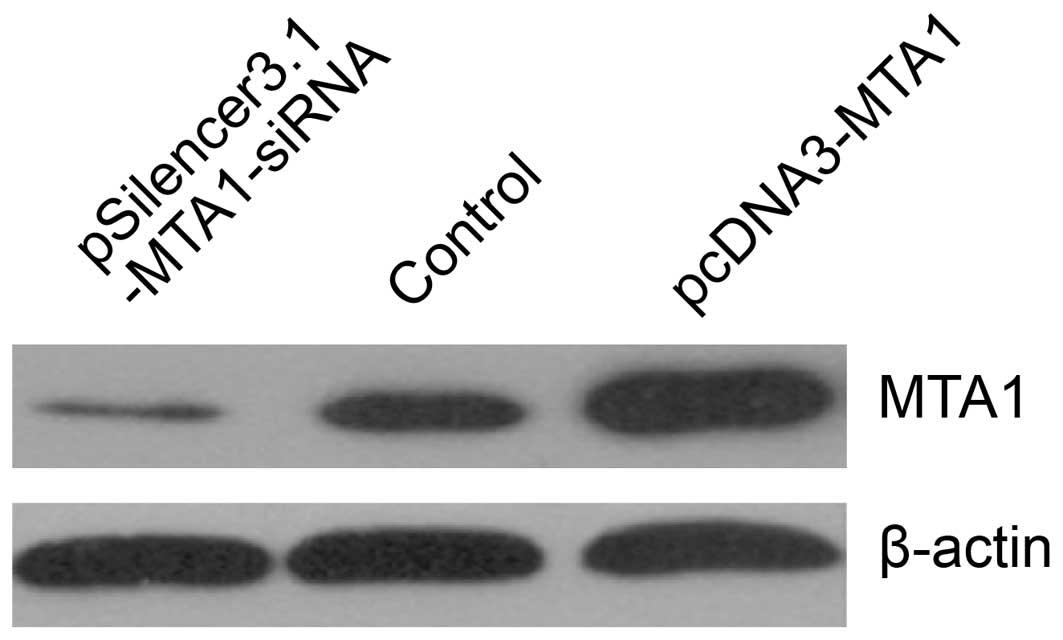
Confirmation of MTA1 expression by RT-PCR
and western blot analysis in the gain-of-function and
loss-of-function studies
We performed gene transfection and RNA interference
to study MTA1 functions. RT-PCR and western blot analysis
confirmed the changes in MTA1 levels after
pcDNA3-MTA1 and pSilencer3.1-MTA1-siRNA transfection.
We found that the expression of MTA1 was either suppressed
or increased at the mRNA and protein level after treatment. The
mRNA levels corresponding to pcDNA3-MTA1, control-siRNA and
MTA1-siRNA, presented by density value, were 2.78±0.046,
1.04±0.119 and 0.32±0.046, respectively. The protein levels were
2.69±0.267, 1.07±0.112 and 0.36±0.069, respectively. Compared with
the control-siRNA-transfected cells, MTA1-siRNA
significantly decreased the expression of MTA1 (P<0.05),
while pcDNA3-MTA1 significantly enhanced MTA1
expression (P<0.05) (Figs. 2 and
3).
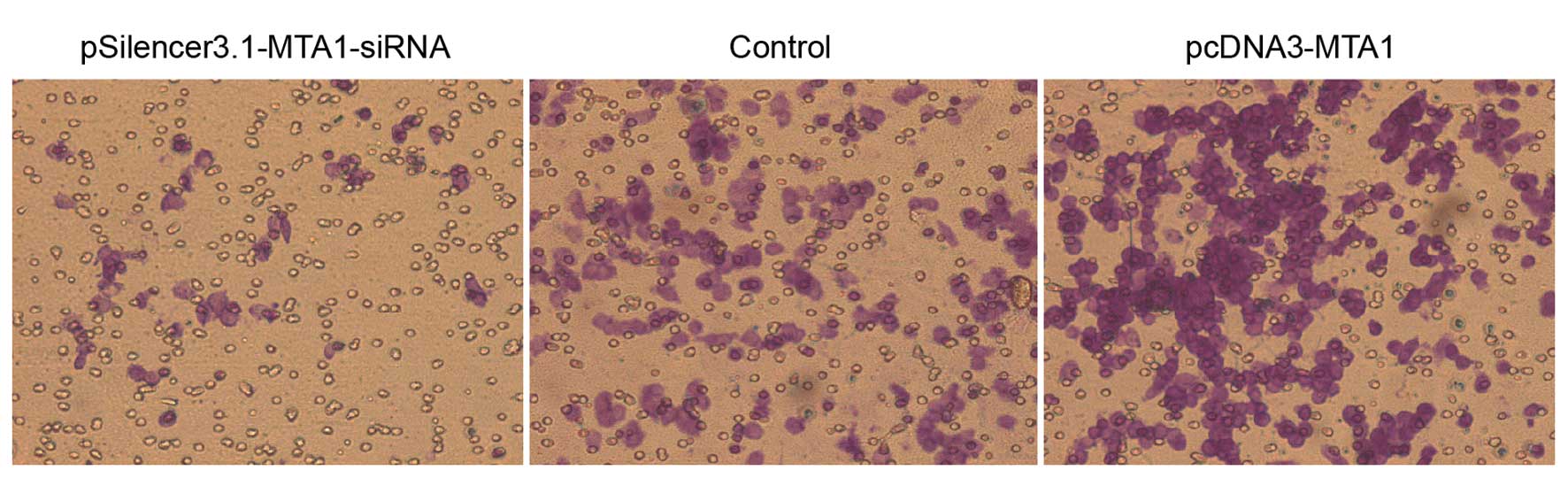
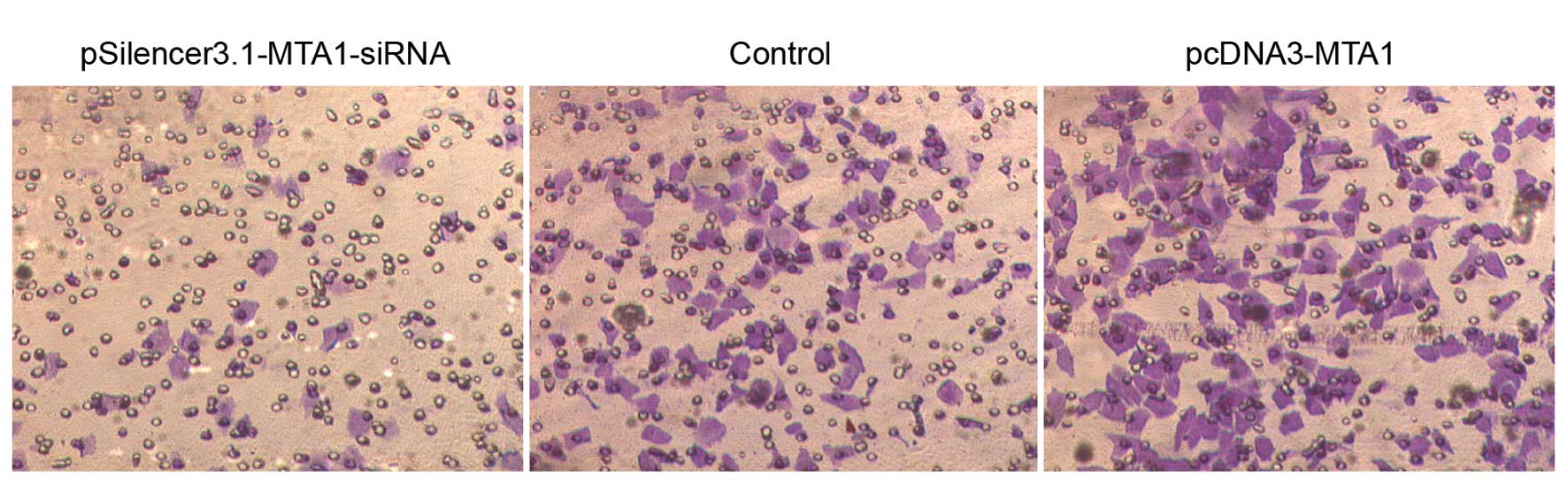
In vitro migration and invasion ability
of HEP-2 cells following transfection of pcDNA3-MTA1 and
pSilencer3.1-MTA1-siRNA
The in vitro migration and invasion ability
of HEP-2 cells transfected with pcDNA3-MTA1 and
MTA1-siRNA were studied using the Boyden chamber model as
described in Materials and methods. In the migration assay,
responding to pcDNA3-MTA1, control-siRNA and
MTA1-siRNA, the number of cells that migrated to the lower
side of the membrane was 549.2±21.51, 352±25.03 and 120.8±17.28,
respectively. The number of cells that migrated to the lower side
of the membrane in the invasion assay was 423.6±14.15, 301.2±25.4
and 115.4±15.52, respectively. Compared with the
control-siRNA-transfected cells, MTA1-siRNA significantly
decreased the migration and invasion ability of HEP-2 cells
(P<0.05), while pcDNA3-MTA1 significantly enhanced the
migration and invasion ability of HEP-2 cells (P<0.05) (Figs. 4 and 5).
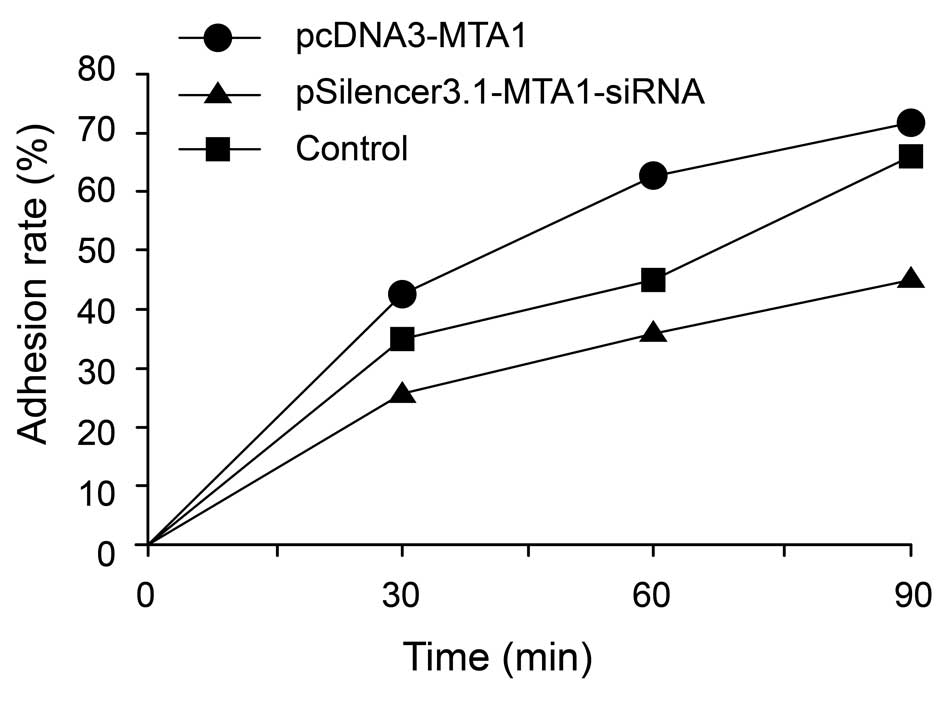
Adhesion assay of pcDNA3-MTA1- and
MTA1-siRNA-treated HEP-2 cell line
The adhesion of cancer cells to the extracellular
matrix and cell surface molecules is a key step during metastasis
in vivo. We evaluated the effects of the MTA1 gene on
the adhesion ability of cancer cells. The results showed that, at
the early stage of adhesion, pcDNA3-MTA1 promoted the
adhesion of HEP-2 cells to the Matrigel matrix, while silencing of
MTA1 by RNAi inhibited the adhesion process. The adhesion
rates of HEP-2 cells transfected with pcDNA3-MTA1,
control-siRNA and MTA1-siRNA at 30 min post-cell seeding
were 41.8±7.16, 35.6±6.08 and 26.6±2.97% (P<0.05,
MTA1-siRNA vs. control and pcDNA3-MTA1 groups),
respectively; 63±10.44, 46±8.34 and 38.6±7.86% (P<0.05,
MTA1-siRNA vs. control and pcDNA3-MTA1 groups) at 60
min post-cell seeding, respectively; and 71.2±6.83, 63.6±7.56 and
45±6.08% (P<0.05, MTA1-siRNA vs. control and
pcDNA3-MTA1 groups) at 90 min post-cell seeding,
respectively (Fig. 6). These
results indicate that MTA1 may exert its effects on
metastasis by regulating the adhesion molecules on the cell
surface.
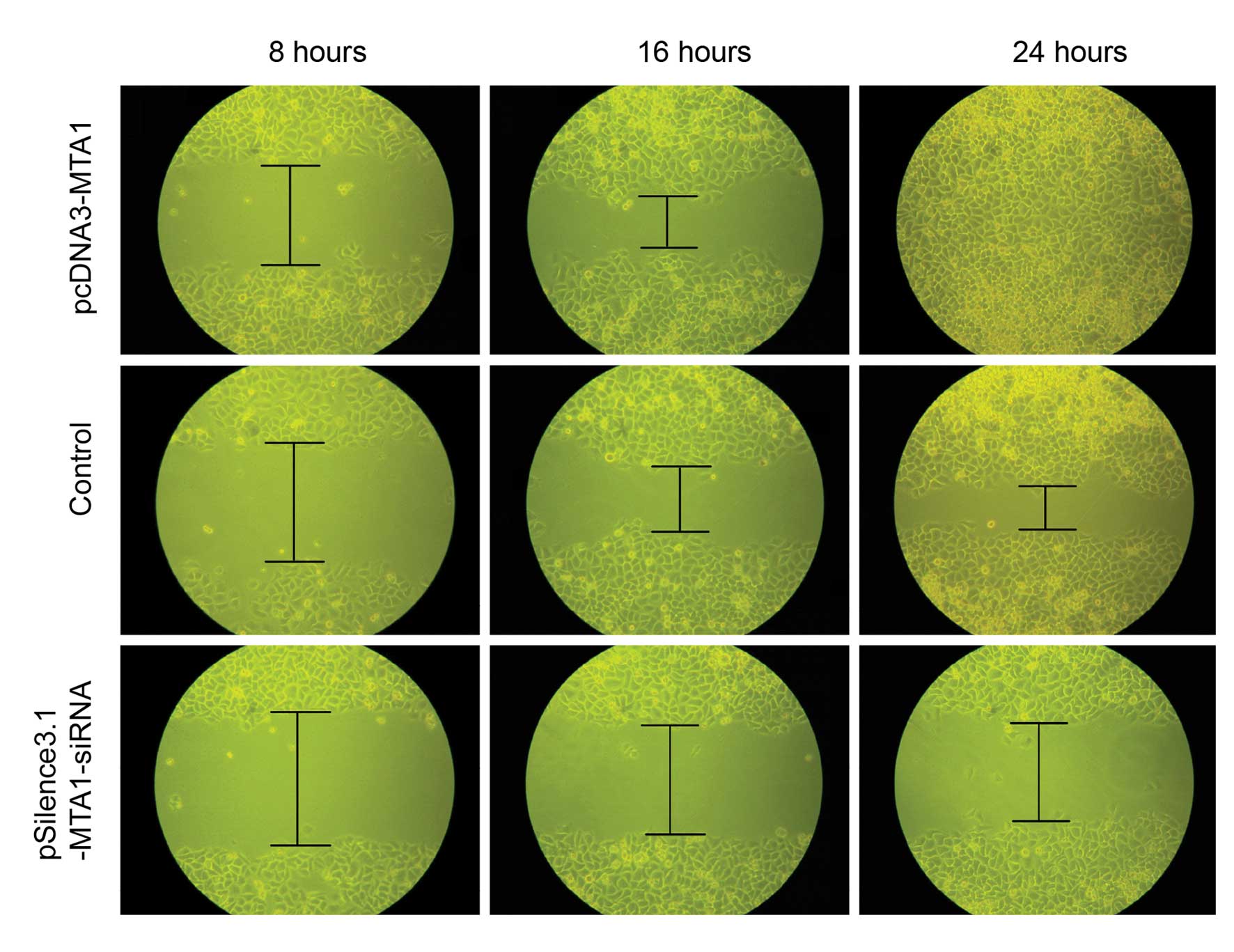
Wound healing assay
For the wound healing assay, equal numbers of
transfected HEP-2 cells were reseeded into 35-mm-diameter culture
wells. The wound healing ability of cells reflects their movement
on the surface to which they are anchored for growth. At 8, 16 and
24 h after wounding, the healing ability of
MTA1-siRNA-transfected cells was significantly poorer than
that of the pcDNA3-MTA1- and control-siRNA-transfected cells
(Fig. 7).
Discussion
Cancer metastasis is the most common factor that
causes cancer patient mortality and is a complex process involving
a wide range of biological behaviors. It is urgent that the
mechanisms underlying cancer metastasis be explored. Carcinogenesis
involves several metastasis-associated molecules, many of which are
altered during the process. Recent studies have revealed that
MTA1 is important in regulating cancer behaviors.
MTA1 expression has been associated with cancer malignancy
in various cancer types, and has been found to increase the
metastatic and invasive potential of carcinoma cells (12,31–33).
LSCC is a type of cancer with a high potential for metastasis and
invasion. To investigate whether MTA1 is responsible, at
least partially, for the metastatic potential of LSCC, MTA1
gene function in the LSCC HEP-2 cell line was biologically studied
using gene expression and RNAi techniques.
To confirm the correlation between MTA1 and
cancerous potential in LSCC cells, the MTA1 level in HEP-2
and HaCaT cells was detected. It was observed that MTA1 was
expressed at relatively low levels in normal human keratinocyte
HaCaT cell lines, while its expression was upregulated in HEP-2
cell lines. This result provided evidence supporting the function
of MTA1 in the development of human LSCC.
For further confirmation, gene transfection and RNA
interference were performed to study MTA1 functions in LSCC.
RT-PCR and western blot analysis showed that the expression of
MTA1 was either suppressed or increased at the mRNA and
protein level after pcDNA3-MTA1 and
pSilencer3.1-MTA1-siRNA transfection. To confirm the
association between MTA1 expression and the metastatic
potential of cancer cells, migration, invasion, adhesion and wound
healing assays were performed after manipulating MTA1
expression. The results showed that overexpression of MTA1
promoted the metastasis potential of HEP-2 cells, while MTA1
silencing by RNAi reversed the malignant phenotypes. The in
vivo metastasis ability of HEP-2 was inhibited by MTA1
silencing. In the cellular biological studies, a causal correlation
between MTA1 expression and the in vitro migration
and invasion ability of LSCC cancer cells was identified.
Silencing of MTA1 also impairs the
angiogenesis of prostate cancer, partially eliminating the
circumstance of cancer growth (34). These studies support the idea that
MTA1 may be a potential target for cancer therapy. In the
current study, silencing of MTA1 reversed the cancerous
behaviors of LSCC, extending the therapeutic value of MTA1
in cancer treatment. MTA1-silencing was observed to result
in a prominent loss of function in cancer cells, while
overexpression of MTA1 only increased cancerous behaviors to
a certain extent. This may indicate that MTA1 is critical in
the life activity of cancer cells.
Molecular pathways were not the focus of the current
study. Therefore, our next aim is to identify the mechanisms of
MTA1 responsible for the change in biological phenotypes and
in the structure-function relationship of MTA1.
In brief, the results of the present study
demonstrate that the expression of MTA1 promotes the
migration and invasion ability of HEP-2 cells, indicating its
importance in the progression of LSCC. Suppressing HDAC activity is
currently the target of chemotherapy (35–37).
MTA1, as an HDAC component of NURD complexes, may be a
potential powerful target for LSCC biotherapy or chemotherapy.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (81071773) and the Major State Basic
Research Development Program (2009CB521807). The authors are
greatly appreciative of their support.
References
|
1
|
Cattaruzza MS, Maisonneuve P and Boyle P:
Epidemiology of laryngeal cancer. Eur J Cancer B Oral Oncol.
32B:293–305. 1996. View Article : Google Scholar
|
|
2
|
Forastiere A, Koch W, Trotti A and
Sidransky D: Head and neck cancer. N Engl J Med. 345:1890–1900.
2001. View Article : Google Scholar
|
|
3
|
Greenlee RT, Hill-Harmon MB, Murray T and
Thun M: Cancer statistics. CA Cancer J Clin. 51:15–36. 2001.
|
|
4
|
Song IH: Cancer metastasis and metastasis
suppressors. Korean J Gastroenterol. 43:1–7. 2004.
|
|
5
|
Stracke ML and Liotta LA: Multi-step
cascade of tumor cell metastasis. In Vivo. 6:309–316.
1992.PubMed/NCBI
|
|
6
|
Toh Y, Pencil SD and Nicolson GL: A novel
candidate metastasis-associated gene, mta1, differentially
expressed in highly metastatic mammary adenocarcinoma cell lines.
cDNA cloning, expression, and protein analyses. J Biol Chem.
269:22958–22963. 1994.
|
|
7
|
Xue Y, Wong J, Moreno GT, Young MK, Côté J
and Wang W: NURD, a novel complex with both ATP-dependent
chromatin-remodeling and histone deacetylase activities. Mol Cell.
2:851–861. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gururaj AE, Singh RR, Rayala SK, Holm C,
den Hollander P, Zhang H, Balasenthil S, Talukder AH, Landberg G
and Kumar R: MTA1, a transcriptional activator of breast cancer
amplified sequence 3. Proc Natl Acad Sci USA. 103:6670–6675. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qian H, Lu N, Xue L, Liang X, Zhang X, Fu
M, Xie Y, Zhan Q, Liu Z and Lin C: Reduced MTA1 expression by RNAi
inhibits in vitro invasion and migration of esophageal squamous
cell carcinoma cell line. Clin Exp Metastasis. 22:653–662. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qian H, Yu J, Li Y, Wang H, Song C, Zhang
X, Liang X, Fu M and Lin C: RNA interference of
metastasis-associated gene 1 inhibits metastasis of B16F10 melanoma
cells in a C57BL/6 mouse model. Biol Cell. 99:573–581. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sasaki H, Yukiue H, Kobayashi Y, Nakashima
Y, Kaji M, Fukai I, Kiriyama M, Yamakawa Y and Fujii Y: Expression
of the MTA1 mRNA in thymoma patients. Cancer Lett. 174:159–163.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sasaki H, Moriyama S, Nakashima Y,
Kobayashi Y, Yukiue H, Kaji M, Fukai I, Kiriyama M, Yamakawa Y and
Fujii Y: Expression of the MTA1 mRNA in advanced lung cancer. Lung
Cancer. 35:149–154. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang H, Stephens LC and Kumar R:
Metastasis tumor antigen family proteins during breast cancer
progression and metastasis in a reliable mouse model for human
breast cancer. Clin Cancer Res. 12:1479–1486. 2006. View Article : Google Scholar
|
|
14
|
Bagheri-Yarmand R, Talukder AH, Wang RA,
Vadlamudi RK and Kumar R: Metastasis-associated protein 1
deregulation causes inappropriate mammary gland development and
tumorigenesis. Development. 131:3469–3479. 2004. View Article : Google Scholar
|
|
15
|
Miyatani T, Kurita N, Mikami C, Kashihara
H, Higashijima J, Yoshikawa K, Nishioka M, Sato H, Iwata T and
Shimada M: Malignant potential of Barrett’s esophagus: special
reference to HDAC-1 and MTA-1 expression. Hepatogastroenterology.
58:472–476. 2011.
|
|
16
|
Park JO, Jung CK, Sun DI, Joo YH and Kim
MS: Relationships between metastasis-associated protein (MTA) 1 and
lymphatic metastasis in tonsil cancer. Eur Arch Otorhinolaryngol.
268:1329–1334. 2011. View Article : Google Scholar
|
|
17
|
Zhu X, Guo Y, Li X, Ding Y and Chen L:
Metastasis-associated protein 1 nuclear expression is associated
with tumor progression and clinical outcome in patients with
non-small cell lung cancer. J Thorac Oncol. 5:1159–1166. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mishra SK, Mazumdar A, Vadlamudi RK, Li F,
Wang RA, Yu W, Jordan VC, Santen RJ and Kumar R: MICoA, a novel
metastasis-associated protein 1 (MTA1) interacting protein
coactivator, regulates estrogen receptor-alpha transactivation
functions. J Biol Chem. 278:19209–19219. 2003. View Article : Google Scholar
|
|
19
|
Yoo YG, Kong G and Lee MO:
Metastasis-associated protein 1 enhances stability of
hypoxia-inducible factor-1alpha protein by recruiting histone
deacetylase 1. EMBO J. 25:1231–1241. 2006. View Article : Google Scholar
|
|
20
|
Li SH, Tian H, Yue WM, Li L, Li WJ, Chen
ZT, Hu WS, Zhu YC and Qi L: Overexpression of metastasis-associated
protein 1 is significantly correlated with tumor angiogenesis and
poor survival in patients with early-stage non-small cell lung
cancer. Ann Surg Oncol. 18:2048–2056. 2011. View Article : Google Scholar
|
|
21
|
Ghanta KS, Li DQ, Eswaran J and Kumar R:
Gene profiling of MTA1 identifies novel gene targets and functions.
PLoS One. 6:e171352011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu L, Mao XY, Fan CF and Zheng HC: MTA1
expression correlates significantly with cigarette smoke in
non-small cell lung cancer. Virchows Arch. 459:415–422. 2011.
View Article : Google Scholar
|
|
23
|
Cui FL, Gong DD, Zhou YJ, Zhu L and Fan Y:
Regulatory effect of MTA1 on the anoikis of human prostate cancer
cells. Zhonghua Nan Ke Xue. 17:427–430. 2011.(In Chinese).
|
|
24
|
Mazumdar A, Wang RA, Mishra SK, Adam L,
Bagheri-Yarmand R, Mandal M, Vadlamudi RK and Kumar R:
Transcriptional repression of oestrogen receptor by
metastasis-associated protein 1 corepressor. Nat Cell Biol.
3:30–37. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kumar R, Wang RA, Mazumdar A, Talukder AH,
Mandal M, Yang Z, Bagheri-Yarmand R, Sahin A, Hortobagyi G, Adam L,
et al: A naturally occurring MTA1 variant sequesters oestrogen
receptor-alpha in the cytoplasm. Nature. 418:654–657. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li DQ, Pakala SB, Reddy SD, Ohshiro K,
Peng SH, Lian Y, Fu SW and Kumar R: Revelation of p53-independent
function of MTA1 in DNA damage response via modulation of the p21
WAF1-proliferating cell nuclear antigen pathway. J Biol Chem.
285:10044–10052. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li DQ, Ohshiro K, Khan MN and Kumar R:
Requirement of MTA1 in ATR-mediated DNA damage checkpoint function.
J Biol Chem. 285:19802–19812. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Deng Y, Zhou D and Zeng L: Expression and
significance of MTA1 and RECK gene in nasopharyngeal carcinoma. Lin
Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 25:534–538. 2011.(In
Chinese).
|
|
29
|
Jiang Q, Zhang H and Zhang P:
ShRNA-mediated gene silencing of MTA1 influenced on protein
expression of ER alpha, MMP-9, CyclinD1 and invasiveness,
proliferation in breast cancer cell lines MDA-MB-231 and MCF-7 in
vitro. J Exp Clin Cancer Res. 30:602011. View Article : Google Scholar
|
|
30
|
Rao Y, Wang H, Fan L and Chen G: Silencing
MTA1 by RNAi reverses adhesion, migration and invasiveness of
cervical cancer cells (SiHa) via altered expression of p53, and
E-cadherin/β-catenin complex. J Huazhong Univ Sci Technolog Med
Sci. 31:1–9. 2011.PubMed/NCBI
|
|
31
|
Hofer MD, Kuefer R, Varambally S, Li H, Ma
J, Shapiro GI, Gschwend JE, Hautmann RE, Sanda MG, Giehl K, et al:
The role of metastasis-associated protein 1 in prostate cancer
progression. Cancer Res. 64:825–829. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Toh Y, Ohga T, Endo K, Adachi E, Kusumoto
H, Haraguchi M, Okamura T and Nicolson GL: Expression of the
metastasis-associated MTA1 protein and its relationship to
deacetylation of the histone H4 in esophageal squamous cell
carcinomas. Int J Cancer. 110:362–367. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kumar R, Wang RA and Bagheri-Yarmand R:
Emerging roles of MTA family members in human cancers. Semin Oncol.
30(5 Suppl 16): 30–37. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kai L, Wang J, Ivanovic M, Chung YT,
Laskin WB, Schulze-Hoepfner F, Mirochnik Y, Satcher RL Jr and
Levenson AS: Targeting prostate cancer angiogenesis through
metastasis-associated protein 1 (MTA1). Prostate. 71:268–280. 2011.
View Article : Google Scholar
|
|
35
|
Denlinger CE, Keller MD, Mayo MW, Broad RM
and Jones DR: Combined proteasome and histone deacetylase
inhibition in non-small cell lung cancer. J Thorac Cardiovasc Surg.
127:1078–1086. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Warrell RP Jr, He LZ, Richon V, Calleja E
and Pandolfi PP: Therapeutic targeting of transcription in acute
promyelocytic leukemia by use of an inhibitor of histone
deacetylase. J Natl Cancer Inst. 90:1621–1625. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Phillips T, Collins T and Davies J:
American Association for Cancer Research - 96th Annual Meeting.
Targeting the cell cycle and HDAC inhibitors. IDrugs. 8:450–453.
2005.PubMed/NCBI
|