Introduction
Breast cancer, the most common spontaneous
malignancy diagnosed in women, is a classical model of hormone
dependency. There is evidence that breast cancer risk is associated
with prolonged exposure to female hormones, as the onset of
menarche, late menopause and hormone replacement therapy are
associated with greater cancer incidence (1). The progression of breast cancer
follows a complex multi-step process that depends on various
exogenous (diet and breast irradiation) and endogenous (age,
hormonal imbalances, proliferative lesions and family history of
breast cancer) factors (2–4). Breast cancer is a complex disease in
which numerous genetic aberrations occur. Cellular and molecular
changes that occur during the development of cancer can be mediated
by a range of endogenous and environmental stimuli. On the basis of
the currently accepted view of breast cancer as a multi-step
process, it is possible that specific abnormalities may be an
essential part of the transformation of a normal cell to an
invasive tumor cell.
Different cytoplasmic proteins are key in the
transformation of a normal cell to an invasive tumor cell and among
these, vimentin is particularly important. It is one of the
cytoplasmic intermediate filament proteins, which are the major
components of the cytoskeleton normally found in embryonic or
mesenchymal stem cells (5,6). However, vimentin is frequently
expressed in neoplastic cells with metastatic properties, including
breast cancer cells (7,8). It is a 57-kDa intermediate filament
protein, which forms a part of the cytoskeleton. Expression of
vimentin and cytokeratins has also been described in breast
carcinomas. Hendrix et al (9) demonstrated that the co-expression of
vimentin and keratin intermediate filaments in human breast cancer
cells results in phenotypic inter-conversion and increased invasive
behavior.
Another important gene, Notch, is also
pivotal in this context. This gene is expressed in a variety of
tissues, indicating that it is involved in multiple signaling
pathways (10–14). It is either overexpressed or
rearranged in human tumors, such as is the case with the 280- to
330-kDa Notch protein (14). The
LIN-12/Notch family of transmembrane receptors is believed
to be central to development by regulating cell fate decisions
(10–13). Notch signals are involved in the
development and maintenance of normal tissues that are
recapitulated in different forms of cancer (14,15).
Notch can either promote or limit tumor growth, depending on the
tumor type, through differentiation, cellular metabolism, cell
cycle progression, angiogenesis and possibly self-renewal and
immune function (16,17,19,20).
The Notch signaling pathway is critical in cell fate decisions,
tissue patterning and morphogenesis, and is hence regarded as a
developmental pathway. However, problems with this pathway can
contribute to cellular transformation and tumorigenesis.
The expression of Notch receptors and their
downstream target genes is upregulated in primary human melanomas
(15,16), and the expression of constitutively
active Notch1 promotes melanoma progression (15,17).
These oncogenic effects correlate with the activation of Wnt
signaling in melanoma cells (15),
which promotes the expression of adhesion molecules such as
N-cadherin (17) through the
transcription factor TCF/LEF (15).
Notch has also been implicated in the pathogenesis of other solid
tumors, such as medulloblastoma (18,19)
and ovarian cancer (20), and the
number of known neoplasms involving some alteration in Notch
signaling is increasing. The aim of the present study was to assess
whether vimentin and Notch gene and protein expression are altered
in breast cancer progression. The importance of vimentin expression
was analyzed by identifying cases of breast cancer with poor
prognosis and comparing vimentin and Notch as biomarkers required
for prognosis in breast cancer patients.
Materials and methods
Cell lines
MCF-10F cells were grown in DMEM/F-12 (1:1) medium
supplemented with antibiotics [100 U/mI penicillin, 100 μg/ml
streptomycin and 2.5 μg/ml amphotericin B (all from Life
Technologies, Grand Island, NY, USA)] and 10 μg/m of 5% equine
serum (Biofluids, Rockville, MD, USA), 0.5 μg/ml hydrocortisone
(Sigma-Aldrich, St. Louis, MO, USA) and 0.02 μg/ml epidermal growth
factor (Collaborative Research, Bedford, MA, USA) (21). An in vitro experimental
breast cancer model (Alpha model) (22), developed by exposing the
immortalized human breast epithelial MCF-10F cell line to low doses
of high linear energy transfer α particle radiation (150 keV/μm)
and subsequent growth in the presence or absence of 17β-estradiol,
was used in this study. This model consisted of human breast
epithelial cells in different stages of transformation: i) a
control cell line (MCF-10F), ii) an Estrogen cell line [(MCF-l0F
continually treated with estradiol at 10−8 M
(Sigma-Aldrich)], iii) a malignant but non-tumorigenic cell line
(Alpha3), iv) a malignant and tumorigenic cell line (Alpha5) and v)
a Tumor2 cell line derived from cells originating from a tumor
after injection of the Alpha5 cell line into nude mice. A total of
21 female CB17 SCID mice (Taconic, Germatown, NY, USA) and nude
mice (Harlam Sprague Dawley, Indianapolis, IN, USA) (age, 1 year)
were used in these studies. Each animal was injected subcutaneously
at two different sites with 8×106 cells in 0.2 ml saline
in the fat pad of the right and left side of the abdominal mammary
gland. The study was approved by the ethics committee of Columbia
University Medical Center (New York, NY, USA)
Pathological analysis
Formalin-fixed, paraffin-embedded, noninvasive and
invasive ductal and lobular carcinomas were obtained from the
archives of the Pathology Department of Dr Gustavo Fricke Hospital,
Viña del Mar, Valparaíso, Chile. Patients had undergone surgery
(total mastectomy with axillary lymph node dissection) between 1997
and 2001. The median patient age at surgery was 56 years (range,
25–92 years). The primary pathological diagnosis was confirmed by
hematoxylin and eosin staining. All operative and pathological
reports were reviewed to confirm disease stage. Sections of 2 μm
were cut and mounted onto polylysine-coated slides, and stained for
vimentin and Notch protein expression. The study was approved by
the ethics committee of Dr. Gustavo Fricke Hospital of Viña del Mar
(Valparaiso, Chile).
Immunoperoxidase staining
Protein expression was evaluated as previously
described (22–24). Exponentially growing cell lines were
plated on a glass chamber slide (Nunc Inc., Naperville, IL, USA) at
a density of 1×104 cells/ml of medium and allowed to
grow for 2–3 days until they reached 70% confluence (21). The cells were fixed with buffered
paraformaldehyde at room temperature, incubated with 1%
H2O2 in methanol to block endogenous
peroxidase and washed twice with buffer solution. Cell cultures
were subsequently covered with normal horse serum for 30 min at RT
and incubated with anti-rabbit monoclonal antibody (vimentin: C-20,
sc 7557 and Notch 4: C-19, sc 8644) (Santa Cruz Biotechnology,
Inc., Santa Cruz, CA, USA) at a 1:500 dilution at 4ºC overnight,
and then incubated for 45 min with diluted biotinylated secondary
antibody solution (Vector Laboratories, Burlingame, CA, USA) and
Vectastin Elite ABC Reagent (Vector Laboratories). The experiments
were repeated three times in cells with identical passages in
vitro. The number of immune-reactive cells (50 cells/field) was
counted in several randomly selected microscopy fields (×400) per
sample using an optical microscope (C×31; Olympus Corporation,
Tokyo, Japan). Ten fields were counted for each cell line.
Inmunofluorescent staining
Protein expression was evaluated by
immunofluorescent staining and confocal microscopy as previously
described (22,23). Cells were viewed on Zeis Axiovert
100 TV microscope (Carl Zeiss, Thornwood, NY, USA) using a 40× 11.3
NA objective lens equipped with a laser scanning confocal
attachment (LSM 410, Carl Zeiss). A semi-quantitative estimation of
the area and the intensity of the staining of the cells present in
the culture dishes were performed based on the relative staining of
the protein expressed by the controls and transformed cells.
Fluorescent-labeled probe preparation for
microarray analysis
The poly(A) mRNA from normal, radiation- and
estrogen-treated breast cell lines was isolated using a
QIA-direct-mRNA isolation kit (Qiagen, Inc., Valencia, CA, USA).
Fluorescent-labeled cDNA was prepared from 1 μg of each of these
poly(A) mRNA samples by using oligo dT-primed polymerization and a
Superscript II reverse transcriptase kit (Life Technologies) in the
presence of either Cy3- or Cy5-labeled dCTP, following the usual
procedure (http://cmgm.stanford.edu/pbrown/protocols/). The
appropriate Cy3- and Cy5-labeled probes were pooled and hybridized
to microarray glass coverslips for 16 h at 65ºC and then washed
with high stringency for analysis.
Affymetrix HG-U133A Plus 2.0 GeneChip
microarray gene expression analysis
The breast cancer model (Alpha model) containing i)
MCF-10F, ii) Estrogen, (iii) Alpha3, iv) Alpha5 and v) Tumor2 cell
lines was analyzed for gene expression using Affymetrix U133A
oligonucleotide microarray (Affymetrix, Santa Clara, CA, USA),
which contains 14,500 genes. Arrays were quantitatively analyzed
for gene expression using the Affymetrix GeneChip Operating
Software, with dual global scaling option in a Genes@Work software
platform of discovery algorithm, Structural Pattern Localization
Analysis by Sequential Histograms, and a false discovery rate of
0.05 (25,26).
Results
Phenotypic and molecular analysis of
vimentin expression in breast cancer progression model
The established breast cancer model (22) has been shown to exhibit important
phenotypic characteristics of breast carcinogenesis. The normal
cell line, MCF-10F, did not exhibit any of the features that
characterize malignant cells, such as anchorage-independent growth
in soft agar, invasion and tumor growth in nude mice (22,24).
The Alpha3 cell line formed colonies in soft agar and had invasive
capabilities, but failed to form tumors in the immuno-suppressed
mice. However, the Alpha5 cell line induced mammary gland tumors in
the animals and metastasis in the liver, lung and kidneys after
injection. This cell line gave rise to the Tumor2 cell line after
removal of the mammary tumor, digestion in in vitro
conditions and culture for many passages.
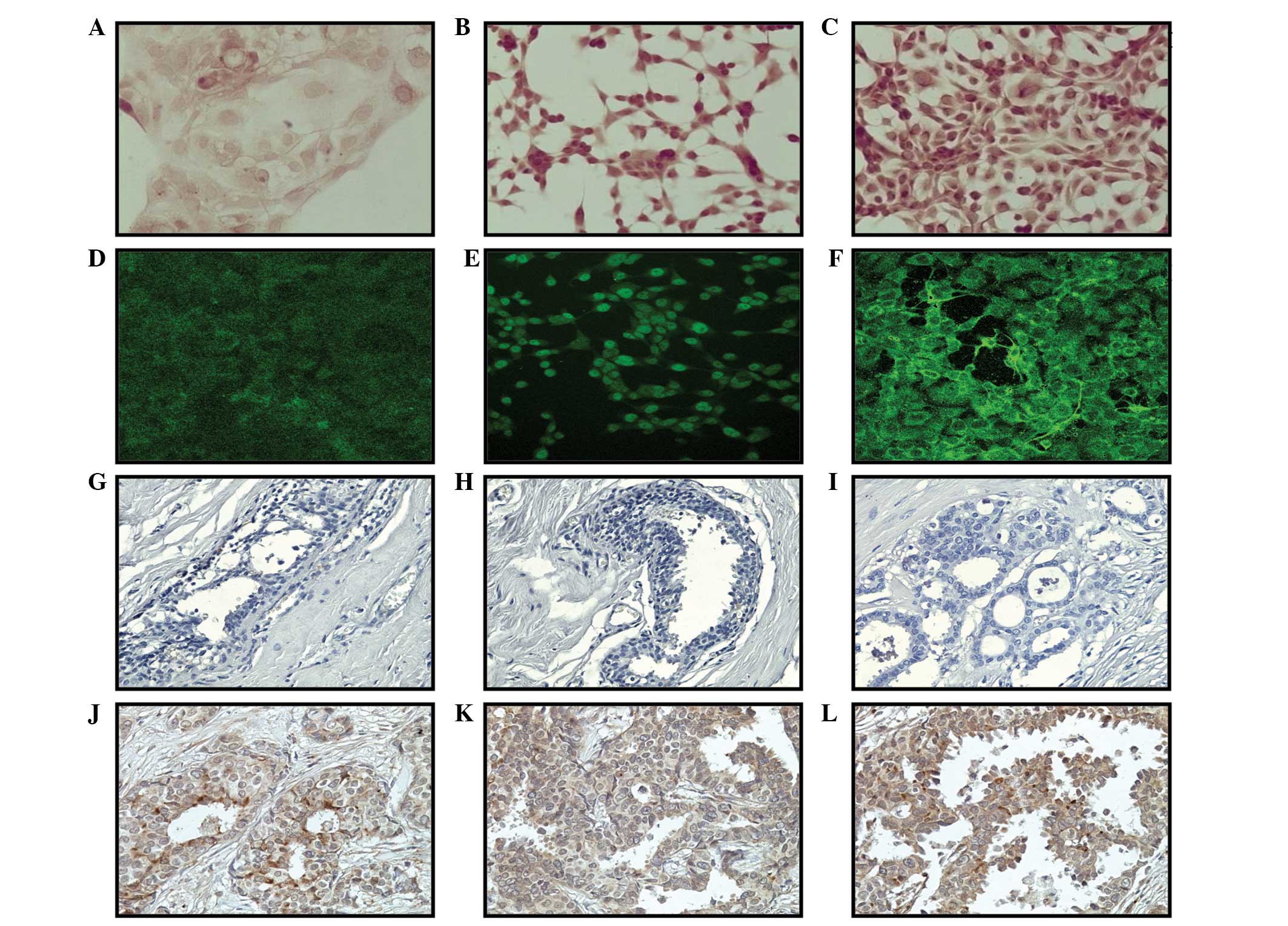
The analysis of immunoperoxidase (Fig. 1A) and immunofluorescence (Fig. 1B) data obtained in relation to the
relative vimentin expression in MCF-10F, Estrogen, Alpha3, Alpha5
and Tumor2 cell lines indicated that such expression was
significantly greater (P<0.05) in the Tumor2, Alpha3 and Alpha5
cell lines, when compared with the MCF-10F and Estrogen cell lines.
Genes that were identified to be differentially expressed between
cell lines of this model were also studied. Histogram plots of the
differential expression of vimentin and Notch genes in these cell
lines were detected by gene chip array. Results of pairwise
comparisons of cell lines examined for vimentin protein expression
were analyzed with the following pairs of cell lines:
MCF-10F/Estrogen, MCF-10F/Alpha3, Estrogen/Alpha5, Alpha3/Alpha5,
Alpha5/Tumor2 and Alpha 3/Tumor2 (Fig.
1C). Results of the pairwise comparisons did not reveal any
alteration in vimentin gene expression between the MCF-10F and
Estrogen cell lines, while there was an almost nine- and five-fold
alteration in the MCF-10F/Alpha3 and Estrogen/Alpha5 combinations,
respectively. There were six- and four- fold changes in gene
expression between the Alpha5 and Tumor2 cell lines, and Alpha3 and
Tumor2 cell lines, respectively.
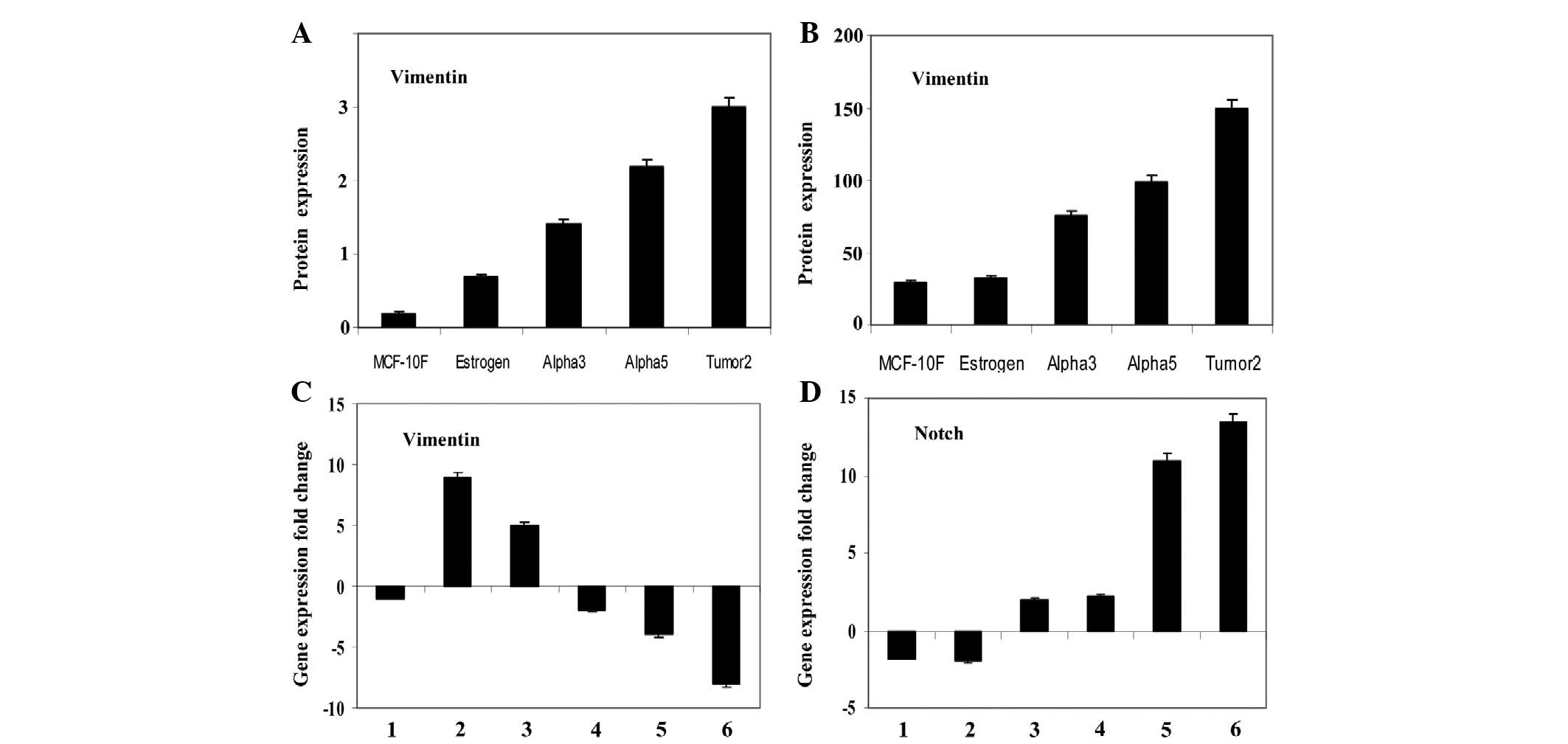 | Figure 1Bars represent the average and
standard error of vimentin protein expression by (A) peroxidase and
(B) immunofluorescent techniques of the MCF-10F, Estrogen, Alpha3,
Alpha5 and Tumor2 cell lines. The primary antibodies used were
mouse monoclonal antibodies (Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA). Fold change of (C) vimentin and (D)
Notch gene expression. Gene expression from scatter plots of
the following pairwise comparative studies of cell lines: MCF-10F/E
(1), MCF-10F/Alpha3 (2), E/Alpha5 (3), Alpha3/Alpha5 (4), Alpha
3/Tumor2 (5) and Alpha5/Tumor2 (6). |
Results of pairwise comparisons of cell lines
examined for Notch gene expression are shown in Fig. 1D. Results of the same pairs of cell
lines were analyzed, revealing no alteration in Notch gene
expression between the MCF-10F and Estrogen cell lines, Estrogen
and Alpha5 cell lines, and Alpha3 and Alpha5 cell lines. By
contrast, there was an almost ten- and fourteen-fold alteration in
the Alpha5/Tumor2 and Alpha3/Tumor2 combinations, respectively,
with higher expression in Alpha3 and Alpha5 than in Tumor2.
Vimentin protein expression in breast
cancer model and breast biopsy specimens
Representative images of vimentin protein
expression, in which greater expression was observed in the Alpha5
and Tumor2 cell lines compared with that in the control MCF-10F
cell line, can be observed in immunoperoxidase (Fig. 2A–C) and immunofluorescence (Fig. 2D–F) studies. Biopsy specimens were
also analyzed for vimentin protein expression to analyze
progression in breast cancer. Fig.
2G–I shows representative tissues of vimentin protein
expression in ducts found in sections of biopsies from breast
cancer patients, as determined by immunoperoxidase staining. This
expression was negative in noninvasive ductal carcinoma and breast
epithelial lesions surrounding the primary tumors, ductal and
lobular hyperplasia, and microcytes. By contrast, this expression
was positive in breast specimens with invasive characteristics, as
shown in Fig. 2J–L. Positive
staining for vimentin was found in 21% of cases.
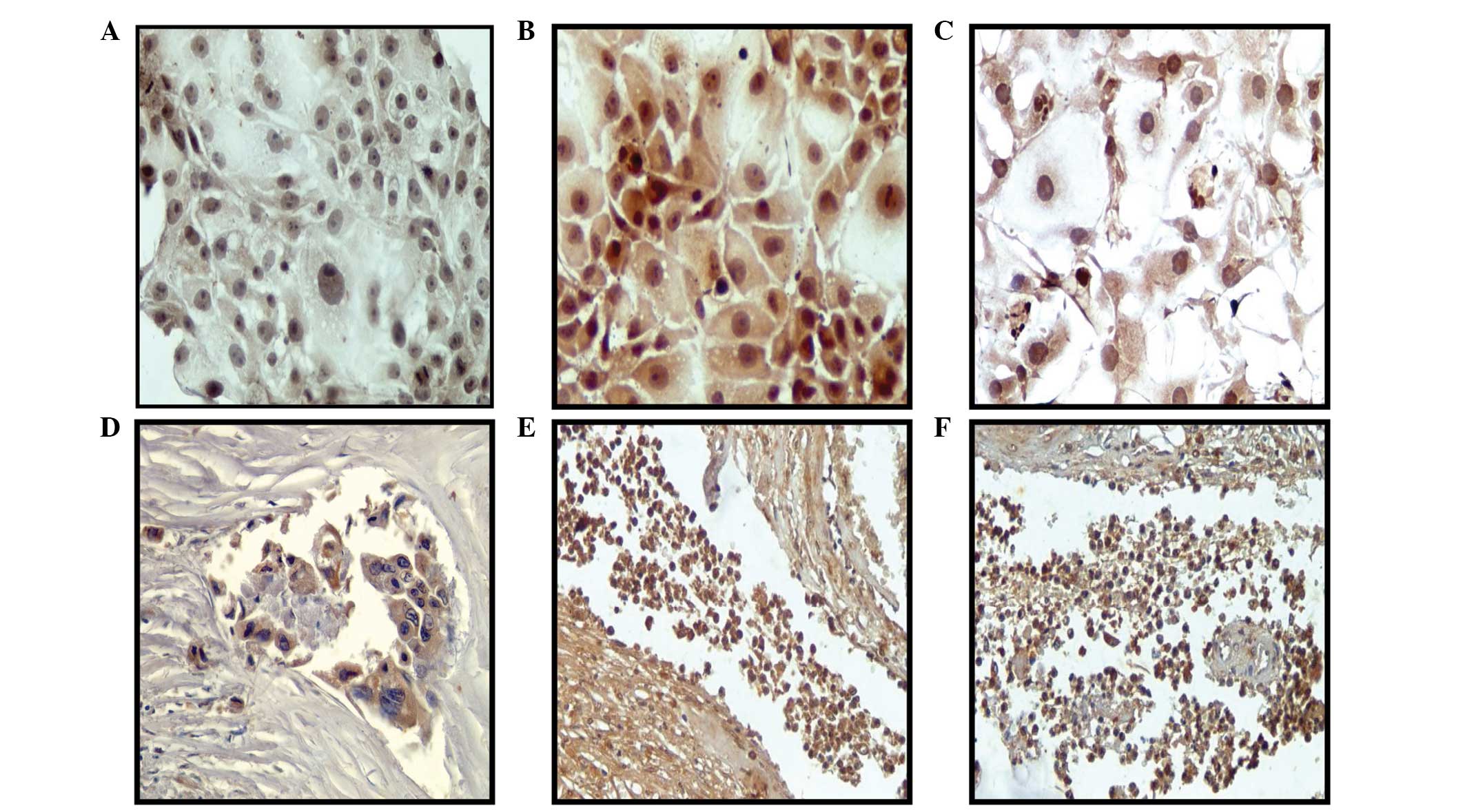
Notch protein expression in breast cancer
model and breast biopsy specimens
In the present study, non-malignant and malignant
cell lines from the model were used to analyze Notch protein
expression. Fig. 3A–C shows higher
Notch protein expression in the Alpha5 and Tumor2 cell lines
compared with that in the control MCF-10F cell line, as determined
by immunoperoxidase staining. Samples from biopsy specimens showed
negative Notch protein expression in noninvasive ductal carcinomas.
However, positive cell expression was observed in those tissues
with cells from invasive ductal carcinomas (Fig. 3D–F), particularly in invasive
isolated tumor cells. Positive staining for Notch was found in 25%
of cases.
Discussion
The main purpose of the present study was to assess
the prognostic value of the markers vimentin and Notch.
Identification of factors involved in cell proliferation and
transformation has been facilitated by studies using various human
epithelial cell lines. The analysis of immunoperoxidase and
immunofluorescence data obtained in relation to the relative
vimentin expression indicated that such expression was
significantly greater in Tumor2 and Alpha5 when compared with
MCF-10F, Estrogen and Alpha3 cell lines.
Results of pairwise comparisons of vimentin
gene expression in the different cell lines indicated that there
was no alteration in vimentin gene expression between the MCF-10F
and Estrogen cell lines, while there was an almost nine- and
five-fold alteration in the MCF-10F/Alpha3 and Estrogen/Alpha5
combinations, respectively. There were six- and four-fold changes
in gene expression between Alpha5 and Tumor2, and Alpha3 and
Tumor2, respectively. Results of the same pairs of cell lines
analyzed for Notch gene expression indicated that there was
no alteration between the MCF-10F and Estrogen, Estrogen and
Alpha5, and Alpha3 and Alpha5 cell lines. By contrast, there was an
almost ten- and fourteen- fold alteration in the Alpha5/Tumor2 and
Alpha3/Tumor2 combinations, respectively, with higher expression in
Alpha3 and Alpha5 than in Tumor2 cells. Vimentin protein expression
in ducts in sections of biopsies from breast cancer patients was
found to be negative for noninvasive ductal carcinoma, but positive
for ductal carcinoma with invasive characteristics.
Vimentin-reactive cells in benign and malignant breast tissue have
been described in many studies (26–29).
These studies reported that vimentin expression appeared to be
associated with poor prognosis in node-negative ductal breast
carcinomas, and that vimentin was preferentially expressed in human
breast carcinomas with low levels of estrogen receptors. Gene
expression patterns of breast carcinomas distinguished tumor
subclasses with clinical implications (30). A possible association was found
between the clinically aggressive behavior of tumors (28,29)
and estrogen receptor negativity (31,32),
high Ki-67 levels (32) and poor
differentiation of tumors with high-grade and positive vimentin
protein expression. Domagala et al (29) reported that vimentin was
preferentially expressed in high-grade ductal and medullary, but
not in lobular, breast carcinomas. Other data showed that more
invasive breast cancer lines expressed vimentin, indicating its
usefulness in identifying cases with poorer prognosis (28,29).
Vimentin is known to be selectively expressed in
aggressive breast cancer cell lines (9). Elevated vimentin expression
levels correlate well with upregulated migration and invasion of
cancer cells (9,26). Sommers et al (27) showed that transfection of
noninvasive human breast cancer cell lines, such as MCF7, with the
vimentin gene led to accelerated invasiveness. The authors also
reported vimentin rather than keratin expression in certain
hormone-independent breast cancer cell lines, and in
oncogene-transformed mammary epithelial cells. The possible
association of vimentin with the clinically aggressive behavior of
tumors described by others (7,28–32)
may be explained by the correlation of vimentin expression with a
lack of steroid receptors and poor differentiation of cancer.
Gilles et al (31) also
found vimentin expression in cervical carcinomas was associated
with invasive and migratory potential.
Thus, we can suggest an improved indicator of breast
cancer progression by adding vimentin to the diagnostic panel when
overall survival is a primary end-point. In the present study,
positive staining for vimentin was found in 21% of cases, which is
in line with previous findings (32). Therefore, vimentin expression
appears to predict survival in ductal breast carcinoma.
Notch protein expression was also higher in the
Alpha5 and Tumor2 cell lines in comparison with that in the control
MCF-10F cell line. When samples from biopsy specimens were analyzed
for Notch protein expression, negative cells were found in
noninvasive ductal carcinomas while positive cells were found in
invasive ductal carcinomas. It has been reported that the Notch
pathway is required for the establishment of embryonic
hematopoietic stem cells (33), and
it has been implicated in the maintenance of several types of
normal cell populations (34–36).
The effects of Notch on cells include increased survival or death,
proliferation or growth arrest and commitment to, or blockage of,
differentiation. These different outcomes are mediated through a
novel signaling pathway in which Notch receptors on the cell
surface give rise to a nuclear transcriptional activation complex.
Studies on Notch are related to the understanding of how this
pathway yields several outcomes. It has been proposed that Notch
may serve as an oncogene or tumor suppressor, a repressor or
inducer of terminal differentiation, or a cancer stem cell factor.
Studies on the multifaceted role of Notch in cancer indicate a
possible therapeutic implication. Notch signaling is frequently
deregulated in breast cancer, and hyperactivation of Notch
contributes to the tumor process. Notch has been shown to be
involved in the controlled proliferation and migration of vascular
endothelial cells, as well as in the integration of Notch and Wnt
signaling, as observed in hematopoietic stem cell maintenance
(34). Guentchev and McKay
(35) observed that Notch
controlled the proliferation and differentiation of stem cells in a
dose-dependent manner. It has also been suggested that Notch acts
as a transducer molecule for developmental processes. Stylianou
et al (36) observed
aberrant activation of Notch signaling in human breast cancer.
Estrogens are known to regulate the proliferation of
breast cancer cells and to alter their phenotypic properties; the
gene networks and pathways through which estrogenic hormones
regulate these events have also been considered (37). We used global gene expression
profiling by Affymetrix GeneChip microarray analysis to identify
genes altered by the presence of estradiol in an MCF-10F human
breast cancer model. Of the >14,000 genes analyzed, over 300
showed a pattern of either up- or downregulation. We observed a
general upregulation of positive proliferation regulators,
including multiple growth factors, genes involved in cell cycle
progression and regulatory factor-receptor loops, and a
downregulation of transcriptional repressors and anti-proliferative
and pro-apoptotic genes, including BCL2 and TGF-β
family growth inhibitory factors. The present study highlights the
diverse gene networks and metabolic and cell regulatory pathways
through which this hormone operates to achieve its widespread
effects on breast cancer cells.
It can be concluded that vimentin and
Notch gene and protein expression are altered in breast
cancer progression, thereby helping to identify cases of breast
cancer with poor prognosis and complementing those biomarkers
required for assessing the prognosis of breast cancer patients.
Acknowledgements
The support given by grants FONDECYT (GMC; no.
1120006) and MINEDUC-Universidad de Tarapacá (GMC) is greatly
appreciated. The authors thank Guiliana Rojas Ordóñez and Georgina
Vargas Marchant for their technical assistance, and Dr Manikandan
Jayapal and Dr Prakash Hande of the National University of
Singapore for the analysis of Affymetrix microarray data.
References
|
1
|
Henderson BE, Ross R and Bernstein L:
Estrogens as a cause of human cancer: the Richard and Hinda
Rosenthal Foundation award lecture. Cancer Res. 48:246–253.
1988.
|
|
2
|
Feigelson HS, Ross RK, Yu MC, Coetzee GA,
Reichardt JK and Henderson BE: Genetic susceptibility to cancer
from exogenous and endogenous exposures. J Cell Biochem Suppl.
25:15–22. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Krieger N: Exposure, susceptibility, and
breast cancer risk: a hypothesis regarding exogenous carcinogens,
breast tissue development, and social gradients, including
black\white differences, in breast cancer incidence. Breast Cancer
Res Treat. 13:205–223. 1989.PubMed/NCBI
|
|
4
|
Dickson RB and Lippman ME: Estrogenic
regulation of growth and polypeptide growth factor secretion in
human breast carcinoma. Endocr Rev. 8:29–43. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Duprey P and Paulin D: What can be learned
from intermediate filament gene regulation in the mouse embryo.
Intl J Dev Biol. 39:443–457. 1995.
|
|
6
|
Stewart M: Intermediate filament structure
and assembly. Curr Opin Cell Biol. 5:3–11. 1993. View Article : Google Scholar
|
|
7
|
Gilles C, Polette M, Zahm JM, Tournier JM,
Volders L, Foidart JM and Birembaut P: Vimentin contributes to
human mammary epithelial cell migration. J Cell Sci. 112:4615–4625.
1999.PubMed/NCBI
|
|
8
|
Whipple RA, Balzer EM, Cho EH, Matrone MA,
Yoon JR and Martin SS: Vimentin filaments support extension of
tubulin-based microtentacles in detached breast tumor cells. Cancer
Res. 68:5678–5688. 2008. View Article : Google Scholar
|
|
9
|
Hendrix MJ, Seftor EA, Seftor RE and
Trevor KT: Experimental co-expression of vimentin and keratin
intermediate filaments in human breast cancer cells results in
phenotypic interconversion and increased invasive behavior. Am J
Pathol. 150:483–495. 1997.
|
|
10
|
Swiatek PJ, Lindsell CE, del Amo FF,
Weinmaster G and Gridley T: Notch1 is essential for
postimplantation development in mice. Genes Dev. 8:707–719. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Weinmaster G, Roberts VJ and Lemke G:
Notch2: a second mammalian Notch gene. Development. 116:931–941.
1992.PubMed/NCBI
|
|
12
|
Kopan R and Weintraub H: Mouse notch:
expression in hair follicles correlates with cell fate
determination. J Cell Biol. 121:631–641. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Uyttendaele H, Marazzi G, Wu G, Yan Q,
Sassoon D and Kitajewski J: Notch4/int-3, a mammary proto-oncogene,
is an endothelial cell-specific mammalian Notch gene. Development.
122:2251–2259. 1996.PubMed/NCBI
|
|
14
|
Girard L, Hanna Z, Beaulieu N, Hoemann CD,
Simard C, Kozak CA and Jolicoeur P: Frequent provirus insertional
mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice
suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes
Dev. 10:1930–1944. 1996. View Article : Google Scholar
|
|
15
|
Balint K, Xiao M, Pinnix CC, Soma A, Veres
I, Juhasz I, Brown EJ, Capobianco AJ, Herlyn M and Liu ZJ:
Activation of Notch1 signaling is required for b-catenin-mediated
human primary melanoma progression. J Clin Invest. 115:3166–3176.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hoek K, Rimm DL, Williams KR, Zhao H,
Ariyan S, Lin A, Kluger HM, Berger AJ, Cheng E, Trombetta ES, et
al: Expression profiling reveals novel pathways in the
transformation of melanocytes to melanomas. Cancer Res.
64:5270–5282. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu ZJ, Xiao M, Balint K, Smalley KS,
Brafford P, Qiu R, Pinnix CC, Li X and Herlyn M: Notch1 signaling
promotes primary melanoma progression by activating
mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt
pathways and up-regulating N-cadherin expression. Cancer Res.
66:4182–4190. 2006. View Article : Google Scholar
|
|
18
|
Marino S: Medulloblastoma: developmental
mechanisms out of control. Trends Mol Med. 11:17–22. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hallahan AR, Pritchard JI, Hansen S, et
al: The SmoA1 mouse model reveals that Notch signaling is critical
for the growth and survival of sonic hedgehog-induced
medulloblastomas. Cancer Res. 64:7794–7800. 2004. View Article : Google Scholar
|
|
20
|
Park JT, Li M, Nakayama K, Mao TL,
Davidson B, Zhang Z, Kurman RJ, Eberhart CG, Shih IeM and Wang TL:
Notch3 gene amplification in ovarian cancer. Cancer Res.
66:6312–6318. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Soule HD, Maloney TM, Wolman SR, Peterson
WD Jr, Brenz R, McGrath CM, Russo J, Pauley RJ, Jones RF and Brooks
SC: Isolation and characterization of a spontaneously immortalized
human breast epithelial cell line, MCF-10. Cancer Res.
50:6075–6086. 1990.PubMed/NCBI
|
|
22
|
Calaf GM and Hei TK: Establishment of a
radiation- and estrogen-induced breast cancer model.
Carcinogenesis. 21:769–776. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Calaf G and Hei TK: Oncoprotein
expressions in human breast epithelial cells transformed by
high-LET radiation. Int J Radiat Biol. 77:31–40. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Calaf G and Russo J: Transformation of
human breast epithelial cells by chemical carcinogens.
Carcinogenesis. 14:483–492. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Califano A: SPLASH: structural pattern
localization analysis by sequential histograms. Bioinformatics.
16:341–357. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zajchowski DA, Bartholdi MF, Gong Y,
Webster L, Liu HL, Munishkin A, Beauheim C, Harvey S, Ethier SP and
Johnson PH: Identification of gene expression profiles that predict
the aggressive behavior of breast cancer. Cancer Res. 61:5168–5178.
2001.
|
|
27
|
Sommers CL, Walker-Jones D, Heckford SE,
Worland P, Valverius E, Clark R, McCormick F, Stampfer M, Abularach
S and Gelmann EP: Vimentin rather than keratin expression in some
hormone-independent breast cancer cell lines and in
oncogene-transformed mammary epithelial cells. Cancer Res.
49:4258–4263. 1989.
|
|
28
|
Domagala W, Lasota J, Bartkowiak J, Weber
K and Osborn M: Vimentin is preferentially expressed in human
breast carcinomas with low estrogen receptor and high Ki-67 growth
fraction. Am J Pathol. 136:219–227. 1990.
|
|
29
|
Domagala W, Wozniak L, Lasota J, Weber K
and Osborn M: Vimentin is preferentially expressed in high-grade
ductal and medullary, but not in lobular breast carcinomas. Am J
Pathol. 137:1059–1064. 1990.PubMed/NCBI
|
|
30
|
Sørlie T, Perou CM, Tibshirani R, et al:
Gene expression patterns of breast carcinomas distinguish tumor
subclasses with clinical implications. Proc Natl Acad Sci USA.
98:10869–10874. 2001.PubMed/NCBI
|
|
31
|
Gilles C, Polette M, Piette J, Delvigne
AC, Thompson EW, Foidart JM and Birembaut P: Vimentin expression in
cervical carcinomas: association with invasive and migratory
potential. J Pathol. 180:175–180. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Heatley MK, Ewings P, Odling Smee W,
Maxwell P and Toner PG: Vimentin expression does not assist in
predicting survival in ductal carcinoma of the breast. Pathology.
34:230–232. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kumano K, Chiba S, Kunisato A, et al:
Notch1 but not Notch2 is essential for generating hematopoietic
stem cells from endothelial cells. Immunity. 18:699–711. 2003.
View Article : Google Scholar
|
|
34
|
Duncan AW, Rattis FM, DiMascio LN, et al:
Integration of Notch and Wnt signaling in hematopoietic stem cell
maintenance. Nat Immunol. 6:314–322. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Guentchev M and McKay RD: Notch controls
proliferation and differentiation of stem cells in a dose-dependent
manner. Eur J Neurosci. 23:2289–2296. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Stylianou S, Clarke RB and Brennan K:
Aberrant activation of notch signaling in human breast cancer.
Cancer Res. 66:1517–1525. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Frasor J, Danes JM, Komm B, Chang KC,
Lyttle CR and Katzenellenbogen BS: Profiling of estrogen up- and
down-regulated gene expression in human breast cancer cells:
insights into gene networks and pathways underlying estrogenic
control of proliferation and cell phenotype. Endocrinology.
144:4562–4574. 2003. View Article : Google Scholar : PubMed/NCBI
|