Introduction
Chromosomal abnormalities are extremely common in
malignancy, including leukemia. Acute myelogeneous leukemia (AML)
has a wide variety of chromosomal rearrangements that involve
specific regions (1–3). The 3q26 region encodes two proteins
involved in AML, ectopic viral integration site 1 (EVI1) and
myelodysplastic syndrome 1 (MDS1). Expression of the EVI1 gene has
been found to correlate with accelerated cell growth of murine
embryonic stem cells. By contrast, the combined effect of these two
genes has been observed to play a transcriptional transactivator
role, resulting in reduced cell growth (4).
The ETV6 gene, located at 12p13, encodes a
transcription factor containing the 5′ helix-loop-helix
dimerization motif and the 3′ ETS DNA-binding domain (5). To date, >40 partner cytobands have
been identified to be associated with translocations involving ETV6
and ≥28 partner genes encoding protein tyrosine kinases,
transcription factors or other proteins (5). The t(3;12)(q26.2;p13) translocation is
a recurrent translocation involving ETV6. This translocation is
relatively rare but specifically observed in myeloid malignancies,
including myelodysplastic syndrome (MDS), AML and chronic
myelogenous leukemia (CML) (6). It
may also be associated with dysplasia of megakaryocytes,
multilineage involvement and disease progression (7). Finally, the 3q26 locus is rearranged
in inversion 3(q21;q26) syndrome, which represents an AML subtype
showing dysmegakaryopoiesis, thrombocytosis and micromegakaryocytes
(8).
At present, mutual translocation of MDS1/EVI1 and
ETV6 has been observed in only two AML-M4 cases; secondary to
myelodysplastic syndrome (MDS) and in CML in blast crisis (9,10). In
the current case report, we describe the molecular and cytogenetic
characterization of a de novo AML-M4 case with
t(3;12)(q26;p13) and trisomy 8. Written informed consent was
obtained from the patient.
Case report
Patient characteristics
A 63-year old female was diagnosed with AML-M4 in
October 2011 due to loss of weight and fever. Hematological
parameters were as follows: White blood cell count,
5.43×109 cells/l; composed of 32.4% neutrophils, 24.4%
lymphocytes, 38.5% monocytes, 0.61% eosinophils and 4.1% basophils.
The platelet count was 1.32×109 cells/l and hemoglobin
levels were 9.61 g/dl. Serum LDH levels were 1,121 U/l (normal,
≤480 U/l). No treatment had been administered prior to the test. In
December 2011, the patient succumbed to unknown causes whilst under
treatment with 100 mg Cytosar.
Methods
Chromosome analysis
Chromosome analysis using GTG-banding was performed
according to standard procedures prior to chemotherapeutic
treatment (11). A total of 20
metaphase cells derived from unstimulated bone marrow culture were
analyzed. Karyotypes were described according to the International
System for Human Cytogenetic Nomenclature (12).
Molecular cytogenetics
Fluorescence in situ hybridization (FISH),
using the LSI BCR/ABL dual color dual fusion translocation and
chromosome enumeration probes (CEPs) for chromosomes 3 and 12 (both
Abbott Laboratories, Des Plaines, IL, USA), were applied according
to the manufacturer’s instructions, together with the TEL/AML1
translocation dual fusion probe (Qbiogene, MP Biomedicales, Santa
Ana, CA, USA) (11). A total of 20
metaphase spreads were analyzed, each using a fluorescence
microscope (Axio Imager Z1; Zeiss, Oberkochen, Germany) equipped
with appropriate filter sets to discriminate between a maximum of
five fluorochromes and the counterstain DAPI. Image capturing and
processing were performed using an ISIS imaging system
(MetaSystems, Altlußheim, Germany).
Reverse transcription-polymerase chain
reaction (RT-PCR)
RT-PCR was performed to investigate the expression
of human ETV6/MDS1/EVI1 fusion transcripts. Total RNA was extracted
from the diagnostic peripheral blood sample using the InviTrap RNA
kit (Invitek, Berlin, Germany) according to the manufacturer’s
instructions. cDNA was prepared from 5 μg total RNA with the
Genequality BCR-ABL kit (AB Analitica, Padova, Italy) according to
the manufacturer’s instructions. The primers used for
ETV6/MDS1/EVI1 were as previously reported (13).
Flow cytometric immunophenotype
Immunophenotyping of leukemic blasts was performed
as described previously (14).
Briefly, flow cytometric analysis was performed using a general
panel of fluorescent antibodies against the following antigens
typical for different cell lineages and cell types: CD1a, CD2, CD3,
CD4, CD5, CD8, CD10, CD11b, CD11c, CD13, CD14, CD15, CD16, CD19,
CD20, CD22, CD23, CD32, CD33, CD34, CD38, CD41a, CD45, CD56, CD57,
CD64, CD103, CD117, CD123, CD138, CD209, CD235a and CD243, as well
as antibodies to κ and λ light Chains, IgD, sIgM and HLA-DR. All
antibodies were purchased from BD Biosciences (San Jose, CA, USA).
Samples were analyzed on a BD FACSCalibur™ flow cytometer.
Autofluorescence, viability and isotype controls were included.
Flow cytometric data acquisition and analysis were conducted by BD
Cellquest™ Pro software.
Results
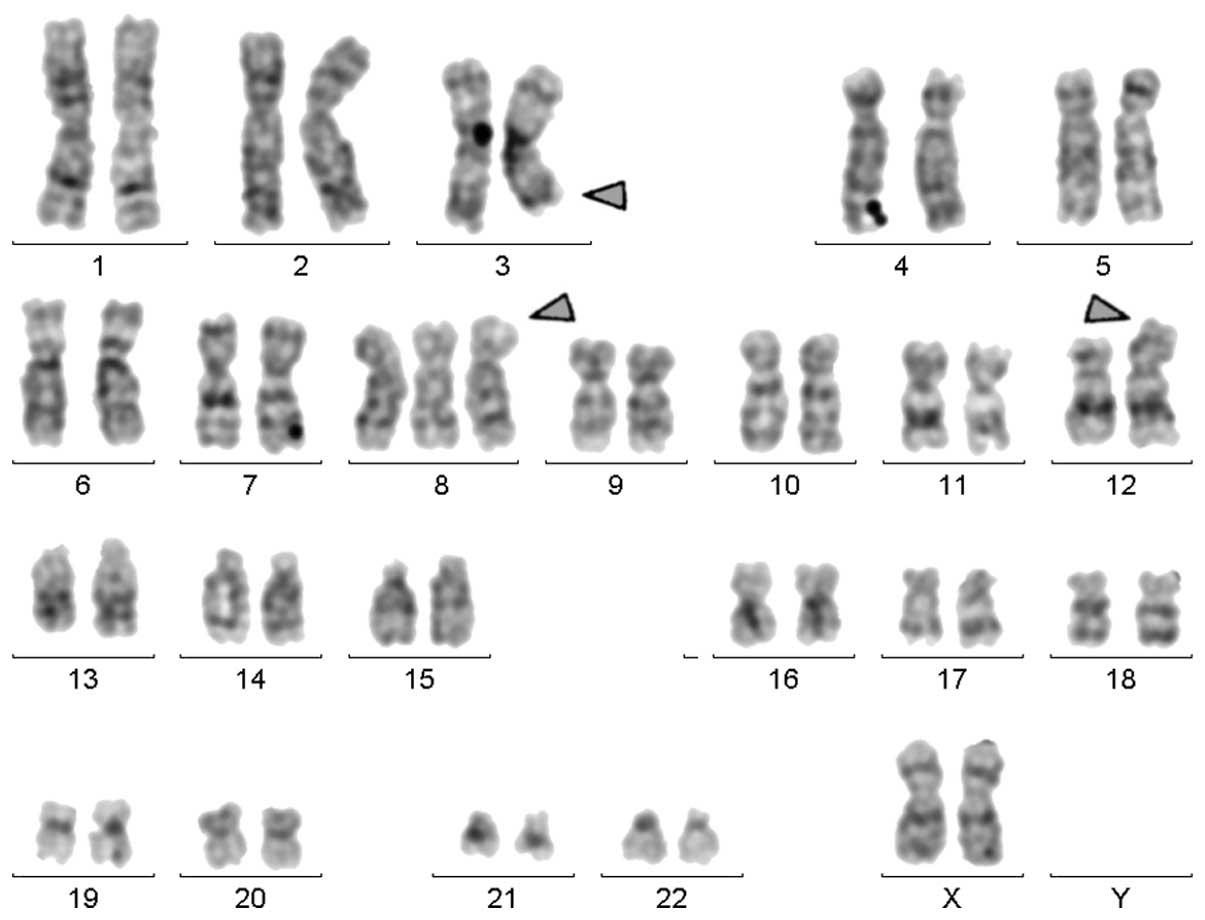
Karyotyping was performed prior to chemotherapy and
the result was 47,XX,+8,t(3;12)[18]/47,XX,+8[2] (Fig. 1). This observation was further
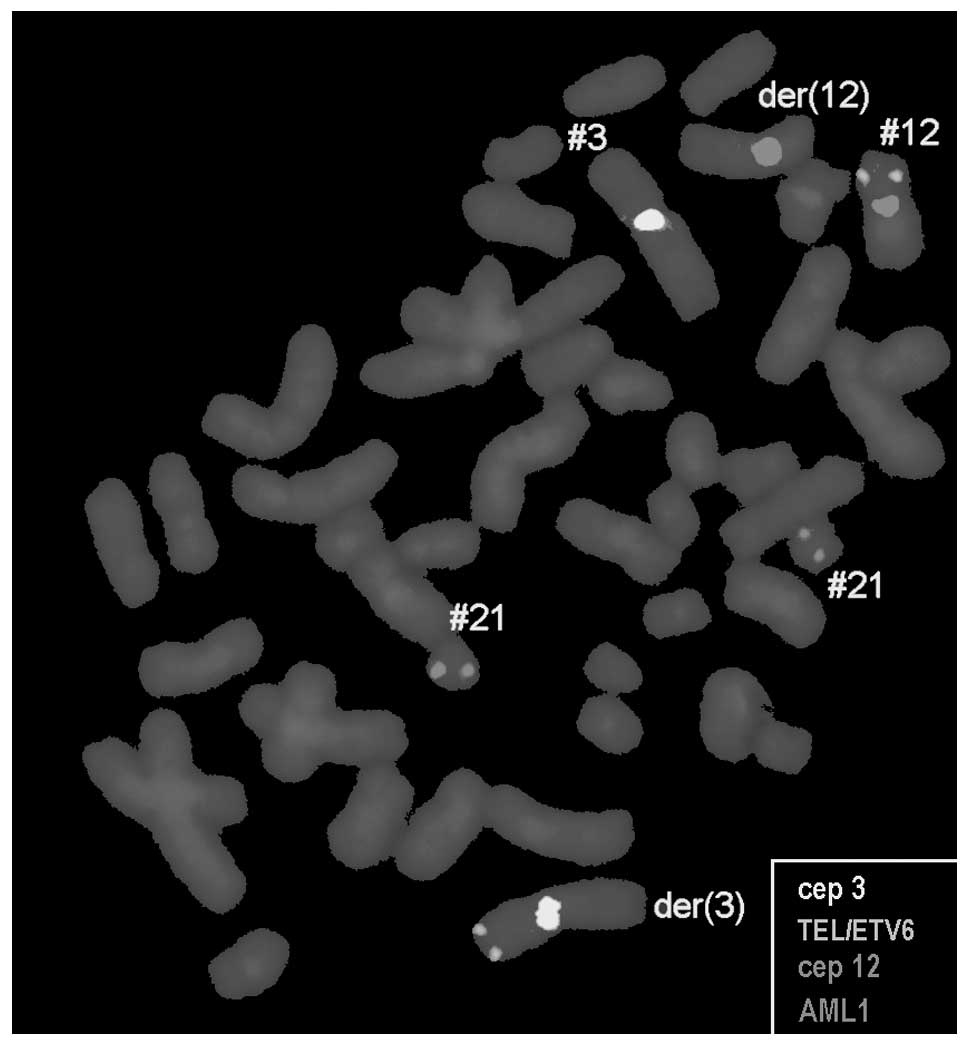
confirmed by molecular cytogenetic studies (Fig. 2). Dual-color-FISH using a CEP probe
specific for chromosomes 3 and 12 was mixed with the TEL/AML1
translocation probe (Fig. 2). Thus,
the following final karyotype was obtained:
47,XX,+8,t(3;12)(q26;p13)[18]/47,XX,+8[2].
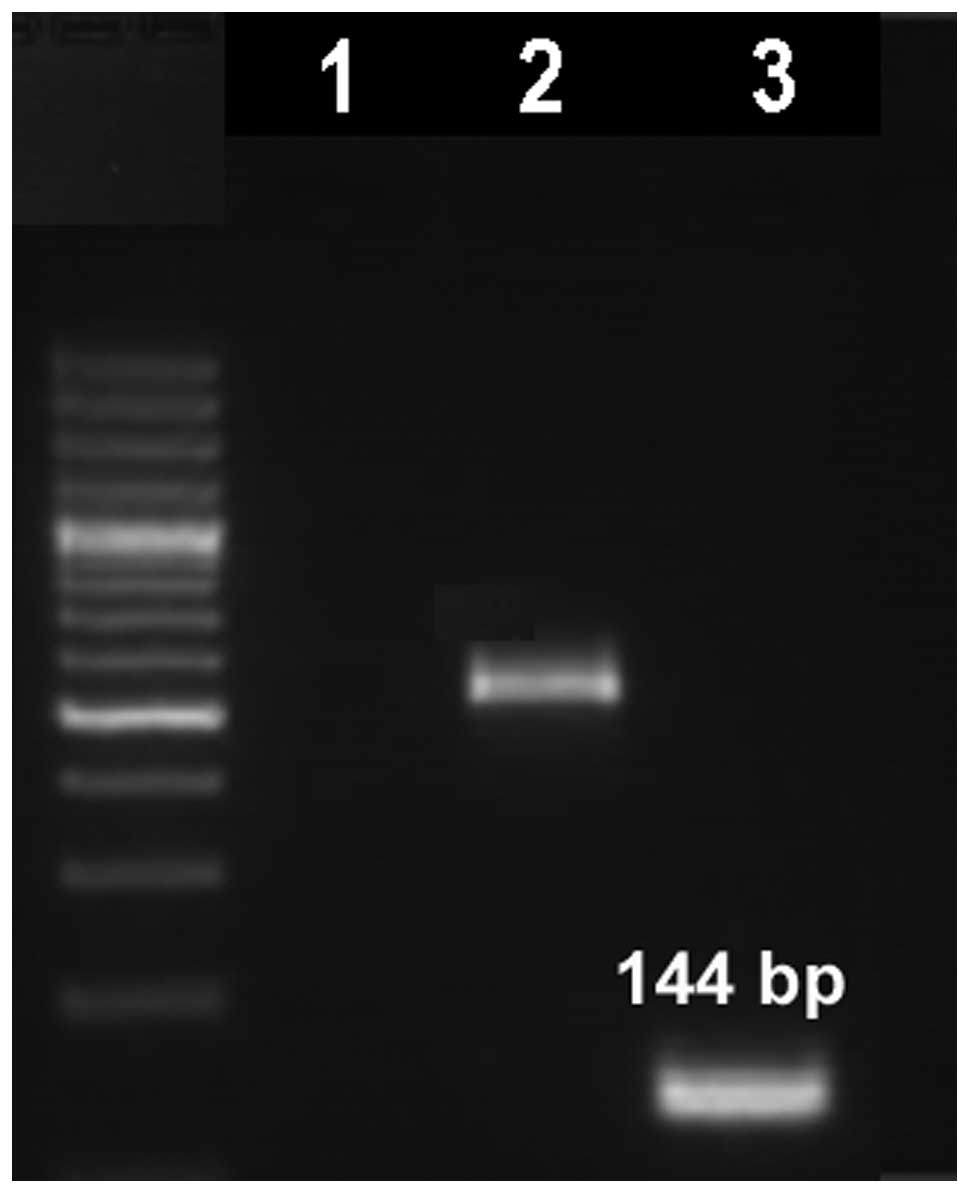
The t(3;12) translocation was further studied by
RT-PCR and the analysis revealed a typical fusion transcript of 144
bp in length, which confirmed the presence of an ETV6/MDS1/EVI1
fusion transcript (Fig. 3).
Immunophenotyping analysis of the peripheral blood
showed that the abnormal cell population was CD45 (96%), CD33
(78%), CD13 (78%), CD18 (91%) and CD11c (55%), and that CD34 (32%),
HLA-DR (58%), CD117 (32%), CD32 (26%), CD16 (54%), CD15 (20%) and
CD235a (29%) were expressed heterogeneously. This immunophenotype
is consistent with AML-M4 according to FAB classification.
Discussion
The t(3;12)(q26;p13) translocation is a rare
cytogenetic abnormality and has been previously reported in 45
cases of myeloid malignancies, including MDS, AML and CML (6). Two cases were described as AML-M4; one
of them was initially a CML case with an additional trisomy 8
(6). To the best of our knowledge,
the present case report is the first to observe a case of a AML-M4
with initial trisomy 8 and secondary developed
t(3;12)(q26;p13).
The common chromosomal abnormalities in AML-M4
include monosomy 5 or del(5q), monosomy 7 or del(7q), trisomy 8,
t(6;9)(p23;q34) and rearrangements involving the MLL gene mapped to
11q23 [del(11)(q23);
t(9;11)(p22;q23), t(11;19)(q23;p13)] and core binding factor B
(CBFβ) mapped to 16q22 [del(16)(q22), inv(16)(p13q22), t(16;16)(p13;q22)] (15). However, in the present case, trisomy
8 was observed. Trisomy 8 is the most frequent numerical aberration
in AML, occurring at a frequency of 10–15%. A previous study
reported that AML patients with trisomy 8 have poor outcomes and
are not responsive to cytarabine-based therapy (15). In addition, a study reported that
trisomy 8 confers an independent prognostic risk in AML (16).
The prognosis of AML with the t(3;12)(q26;p13)
translocation has been reported to be poor, with a survival of only
a few months. This is associated with treatment refractoriness and
may reflect the fact that this AML is secondary to MDS with the
presence of chromosome 7 abnormalities (4). This has a poor prognosis in general
and is observed in the majority of t(3;12)(q26;p13) cases with
chromosome 7 rearrangements. In three cases, resistance to therapy
included treatment with allogeneic bone marrow transplantation
(6,7,17).
Notably, all three patients had an early relapse within a few
months from transplantation (4).
EVI1 is a transcription factor with two zinc finger
motifs and its acquisition is known to be a poor prognostic factor
for AML (18). It is located on
3q26 and its aberrant expression is mainly mediated by the
t(3;3)(q21;q26), inv(3)(q21q26), t(3;12)(q26;p13) and
t(3;21)(q26;q22) genomic aberrations (19). These abnormalities lead to 3q21q26
syndrome, which is associated with thrombocytosis, megakaryocytic
dysplasia, resistance to chemotherapy and poor prognosis (4–20).
The MDS1/EVI1 fusion generates a protein domain with
homology to the positive regulatory domain of PRDI-BF1/Blimp-l, a
transcriptional repressor of the interferon β gene and an inducer
of genes that play a role in B-lymphocyte maturation (21).
In conclusion, the current case report presents a
novel cytogenetic case of AML-M4 with initial trisomy 8 and a
secondary t(3;12)(q26;p13), the latter having more proliferative
capacity than cells with pure trisomy 8. The patient succumbed to
unknown causes whilst under treatment. Therefore, we conclude that
trisomy 8 with t(3;12)(q26;p13) has a negative prognosis.
Acknowledgements
The authors thank Professor I. Othman, the Director
General of the Atomic Energy Commission of Syria (AECS) and Dr N.
Mirali, Head of the Department of Molecular Biology and
Biotechnology, for their support. This study was supported by the
AECS and, in part, by the Stefan-Morsch-Stiftung Foundation.
References
|
1
|
Rubnitz JE: Childhood acute myeloid
leukemia. Curr Treat Options Oncol. 9:95–105. 2008. View Article : Google Scholar
|
|
2
|
Byrd JC, Mrózek K, Dodge RK, Carroll AJ,
Edwards CG, Arthur DC, Pettenati MJ, Patil SR, Rao KW, Watson MS,
et al; Cancer and Leukemia Group B (CALGB8461). Pretreatment
cytogenetic abnormalities are predictive of induction success,
cumulative incidence of relapse, and overall survival in adult
patients with de novo acute myeloid leukemia: results from Cancer
and Leukemia Group B (CALGB 8461). Blood. 100:4325–4336. 2002.
View Article : Google Scholar
|
|
3
|
Grimwade D, Walker H, Oliver F, Wheatley
K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A and
Goldstone A: The importance of diagnostic cytogenetics on outcome
in AML: analysis of 1,612 patients entered into the MRC AML 10
trial. The medical research council adult and children’s leukaemia
working parties. Blood. 92:2322–2333. 1998.PubMed/NCBI
|
|
4
|
Voutsadakis IA and Maillard N: Acute
myelogenous leukemia with the t(3;12)(q26;p13) translocation: case
report and review of the literature. Am J Hematol. 72:135–137.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sitailo S, Sood R, Barton K and Nucifora
G: Forced expression of the leukemia-associated gene EVI1 in ES
cells: a model for myeloid leukemia with 3q26 rearrangements.
Leukemia. 13:1639–1645. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mitelman F, Johansson B and Mertens F:
Mitelman database of chromosome aberrations and gene fusions in
cancer. 2013, http://cgap.nci.nih.gov/Chromosomes/Mitelman.
Accessed February 19, 2013
|
|
7
|
Raynaud SD, Baens M, Grosgeorge J, Rodgers
K, Reid CD, Dainton M, Dyer M, Fuzibet JG, Gratecos N, Taillan B,
et al: Fluorescence in situ hybridization analysis of
t(3;12)(q26;p13): a recurring chromosomal abnormality involving the
TEL gene (ETV6) in myelodysplastic syndromes. Blood. 88:682–689.
1996.
|
|
8
|
Fonatsch C, Gudat H, Lengfelder E, Wandt
H, Silling-Engelhardt G, Ludwig WD, Thiel E, Freund M, Bodenstein H
and Schwieder G: Correlation of cytogenetic findings with clinical
features in 18 patients with inv(3)(q21q26) or t(3;3)(q21;q26).
Leukemia. 8:1318–1326. 1994.
|
|
9
|
Mozziconacci MJ, Brunel V, Sainty D,
Amoulet C, Gabert J, Simonetti J and Lafage-Pochitaloff M:
Translocation (3,12)(q26.2;p13.1) in myeloid malignancies: a new
entity. Nouv Rev Fr Hematol. 37:481995.
|
|
10
|
Nishimura Y, Wada H, Mori A, Takatsuka H,
Tamura A, Fujimori Y, Okamoto T, Takemoto Y and Kakishita E:
Detection of ETV6/MDS1/Evi-1 chimeric transcripts in a patient with
acute myelocytic leukemia and t(3;12)(q26;p13). Int J Hematol.
72:108–109. 2000.PubMed/NCBI
|
|
11
|
Al-Achkar W, Wafa A and Nweder MS: A
complex translocation t(5;9;22) in Philadelphia cells involving the
short arm of chromosome 5 in a case of chronic myelogenous
leukemia. J Exp Clin Cancer Res. 26:411–415. 2007.
|
|
12
|
Shaffer L, Slovak M and Cambell L: ISCN
(2009): An International System for Human Cytogenetic Nomenclature.
Karger Publishers; Basel: pp. 15–33. 2009
|
|
13
|
Peeters P, Wlodarska I, Baens M, Criel A,
Selleslag D, Hagemeijer A, Van den Berghe H and Marynen P: Fusion
of ETV6 to MDS1/EVI1 as a result of t(3;12)(q26;p13) in
myeloproliferative disorders. Cancer Res. 57:564–569.
1997.PubMed/NCBI
|
|
14
|
Chen Z and Sandberg AA: Molecular
cytogenetic aspects of hematological malignacies: clinical
implications. Am J Med Genet. 115:130–141. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Byrd JC, Lawrence D, Arthur DC, et al:
Patients with isolated trisomy 8 in acute myeloid leukemia are not
cured with cytarabine-based chemotherapy: results from cancer and
leukemia group B 8461. Clin Cancer Res. 4:1235–1241. 1998.
|
|
16
|
Pedersen B: MDS and AML with trisomy 8 as
the sole chromosome aberration show different sex ratios and
prognostic profiles: a study of 115 published cases. Am J Hematol.
56:224–229. 1997. View Article : Google Scholar
|
|
17
|
Massaad L, Prieur M, Leonard C and
Dutrillaux B: Biclonal chromosome evolution of chronic
myelomonocytic leukemia in a child. Cancer Genet Cytogenet.
44:131–137. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Barjesteh van Waalwijk van
Doorn-Khosrovani S, Erpelinck C, van Putten WL, Valk PJ, van der
Poel-van de Luytgaarde S, Hack R, Slater R, Smit EM, Beverloo HB,
Verhoef G, et al: High EVI1 expression predicts poor survival in
acute myeloid leukemia: a study of 319 de novo AML patients. Blood.
101:837–845. 2003.PubMed/NCBI
|
|
19
|
Ogawa S, Kurokawa M, Mitani K, Yazaki Y
and Hirai H: Overexpression of Evi-1 and dysmegakaryopoiesis in
human leukemias: reply to Carapeti, Goldman and Cross, Leukemia
1996; 10: 1561. Leukemia. 10:18491996.PubMed/NCBI
|
|
20
|
Iwase S, Furukawa Y, Horiguchi-Yamada J,
Nemoto T, Takahara S, Kawano T, Sekikawa T, Ito K, Yamazaki Y,
Kikuchi J, et al: A novel variant of acute myelomonocytic leukemia
carrying t(3;12)(q26;p13) with characteristics of 3q21q26 syndrome.
Int J Hematol. 67:361–368. 1998. View Article : Google Scholar
|
|
21
|
Fears S, Mathieu C, Zeleznik-Le N, Huang
S, Rowley JD and Nucifora G: Intergenic splicing of MDSI and EVIl
occurs in normal tissues as well as in myeloid leukemia and
produces a new member of the PR domain family. Proc Natl Acad Sci
USA. 93:1642–1647. 1996. View Article : Google Scholar
|