Introduction
Cervical cancer is the second most frequent cause of
cancer-related mortality in females worldwide and the most common
type of cancer among females in the majority of developing
countries (1–3). Currently, clinical management of
preinvasive cervical cancer largely relies on histological
examination to confirm cervical intraepithelial neoplasia (CIN) and
its grading. Generally, higher grade (CIN II–III) cases require
active treatment, including cervial conization or cervical loop
electrosurgical excision procedure. Although the histological
features of CIN are well understood, inconsistent use and
misinterpretation of such features may occur due to intra- and
interobserver variability (4,5). On
the basis of morphology alone, it can be difficult to distinguish
between squamous metaplasia coupled with hyperplasia (SMH) and CIN
I, and between CIN I and CIN II–III. Moreover, small biopsy size,
tangential sectioning, thermal artifact, coexistent inflammatory or
reactive lesions and application of subjective criteria all
increase the difficulty for the diagnosis of CIN (6). Therefore, objective diagnostic methods
in addition to histology are required to accurately diagnose CIN in
histological specimens.
Human papillomavirus (HPV) plays an etiologic role
in cervical carcinogenesis and is detectable in preinvasive and
invasive cervical epithelial neoplasms (7,8). The
most common high-risk HPV subtypes include types 16 and 18, which
account for ~70% of HPV species detected in cervical cancer.
Low-risk types, 6 and 11, account for ~90% of HPV species present
in genital warts (1). Infections
with low-risk types have been known to cause genital warts and
low-grade cervical abnormalities.
p16 is a cyclin-dependent kinase inhibitor that
regulates the transition from G1 to S phase of the cell cycle and
normally functions as a tumor suppressor (9). Although p16 levels are reduced in a
variety of malignant tumors, this gene product has been shown to be
upregulated (or overexpressed) in the majority of high-grade
cervical dysplasias and carcinomas induced by high-risk HPV
subtypes (10). p53, as a tumor
suppressor, is one of the major factors controlling cell
proliferation. As the ‘guardian of the genome’, it arrests the cell
cycle in response to DNA damage or directs the damaged cell to an
apoptotic pathway.
To investigate HPV infection and the protein
expression of p16INK4A and p53, and to evaluate their
potential roles in the pathological diagnosis and grading of
cervical CIN, HPV DNA and p16INK4A and p53 expression
were examined in a panel of clinical tissues samples (including 40
SMH, 24 cervical condyloma, 120 CIN and 19 cervical cancer) using
polymerase chain reaction (PCR) or immunohistochemistry (IHC). In
addition, the correlation between p16INK4A and p53 and
the degree of CIN with HPV were analyzed.
Materials and methods
Tissue samples
Samples were obtained from formalin-fixed,
paraffin-embedded blocks of cervical biopsies from 203 patients
accessioned at the Department of Pathology, The Fifth People’s
Hospital of Shanghai, Fudan University (Shanghai, China), between
2006 and 2012. The age of patients ranged between 12 and 77 years,
with a median age of 48 years. None of the patients had been
treated for cervical abnormalities prior to the biopsy.
The specimens included 40 cases of SMH, 24 cases of
cervical condyloma, 120 cases of CIN (CIN I, 37 cases; CIN II–III,
83 cases) and 19 cases of cervical cancer. All slides were reviewed
by at least three pathologists, discrepancies were resolved by
reevaluation and discussion, and concurrence was achieved. Only one
final diagnosis was determined for each case. Representative images
of hematoxylin and eosin (H&E) staining from tissues with
increasing grades of cervical lesions are shown in Fig. 1 [images were viewed under a light
microscope (BX45, Olympus, Tokyo, Japan)]. The study was approved
by the ethics committee of the Fifth People’s Hospital of Shanghai,
Fudan University (Shanghai, China). Written informed consent was
obtained from the patient’s families.
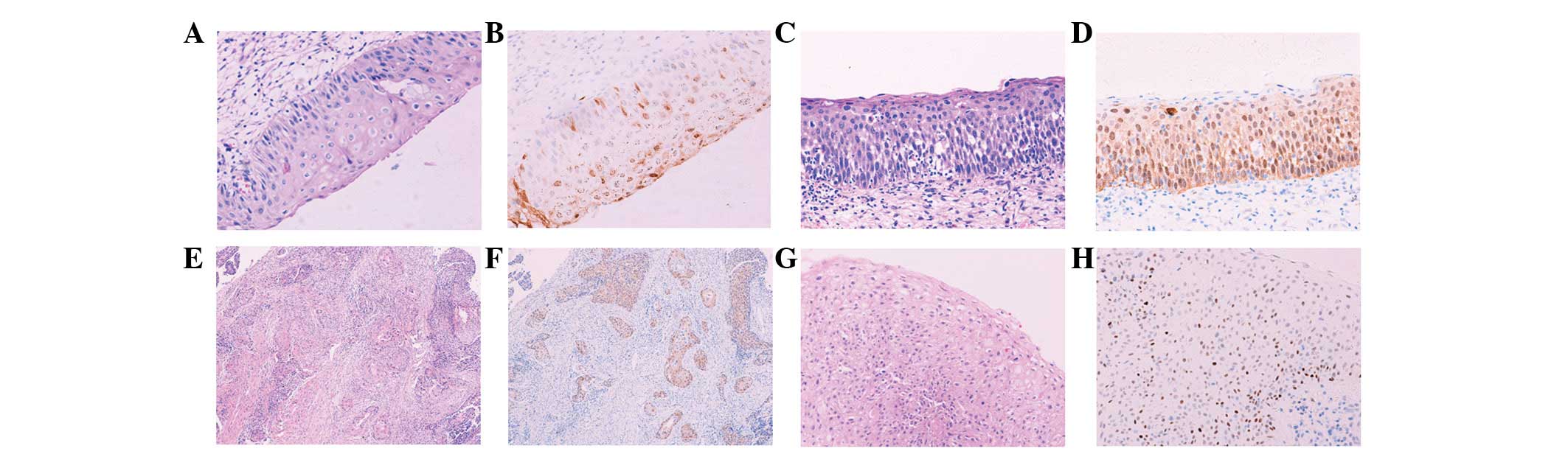 | Figure 1H&E and immunohistochemical
staining of various tissue sections. Images were captured under a
light microscope at ×200 magnification with the exception of panels
E and F (magnification, ×100). Tissue sections presented are (A and
B) CIN I, (C and D) CIN II, (E and F) cervical cancer and (G and H)
cervical condyloma. (A, C, E and G) H&E staining showing
morphologies of all tissues and (B, D, F and H) immunohistochemical
staining, showing differential expression of p16INK4A
protein. (B) Spotty and (D and F) band-like patterns and (H) strong
expression of p53 protein in the cervical condyloma. H&E,
hematoxylin and eosin; CIN, cervical intraepithelial neoplasia. |
DNA extraction
Only 104 specimens of DNA were detected. In total,
10 formalin-fixed, paraffin-embedded tissues were sectioned
(5-μm-thick). The tissue sections were first deparaffinized in
xylene and grading alcohol, and then digested by proteinase K at
1.25 mg/ml in 400 μl lysis buffer [20 mM Tris (pH 8.0), 10 mM EDTA
(pH 8.0) and 2% SDS (pH 7.2)] at 56ºC overnight. The proteinase K
was heat inactivated at 95ºC for 10 min, one-third of the volume of
saturated sodium acetate (pH 5.2) was added and samples were
centrifuged for 15 min at 18,000 × g. Supernatant was transferred
into a fresh tube, two volumes of ice-cold 100% ethanol were added
and the sample was centrifuged for 15 min at 18,000 × g. Next, the
supernatant was discarded and the pellet was washed twice with 75%
ethanol. Following air drying at room temperature for 15 min, the
DNA pellet was dissolved in distilled deionized water and the DNA
solution was stored at −20ºC.
To ensure integrity of the DNA, a 5-μl DNA aliquot
was amplified for the β-globin housekeeping gene by PCR with Taq
polymerase (Sangon Biotech Shanghai Co., Ltd., Shanghai, China).
Primers specific for the β-globin gene were as follows: PC04,
5′-CAA GAG CCA AGG ACA GGT AC-3′; and GH20, 5′-CAA CTT CAT CCA CGT
TCA CC-3′. PCR was performed in a 50-μl reaction with 40 cycles of
amplification. Each cycle included a denaturation step at 94ºC for
30 sec, primer annealing step at 53ºC for 30 sec and chain
elongation step at 72ºC for 40 sec, followed by the final
elongation step at 72ºC for 5 min. DNA products were visualized in
2.0% agarose gel.
HPV detection and typing
The current study focused on the detection of the
most common HPV subtypes; high-risk HPV16/18 and low-risk HPV6/11.
In total, three various primer sets were used for the PCR
detection. For PCR of the HPV6/11 DNA to amplify a 334-bp HPV L1
gene fragment, the following primers were used: 5′-TGC AAG AAT GCA
CTG ACC AC-3′ and 5′-TGC ATG TTG TCC AGC AGT GT-3′ (11). PCR reactions were performed in 50-μl
volumes containing 5 μl DNA and 2.5 U Taq DNA polymerase using the
following conditions: Initial 5-min denaturation step at 94ºC,
followed by 40 cycles of amplification in a PCR processor (PTC-100;
MJ Research Inc., St. Bruno, QC, Canada). Each cycle included a
denaturation step at 94ºC for 30 sec, primer annealing step at 51ºC
for 30 sec and chain elongation step at 72ºC for 40 sec, followed
by a final elongation step at 72ºC for 5 min. For the detection of
HPV16 and HPV18, the following specific primers were used: HPV16,
5′-TGA GCA ATT AAA TGA CAG CTC AGA-3′ and 5′-GA GAA CAG ATG GGG CAC
ACA AT-3′; and HPV18, 5′-GAC CTT CTA TGT CAC GAG CAA TTA-3′ and
5′-GC ACA CCA CGG ACA CAC AAA G-3′ (12). The HPV16 primers amplified a 212-bp
fragment and the HPV18 primers amplified a 236-bp fragment.
HPV16/18 PCR reactions were performed under conditions identical to
PCR for HPV6/11, with the exception of the annealing temperature of
54ºC. All PCR products were subjected to electrophoresis in a 2%
agarose gel at 100 V for 30 min. The 334- or 212/236-bp product was
visualized with ethidium bromide staining. Additional confirmation
of the amplified HPV-specific sequence was performed by DNA
sequencing (Shanghai GeneCore BioTechnologies Co., Ltd., Shanghai,
China). Of note, the absence of HPV6/11 or HPV16/18 PCR products
did not exclude the presence of other HPV subtypes, which was not
assessed in the present study.
Immunohistochemical staining
For the detection of p16INK4A and p53
protein expression, IHC was performed using the Chemmate™ EnVision™
detection kit (DakoCytomation, Glostrup, Denmark). The sections
were dewaxed in xylene, rehydrated in a series of gradient alcohol
solutions and rinsed in water. Sections were then treated by two
immersions in a 3% hydrogen peroxide bath in absolute methanol (5
min each) to inhibit endogenous peroxidase activity, followed by
rinsing in water. Antigen retrieval was accomplished by heating the
specimen in 0.01 M citrate buffer (pH 6.0) in a high-intensity
microwave oven for 25 min. Monoclonal antibodies against p16 (clone
16P07; 1:100) and p53 (clone DO-7; 1:100) (Shanghai Changdao
Biotech Co., Ltd., Shanghai, China) were applied for 12 h at 4ºC.
The sections were sequentially incubated with secondary antibody at
37ºC for 30 min and carefully rinsed with several changes of TBS
between each step. The sections were then incubated with
3,3′-Diminobenzidene (DakoCytomation) and lightly counterstained
with Harris hematoxylin for 60 sec. Sections of the
p16INK4A-positive cervical cancer and p53-positive
breast carcinoma were included to serve as the positive controls
for p16INK4A and p53 IHC, respectively. Negative control
sections were processed by eliminating the use of respective
primary antibodies.
Evaluation of immunohistochemical
staining
Nuclear and/or cytoplasmic staining in >10% of
atypical cells was interpreted as positive for p16INK4A;
cytoplasmic staining alone was considered non-specific and
interpreted as negative.
Brown staining in the nuclei of >10% of atypical
cells was indicated as positive for p53. For cases with condyloma,
CIN and/or cervical cancer, p16INK4A and p53 were
evaluated in the areas exhibiting the highest grade of atypia or
dysplasia. The immunostained slides were reviewed by at least three
pathologists and a consensus was achieved. Staining patterns were
found to correlate with the respective H&E diagnoses. The two
staining patterns observed in p16 staining were band-like and
spotty. The staining pattern was recorded as band-like when >90%
of contiguous squamous cells stained positive or spotty when
>10% of scattered squamous cells stained positive (6).
Statistical analysis
Statistical analysis was performed using
χ2, Fisher’s exact or Spearman’s rho tests. SPSS
software (version 13.0; SPSS, Inc., Chicago, IL, USA) was used for
all statistical analyses. For all tests, P<0.05 was considered
to indicate a statistically significant difference.
Results
Detection of HPV-specific DNA by PCR
DNA prepared from previously collected cervical
tissue samples were pre-tested for their integrity and PCR
performance. The β-globin housekeeping gene DNA was detected in all
104 samples (including 12 SMH, 10 cervical condyloma, 65 CIN and 17
cervical cancer). Representative DNA products amplified with HPV
subtype-specific primers are shown in Fig. 2. Each PCR reaction produced only one
correct band with the predicted size, indicating the specificity of
PCR amplifications. Positive DNA products were confirmed for their
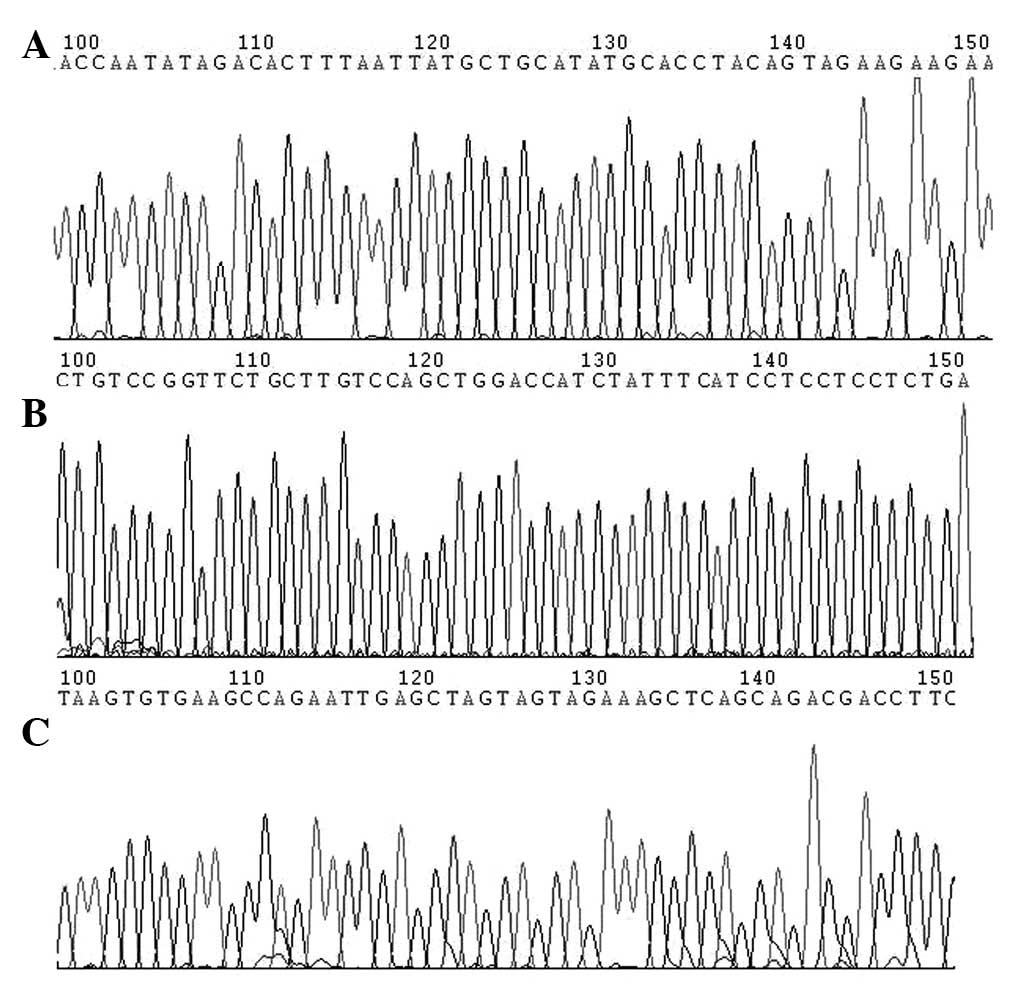
correct nucleotide sequences by DNA sequencing (Fig. 3).
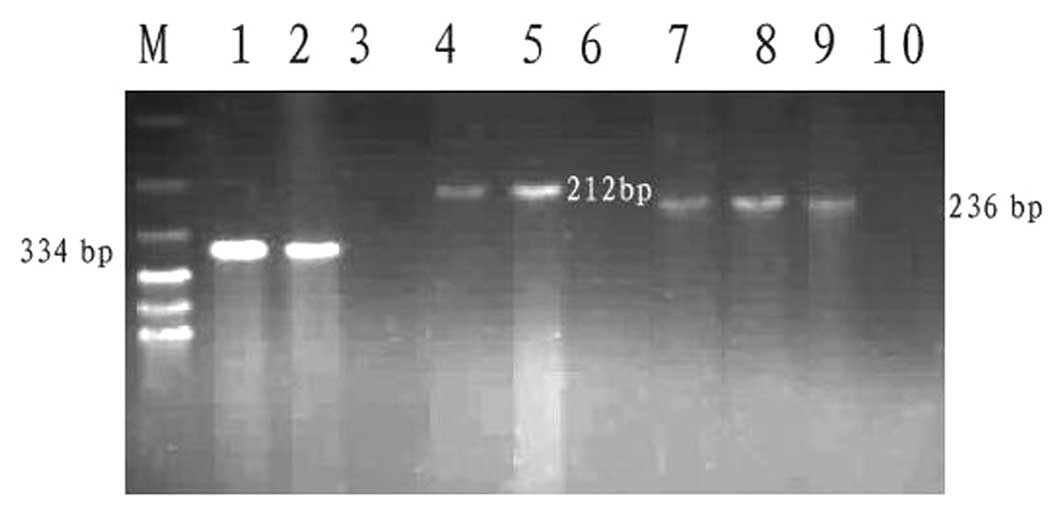 | Figure 2Images of agarose gel with polymerase
chain reaction products for various HPV DNA species. DNA bands in
the following lanes present an amplified product specific for viral
DNA: M, 100-bp DNA ladder (molecular weight marker); 1, HPV6/11
subtype; 2, HPV6/11-positive control; 3, HPV6/11-negative control;
4, HPV16 subtype; 5, HPV16-positive control; 6, HPV16-negative
control; 7 and 8, HPV18 subtype; 9, HPV18-positive control; and 10,
HPV18-negative control. HPV, human papillomavirus. |
Results of all PCR detection are summarized in
Table I. DNA sequences for the
HPV6/11 subtype was amplified from 10/104 (9.6%) cases and its
frequency in the cervical condyloma group was significantly higher
than those in other groups (P<0.05). The HPV16/18 subtype was
identified in 51/104 (49.0%) cases. Of the 51 HPV16/18-positive
cases, 31 were HPV16, 15 were HPV18 and double-positive expression
of HPV16/18 was identified in five cases. The HPV16/18 frequency in
tissues increased in the following order: SMH, cervical condyloma,
CIN I, CIN II–III and cervical cancer. Of those infected by
HPV16/18, 48 (94.1%) cases were diagnosed with CIN or cervical
cancer.
 | Table IPolymerase chain reaction-detected HPV
subtype-specific DNA in various tissues. |
Table I
Polymerase chain reaction-detected HPV
subtype-specific DNA in various tissues.
| | Positive cases, n
(%) |
|---|
| |
|
|---|
| Histological
classification | Total cases, n | HPV 16/18
subtype | HPV 6/11 subtype |
|---|
| SMH | 12 | 1 (8.3) | 0 (0) |
| Condyloma | 10 | 2 (20.0) | 9 (90.0) |
| CIN I | 18 | 10 (55.6) | 1 (5.6) |
| CIN II–III | 47 | 26 (55.3) | 0 (0) |
| Cervical cancer | 17 | 12 (70.6) | 0 (0) |
| Total | 104 | 51 (49.0) | 10 (9.6) |
HPV16/18 detection frequencies were not
significantly different between the CIN II–III and CIN I groups, or
between the cervical cancer and CIN II–III groups (all P>0.05).
However, the HPV16/18 frequency was significantly higher in the CIN
I group than that in the SMH group (P=0.018).
p16INK4A expression by
immunohistochemical staining
The p16INK4A expression was located in
the nucleus and/or cytoplasm and exhibited two staining patterns,
scattered spotty and contiguous band-like expression. The SMH and
cervical condyloma groups were negative for p16INK4A
expression or exhibited predominately spotty expression. The CIN I
group exhibited similar spotty expression to the SMH and condyloma
groups, or contiguous band-like expression (Fig. 1A and B). By contrast, the CIN II–III
and cervical cancer groups showed mainly contiguous band-like
patterns (Fig. 1C–F).
p16INK4A expression was not observed in squamous and
glandular epithelia adjacent to the dysplastic lesions coexisting
in the same tissue sections.
The results of the p16INK4A staining are
summarized in Table II.
p16INK4A-positive expression was observed in 151/203
(74.4%) cases examined. Of the 151 cases, 114 (75.5%) revealed the
band-like positive pattern and 37 (24.5%) showed the spotty,
positive pattern. The p16INK4A positive expression rates
were significantly higher in the CIN I group (81.1%,) than in the
SMH group (32.5%) (P<0.0001), and significantly higher in the
CIN II–III group (95.8%) than that in CIN I group (P=0.005).
However, the p16INK4A-positive expression was not
significantly different between the cervical cancer and CIN II–III
groups (P=0.738). The expression patterns of p16INK4A
changed from the scattered spotty to contiguous band-like pattern
with increasing grade of cervical lesions. In addition, a
significant difference was achieved in the p16INK4A
staining pattern (band-like vs. spotty or negative) among cervical
tissues with various stages of lesion progression (P<0.0001;
Table II).
 | Table IIExpression of p16INK4A and
p53 with the degree of CIN in 203 cervical lesions. |
Table II
Expression of p16INK4A and
p53 with the degree of CIN in 203 cervical lesions.
| | Expression of
p16INK4A | |
|---|
| |
| |
|---|
| Histological
classification | n | Band, n | Spotty, n | Total, n (%) | Expression of p53,
n (%) |
|---|
| SMH | 40 | 3 | 10 | 13 (32.5) | 8 (20.0) |
| Condyloma | 24 | 2 | 8 | 10 (41.7) | 18 (75.0) |
| CIN I | 37 | 15 | 15 | 30 (81.1) | 18 (48.6) |
| CIN II–III | 83 | 76 | 4 | 80 (95.8) | 40 (48.2) |
| Cervical
cancer | 19 | 18 | 0 | 18 (94.7) | 12 (63.2) |
| Total | 203 | 114 | 37 | 151 (74.4) | 96 (47.3) |
p53 expression and its correlation with
p16INK4A protein by IHC
The p53 expression was localized in the nuclei and
positive expression was identified in 47.3% (96/203) of the cases
examined. The strong p53 expression was observed in cases of
cervical condyloma with positive HPV6/11 (Fig. 1G–H). However, p53 staining was not
observed in the normal epithelium coexisting in the same tissue
sections.
The rate of p53 positive expression in cervical
condyloma was significantly higher than that in the other groups
(P<0.05), with the exception of the cervical cancer group.
Results of p53 expression during lesion progression are shown in
Table II.
In addition, the correlation between
p16INK4A and p53 protein expression were analyzed by the
Spearman’s rho test (Table III)
and no correlation was identified between the expression of the two
proteins (r=0.6669; P=0.2189).
 | Table IIICorrelation between
p16INK4A and p53 expression with no regard to tissue
type. |
Table III
Correlation between
p16INK4A and p53 expression with no regard to tissue
type.
| p16INK4A
expression | Negative, n | Spotty, n | Band (+), n | Band (++), n | Band (+++), n | Total, n |
|---|
| p53 negative | 37 | 18 | 12 | 17 | 23 | 107 |
| p53 positive | 15 | 19 | 12 | 20 | 30 | 96 |
| Total | 52 | 37 | 24 | 37 | 53 | 203 |
Correlation between p16INK4A
or p53 expression and HPV infection
Table IV shows that
p16INK4A expression was found to correlate with HPV
infection in every grade of cervical lesion. The
p16INK4A-positive expression was observed in 47 cases in
which HPV16/18 was also found to be positive. Of the 47 lesions, 42
cases (89.4%) showed p16INK4A band-like expression
patterns and the remainder (10.6%) showed spotty expression
patterns or were negative for p16INK4A. The presence of
spotty and band-like expression was found to correlate markedly
with HPV16/18 infection (r=1.0000; P<0.0001). In addition, the
presence of the band-like expression alone was found to correlate
with HPV16/18 infection (r=0.9747; P=0.0048). However, no
significant correlation was identified between spotty or negative
p16INK4A expression and HPV16/18 infection (r=0.2294;
P=0.7015). Of the 10 cases that positively expressed HPV6/11, 5
(50.0%) were p16INK4A-positive with spotty staining
pattern. No correlation was observed between p16 INK4A
staining and HPV6/11 infection (r=−0.5643; P=0.3217).
 | Table IVExpression patterns of
p16INK4A in 51 HPV16/18-positive and 10 HPV6/11-positive
cases. |
Table IV
Expression patterns of
p16INK4A in 51 HPV16/18-positive and 10 HPV6/11-positive
cases.
| HPV subtype | n | SMH | Condyloma | CIN I | CIN II–III | Cervical
cancer |
|---|
|
|
|
|
|
|---|
| Spotty, n | Band, n | Spotty, n | Band, n | Spotty, n | Band, n | Spotty, n | Band, n | Spotty, n | Band, n |
|---|
| 16/18 | 47 | 0 | 0 | 1 | 0 | 4 | 4 | 0 | 26 | 0 | 12 |
| 6/11 | 5 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 52 | 0 | 0 | 6 | 0 | 4 | 4 | 0 | 26 | 0 | 12 |
Widespread expression of p53 was observed in lesions
infected with HPV6/11 (Fig. 3G and
H) and it was more intense than in those infected with
HPV16/18. Of the 51 cases infected by HPV16/18, 23 were p53
positive and of the 10 cases infected by HPV6/11, 8 were p53
positive. The presence of HPV infection (HPV16/18 or HPV6/11) was
found to significantly correlate with the immunoreactivity of p53
(P=0.044).
Discussion
To date, >100 types of HPV have been identified
from clinical samples. In total, ~40 different HPV genotypes are
considered to be sexually transmitted, which may be regarded as
causal agents for cervical cancer and CIN. The majority of previous
studies have hypothesized that high-grade CIN (CIN II and III) and
cervical cancer markedly correlate with the persistence of
high-risk HPV types (13,14).
In the present study, HPV16 and HPV18 subtypes were
detected with a frequency of 49.0%, mainly in the CIN and cervical
cancer groups. HPV16 was predominant in 60.8% of the
HPV16/18-positive tissues. No statistically significant difference
in HPV16/18 detection frequencies was observed between the CIN I
and CIN II–III groups, or between the CIN II–III and cervical
cancer groups. However, the frequency in the CIN I group was
significantly higher than that in the SMH group (P=0.018).
Therefore, HPV16/18 detection was also useful to distinguish CIN
and cervical squamous cell carcinoma from SMH.
HPV6/11 was detected in 90.0% of condyloma cases,
but only in 5.6% of the CIN I cases, where 55.6% were positive for
HPV16/18. Therefore, the difference in the distribution of HPV
subtypes was significant between the cervical condyloma and CIN I
groups. Since it is well known that prognoses for high- and
low-risk HPVs are significantly different, future studies are
required to confirm that cervical condyloma and CIN I belong to
low-grade squamous intraepithelial lesions in the Bethesda System
(2001).
The p16 gene product normally acts to inhibit
progression through the cell cycle by binding to cyclin-dependent
kinase 4/6, therefore, preventing the phosphorylation and
subsequent inactivation of retinoblastoma protein. A previous study
by Sakaguchi et al was the first to correlate
p16INK4A overexpression with cellular malignant
behaviors (15). In addition, a
previous study by Sano et al was the first to describe the
use of p16INK4A as a diagnostic marker in the pathology
of CIN (16). The majority of
studies have since reported a significant correlation between
diffuse or contiguous band-like patterns of p16INK4A
staining in CIN II–III and the presence of high-risk HPV types. In
addition, studies have reported a correlation between negative to
scattered spotty p16INK4A staining patterns in CIN I and
the presence of low-risk HPV types (17–19).
However, in contrast to the majority of studies, certain studies
have shown that a significant number of high-grade CIN and squamous
cell carcinomas are negative for p16INK4A (20,21),
and another study observed diffused or band-like
p16INK4A staining patterns in specific cases of CIN I
and SMH (22). Possible
explanations for such heterogeneity in p16INK4A staining
may include the inherent and unavoidable intra- and inter-observer
variability in the morphological categorization of CIN, lack of
standardization in the scoring of p16INK4A
immunoexpression (nuclear and/or cytoplasmic staining; and positive
vs. negative distribution within the epithelium), various antibody
clones used for IHC and the existence of various HPV geographical
variations (23).
In the present study, p16INK4A expression
increased gradually with the progression of lesions, and
immunostaining patterns changed from the scattered spotty to
contiguous band-like patterns. These results indicated a marked
correlation between p16INK4A immunostaining and degree
of epithelial atypia. In addition, the expression rate of
p16INK4A in the CIN II–III group was found to be
significantly higher than that in the CIN I group, suggesting the
potential use of p16INK4A to discriminate between CIN
II–III and CIN I. On the other hand, the expression rate of
p16INK4A was significantly higher (P<0.0001) in CIN
I, compared with that in SMH. In addition, the immunostaining
patterns for p16INK4A were not only spotty, but also
band-like in specific cases, suggesting that p16INK4A
may also be a potential marker to distinguish between CIN I and
SMH.
Previous studies have shown that high-risk HPV may
induce p16INK4A overexpression (16,24).
One mechanism of which may be due to the negative feedback loop
where p16INK4A acts as a cell cycle inhibitor and may be
induced in response to the HPV infection to reduce the
retinoblastoma function (such as inactivation by HPV E7). However,
Kuo et al previously hypothesized that HPV-related mucosal
dysplasia in various anatomical locations may lead to various
molecular pathways (25). To date,
the majority of studies have reported that immunoreactivity for
p16INK4A may be used as a surrogate marker of HR-HPV and
Kong et al hypothesized that p16INK4A IHC was
superior for the detection of HR-HPV by in situ
hybridization (26). In the current
study, a band-like pattern of p16 immunostaining was found to
markedly correlate with HPV16/18 infection, suggesting that a
band-like expression of p16INK4A may be induced by
HPV16/18 DNA. It appears that the higher rate of the HPV16/18
expression correlates with stronger intensity of
p16INK4A staining.
Mutations in the p53 gene are the most frequent
mutations encountered in types of human cancer. The majority of
studies have found that the presence of p53 in cancer cells
correlates with the consequence of p53 mutation due to failure of
the mutated protein to transactivate its own negative regulator
MDM2. The p53 oncoprotein expression has been detected in
carcinomas from various sites, including breast, colon, bladder and
lung. However, the correlation between HPV and p53 IHC staining in
cervical lesions has been controversial (27,28).
In the present study, no significant correlation was observed
between p53 expression and cervical lesions. In addition, no
correlation was identified between HPV types and the
immunoreactivity of p53. However, the presence of the HPV infection
by HPV6/11 or HPV16/18 subtype was found to significantly (P=0.044)
correlate with the immunoreactivity of p53, which is consistent
with a study by Giannoudis et al who hypothesized that p53
was more widely expressed in cervical lesions infected with
low-risk HPV than those with intermediate or high-risk HPV types
(29).
In conclusion, the management of preinvasive
cervical diseases remains largely dependent on the confirmation of
CIN. However, the current classification of CIN evaluated by
histology exhibits a significant overlap in the morphological
criteria between SMH and CIN I, as well as between CIN II and CIN
II–III. The results of the current study suggested that
p16INK4A immunostaining may be used as an additional
marker for a more accurate determination of CIN in the cervical
lesions.
Acknowledgements
The authors would like to thank Weiqing Shu and Hui
Liu for advice on immunohistochemical staining, and Dr Ju Yang and
Yongjuan Liu for useful scientific discussions.
References
|
1
|
Bauer HM and Ault K: Human papillomavirus:
Current prevalence and future Protection. Sex Trans Dis.
33:509–511. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chih HJ, Lee AH, Colville L, Binns CW and
Xu D: A review of dietary prevention of human
papillomavirus-related infection of the cervix and
cervicalintraepithelial neoplasia. Nutr Cancer. 65:317–328. 2013.
View Article : Google Scholar
|
|
3
|
Qian JH, Xie X and Yie DF: Epidemiologic
features of human paplillomavirus infection in cervix. Chin J
Obstet Gynecol. 38:712–714. 2006.
|
|
4
|
Martin CM and O’Leary JJ: Histology of
cervical intraepithelial neoplasia and the role of biomarkers. Best
Pract Res Clin Obstet Gynaecol. 25:605–615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dascau V, Furau G, Furau C, Paiusan L, et
al: Cervical intraepithelial neoplasia in the ‘dr. Salvator vuia’
clinical obstetrics and gynecology hospital - arad during the
2000–2009 period. Maedica (Buchar). 7:138–142. 2012.
|
|
6
|
Walts AE, Lechago LJ and Bose S: P16 and
Ki67 immunostaining is a useful adjunct in the assessment of
biopsies for HPV-associated anal intraepithelial neoplasia. Am J
Surg Pathol. 30:795–801. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Monsonego J, Pintos J, Semaille C, Beumont
M, et al: Human papillomavirus testing improves the accuracy of
colposcopy in detection of cervical intraepithelial neoplasia. Int
J Gynecol Cancer. 16:591–598. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Massad LS, Einstein MH, Huh WK and Katki
HA: 2012 updated consensus guidelines for the management of
abnormal cervical cancer screening tests and cancer precursors. J
Low Genit Tract Dis. 17(5 Suppl 1): S1–S27. 2013. View Article : Google Scholar
|
|
9
|
Tsoumpou I, Arbyn M, Kyrgiou M, Wentzensen
N, et al: p16(INK4a) immunostaining in cytological and histological
specimens from the uterine cervix: a systematic review and
meta-analysis. Cancer Treat Rev. 35:210–220. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sano T, Oyama T, Kashiwabara K, Fukuda T
and Nakajima T: Expression status of p16 protein is associated with
human papillomavirus oncogenic potential in cervical and genital
lesions. Am J Pathol. 153:1741–1748. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sotlar K, Diemer D, Dethleffs A, Hack Y,
et al: Detection and typing of human papillomavirus by E6 nested
multiplex PCR. J Clin Microbiol. 42:3176–3184. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gheit T and Tommasino M: Detection of
high-risk mucosal human papillomavirus DNA in human specimens by a
novel and sensitive multiplex PCR method combined with DNA
microarray. Methods Mol Biol. 665:195–212. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shirendeb U, Hishikawa Y, Moriyama S, Win
N, et al: Human papillomavirus infection and its possible
correlation with p63 expression in cervical cancer in Japan,
Mongolia, and Myanmar. Acta Histochem Cytochem. 42:181–190. 2009.
View Article : Google Scholar
|
|
14
|
Lax S: Histopathology of cervical
precursor lesions and cancer. Acta Dermatovenerol Alp Panonica
Adriat. 20:125–133. 2011.PubMed/NCBI
|
|
15
|
Sakaguchi M, Fujii Y, Hirabayashi H, Yoon
HE, et al: Inversely correlated expression of p16 and Rb protein in
non-small cell lung cancers: an immunohistochemical study. Int J
Cancer. 65:442–445. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sano T, Oyama T, Kashiwabara K, Fukuda T
and Nakajima T: Immunohistochemical overexpression of p16 protein
associated with intact retinoblastoma protein expression in
cervical cancer and cervical intraepithelial neoplasia. Pathol Int.
48:580–585. 1998. View Article : Google Scholar
|
|
17
|
Koshiol J, Lindsay L, Pimenta JM, Poole C,
et al: Persistent human papillomavirus infection and cervical
neoplasia: a systematic review and meta-analysis. Am J Epidemiol.
168:123–137. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Carozzi F, Gillio-Tos A, Confortini M, et
al: Risk of high-grade cervical intraepithelial neoplasia during
follow-up in HPV-positive women according to baseline p16-INK4A
results: a prospective analysis of a nested substudy of the NTCC
randomised controlled trial. Lancet Oncol. 14:168–176. 2013.
View Article : Google Scholar
|
|
19
|
Lesnikova I, Lidang M, Hamilton-Dutoit S
and Koch J: p16 as a diagnostic marker of cervical neoplasia: a
tissue microarray study of 796 archival specimens. Diagn Pathol.
4:222009. View Article : Google Scholar
|
|
20
|
Lee S, Kim H, Kim H, Kim C and Kim I: The
utility of p16INK4a and Ki-67 as a conjunctive tool in uterine
cervical lesions. Korean J Pathol. 46:253–260. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lin ZH, Liu MZ, Zhao YW, Wu QY, et al:
Study of p16 expression and DNA ploidy in HPV-negative cervical
cancers and precursors. Chin J Pathol. 35:412–416. 2006.(In
Chinese).
|
|
22
|
Soma M and Kamaraj S: Detection of human
papillomavirus in cervical gradings by immunohistochemistry and
typing of HPV 16 and 18 in high-grades by polymerase chain
reaction. J Lab Physicians. 2:31–36. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Samir R, Asplund A, Tot T, Pekar G and
Hellberg D: High-risk HPV infection and CIN grade correlates to the
expression of c-myc, CD4+, FHIT, E-cadherin, Ki-67, and p16INK4a. J
Low Genit Tract Dis. 15:280–286. 2011.PubMed/NCBI
|
|
24
|
Yim EK and Park JS: Biomarkers in cervical
cancer. Biomark Insights. 7:215–225. 2007.
|
|
25
|
Kuo KT, Chang HC, Hsiao CH and Lin MC:
Increased Ki-67 proliferative index and absence of P16INK4 in
CIN-HPV related pathogenic pathways different from cervical
squamous intraepithelial lesion. Br J Ophthalmol. 90:894–899. 2006.
View Article : Google Scholar
|
|
26
|
Kong CS, Balzer BL, Troxell ML, Patterson
BK and Longacre TA: p16INK4A immunohistochemistry is superior to
HPV in situ hybridization for the detection of high-risk HPV in
atypical squamous metaplasia. Am J Surg Pathol. 31:33–43. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bragança JF, Sarian LO, Pitta DR, Maito
AB, et al: Expression of p16(INK4a) and cervical infection with
high-risk human papillomavirus are not related to p53 activity in
cervical intraepithelial neoplasia. Int J Gynecol Cancer.
18:1060–1064. 2008.PubMed/NCBI
|
|
28
|
Türkçüoğlu I, Tezcan S, Kaygusuz G,
Atabekoğlu CS, et al: The role of p53, Bcl-2 and Ki-67 in
premalignant cervical lesions and cervical cancer. Eur J Gynaecol
Oncol. 28:290–293. 2007.PubMed/NCBI
|
|
29
|
Giannoudis and Herrington CS: Differential
expression of p53 and p21 in low grade cervical squamous
intraepithelial lesions infected with low, intermediate, and high
risk human papillomaviruses. Cancer. 89:1300–1307. 2000. View Article : Google Scholar
|