Introduction
Gliomas are the most common and aggressive type of
brain tumor. They account for 32–45% of all primary brain tumors
(1,2) and 70–80% of all malignant brain tumors
(2,3). A high disease incidence, mortality
rate and disability rate place gliomas among the most fatal tumors.
For the last two decades, important advances have been made in
neurological and clinical surgery techniques. Gross total tumor
removal, chemotherapy and radiotherapy have become standard
treatments for newly diagnosed patients (4). Despite the most extensive treatment
protocols, including clinical surgery, radiation treatment and
chemotherapy, patients have a median survival time of only 12–15
months (5). This is due to the
malignant behavior of the tumor and resistance to current
therapeutic approaches (6).
Patients usually undergo another surgery following a short period
of remission. Additionally, a large number of patients succumb to
glioma prior to a second surgery. More effective individualized
treatment has become the focus of research on the treatment of
glioma. Parsons et al presented researchers with a new
direction for the study of glioma treatment following the
observation that mutation of the isocitrate dehydrogenase 1 (IDH1)
gene is frequent in glioma (7).
IDH1 mutation may represent a new gene subtype of glioma and be an
effective target for tumor therapy. IDH plays an important role in
the tricarboxylic acid (TCA) cycle. Through evolution, humans have
developed three IDH enzymes: NADP-dependent enzymes IDH1 and IDH2,
and NAD-dependent enzyme IDH3. IDH1 is present in the cytoplasm and
peroxisomes while IDH2 and IDH3 are found in the mitochondria.
NAD+-specific IDH catalyzes a rate-limiting step in the
TCA cycle. The affinity of yeast IDH for isocitrate is enhanced by
AMP and reduced by NADH (8,9). The enzyme IDH1 catalyzes the citric
acid oxidation of grass succinic acid and the subsequent oxidative
decarboxylation generates α-ketoglutarate and produces NADPH
(10). IDH enzymes are of great
importance in the generation of biological energy and synthesis of
metabolic pathways. Mutated IDH1 consumes rather than produces
NADPH (10), thus markedly reducing
NADPH levels. An increasing number of studies have shown that
patients carrying mutated IDH1 genes have an improved prognosis.
However, the mechanism by which the mutated IDH1 gene improves
prognosis remains unclear.
In the present study, three cell lines stably
expressing wild-type IDH1 (wIDH1), mutated IDH1 (mIDH1) and
enhanced green fluorescent protein (EGFP) were constructed for the
study of their effects on the biological behavior of glioma cells.
The results aim to elucidate the mechanisms underlying these
effects and provide clinicians with an overview of the current
understanding of IDH1 mutation at the molecular level. This
understanding is likely to lead to new therapeutic targets and more
individualized treatment approaches for glioma.
Materials and methods
Materials
IDH1 and mIDH1 monoclonal antibodies were purchased
from YiKe Company, (ExCell Bio, Shanghai, China). Cell division
control protein 2 homolog (CDC2) and bromodomain-containing protein
2 (Brd2) monoclonal antibodies were obtained from Signalway
Antibody (College Park, MD, USA), and matrix metalloproteinase-2
(MMP-2) and −9 (MMP-9) monoclonal antibodies and fluorescent
labeling goat anti-rabbit IgG (H+L) were purchased from BioWorld
Technology, Inc. (Tulare County, CA, USA). High fidelity Platinum
Taq DNA polymerase, dNTP mix, DNA marker, primers and the
DNA Gel Extraction kit were purchased from Beijing Aoke
Biotechnology Co., (Beijing, China). Restricted incision enzymes
BamHI and SacI, G418 and T4 DNA ligase were purchased
from Gibco-BRL (Carlsbad, CA, USA). Lipofectamine 2000 transfection
reagent was purchased from Invitrogen Life Technologies (Carlsbad,
CA, USA). The U87 human glioma cell line was obtained from the
Central Laboratory of Xi’an Jiaotong University (Xi’an, China). U87
cells were propagated in Dulbecco’s modified Eagle’s medium (DMEM),
supplemented with 10% fetal bovine serum (FBS) and antibiotics, in
a humidified incubator containing 5% CO2 at 37°C.
Construction of the vectors
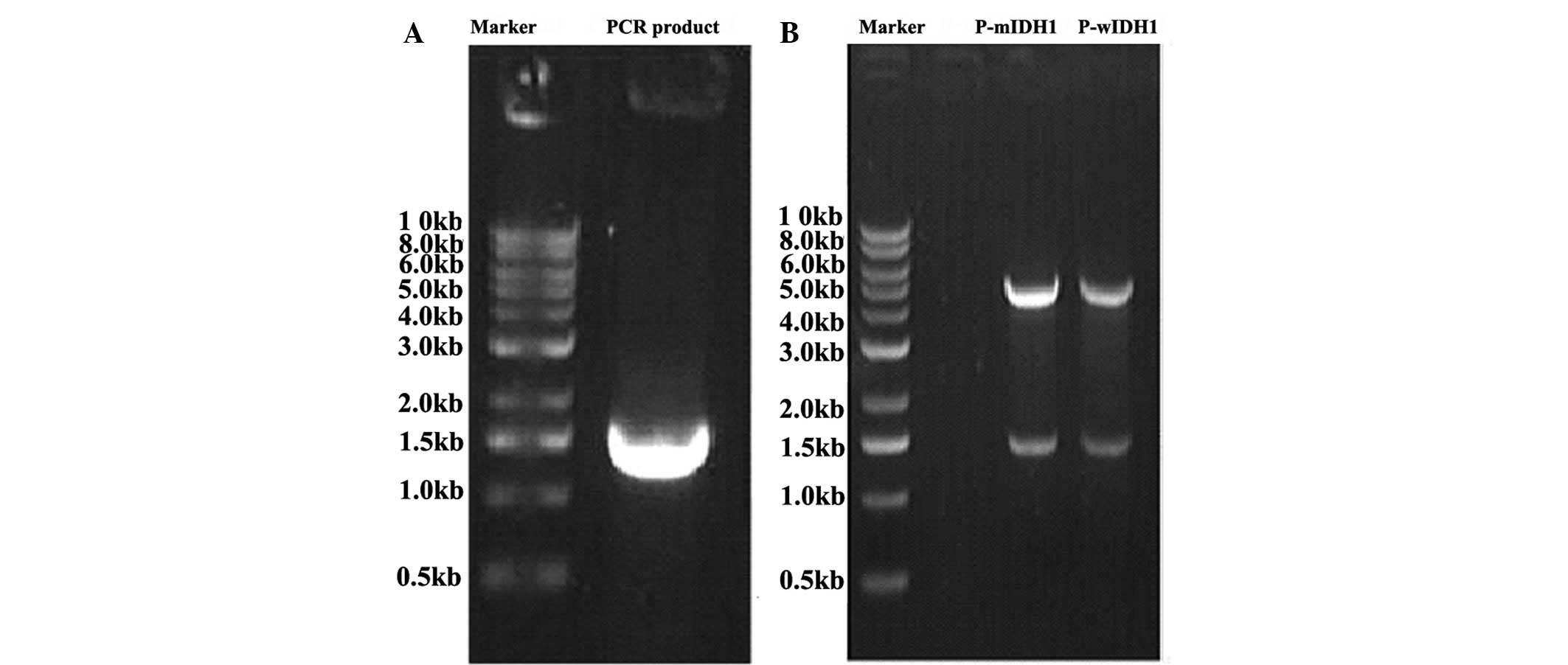
The IDH1 gene was amplified with the high-fidelity
Platinum Taq DNA polymerase and detected by 0.8% agarose gel
electrophoresis (Fig. 1A). The
primers were designed as follows: 5′-ATCGAGCTCA
GGAACTGGGGTGATAAGA-3′ (sense primer) and
5′-CGCGGATCCTTCACAAAGGTGGCAATAAC-3′ (anti- sense primer). The final
polymerase chain reaction products were cloned into the vector
p-enhanced green fluorescent protein gene (EGFP)-C1 and then
transferred into DH5α. The recombinant plasmid, named p-EGFP-wIDH1,
was treated with the gene site-directed mutagenesis kit following
the manufacturer’s instructions and amplified by the same method to
obtain p-EGFP-mIDH1. The successful construction of p-EGFP-wIDH1
and p-EGFP-mIDH1 was confirmed by agarose gel electrophoresis
(Fig. 1B) and sequence
detection.
Stable transfection and
characterization
U87 cells were transfected with vectors
p-EGFP-wIDH1, p-EGFP-mIDH1 or p-EGFP-C1 using Lipofectamine 2000
according to the stable transfection procedure. The stably
transfected U87 cells were selected by culturing with DMEM
containing FBS and 500 μg/ml G418. During the first six days, the
FBS content contained in the medium was 10%, whereas seven days
following transfection, the content was increased to 15%. The
selection lasted for approximately six weeks, until single-cell
colonies were formed. Subsequently, the colonies were cultured in
DMEM containing 10% FBS and 300 μg/ml G418. The stable cell lines
were named U87-EGFP-wIDH1, U87-EGFP-mIDH1 and U87-EGFP.
Cell morphology observation
Following digestion and culturing of the transfected
cells in six-well plates for 24 h under routine conditions, the
cells were assayed by fluoroscopy and their morphologies were
observed (GPJ9-TS100-F, Nikon, Tokyo, Japan).
Proliferation by cell counting and Cell
Counting Kit (CCK)-8 assay
Cells were adjusted to a density of 2×104
cells/ml and seeded and cultured under routine conditions in
six-well plates (4×104 cells per well). Each group of
cells was divided into three parallel samples and trypsinized on
days 1, 3 and 5 of cell culture. The cell number of each parallel
sample was determined by direct cell counting, using a
hemocytometer, and the relative growth rate was calculated.
Cell-cycle distribution analysis
Cells were cultured in 6-well plates for 24 h,
harvested and fixed in ice-cold 70% (v/v) ethanol for 24h at 4°C.
They were then washed twice with ice-cold phosphate-buffered saline
(PBS) and centrifuged at 90 × g for 5 min, followed by treatment
with 1 mg/ml RNase for 30 min at 37°C. Following staining with 40
μl propidium iodide (0.1 μg/ml), >10,000 cells per sample were
subjected to flow cytometric analysis.
Flow cytometry for apoptosis
determination by the Annexin V-red fluorescent protein (RFP)
method
Cells were cultured under routine conditions in a
6-well plate. To determine the number of apoptotic cells, Annexin
V-RFP assays were performed using an apoptosis detection kit
(Annexin V-RFP Apoptosis Detection kit, Shanghai Ruisai Company,
Shanghai, China). At 24 and 48 h time points of the incubation
period, cells were harvested and treated according to the
manufacturer’s instructions of the Annexin V-RFP kit, and analyzed
within 30 min by flow cytometry (BD FACSCanto™ Flow Cytometer, BD
Biosciences, New Jersey, NY, USA).
Transwell assay for migration and
invasion ability determination
Cells (2,000 per well) were plated in the upper
chamber of a Transwell insert (BD Biosciences, San José, CA, USA)
in serum-free DMEM. FBS (10%) served as a chemotactic agent and
cells were allowed to migrate for 12 h. The migrated cells were
subsequently fixed and stained. Values for migration were obtained
by counting the migrated cells under an inverted microscope.
Results were expressed as the number of cells identified per random
microscope field [mean ± standard deviation (SD)]. To determine the
values for invasion, 1×104 cells were seeded in an 8-μm
pore polycarbonate membrane chamber inserted in a Transwell insert
coated with Matrigel. FBS (15%) served as a chemotactic agent.
After 24 h, the migrated cells were fixed, stained and subjected to
microscopic inspection. Values for invasion were obtained by cell
counting under the inverted microscope.
Immunofluorescence assay
Cells were seeded on the cover slips in a 24-well
plate. After 24 h, cover slips were fixed in 4% paraformaldehyde,
rinsed and blocked with 1% bovine serum albumin (BSA). The first
antibody (diluted with 1% BSA) was added and incubated overnight at
4°C. Next, cover slips were washed twice with PBS and secondary
antibody was added and incubated for 30 min at 37°C. Subsequently,
the slides were submitted for fluorescence microscopy
observation.
Western blot analysis
Total protein from the three cell lines was
separated electrophoretically in 4–12% SDS-PAGE gels (Ronbio
Scientific, Shanghai, China) and transferred to nitrocellulose
membranes. Following blocking with 5% non-fat milk for 1 h, the
membranes were incubated with the primary antibodies (wild IDH1,
mutated IDH1, CDC2, Brd2, MMP-2, MMP-9, β-actin and GAPDH) at 4°C
overnight, followed by a 1-h incubation with horseradish
peroxidase-conjugated goat-anti-mouse and goat-anti-rabbit
secondary antibodies. The membranes were washed thoroughly with
Tris-buffered saline containing Tween 20, following each treatment
with the antibodies. The bands were visualized by a
chemiluminescence method. Data collection and processing were
performed using a LAS-3000 luminescent image analyzer (Fujifilm,
Tokyo, Japan).
Statistical analysis
The relative variation rate was statistically
analyzed using SPSS 13.0 software (SPSS Inc., Chicago, IL, USA).
Data are presented as the mean ± standard deviation. Comparison
between groups was performed by one-way analysis of variance. The
Student-Newman-Keuls test was used to evaluate the differences
between multiple groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Stable cell line determination and
morphologies of the three stable cell lines
Positive GFP signals were observed in the three
stable cell lines (U87-EGFP-wIDH1, U87-EGFP-mIDH1 and U87-EGFP),
indicating that the target genes had been successfully transfected.
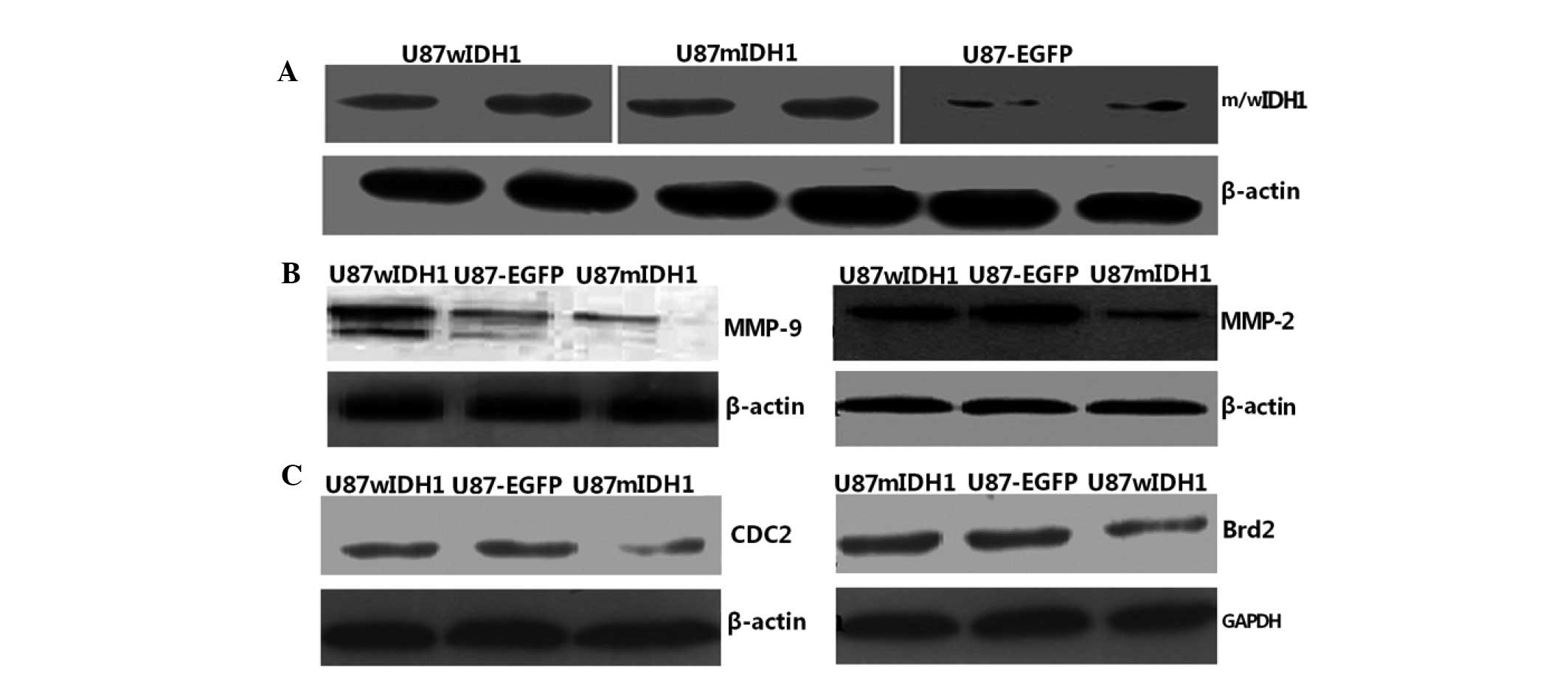
Western blotting indicated that the wIDH1 and mIDH1 proteins had
been successfully expressed (Fig.
2A). The U87-EGFP-wIDH1 cells exhibited a homogeneous
morphology during the primary selection period. These cells
characteristically possessed relatively compact, small nuclei and
abundant cytoplasm. In addition, the majority of U87-EGFP-wIDH1
cells exhibited a spindle-shaped form, as observed in the original
U87 cell line. However, the U87-EGFP-mIDH1 cell line tended to
exhibit larger nuclei of various sizes during primary selection and
appeared to be more immature. The three cell lines demonstrated no
marked differences in morphology when primary selection was
complete.
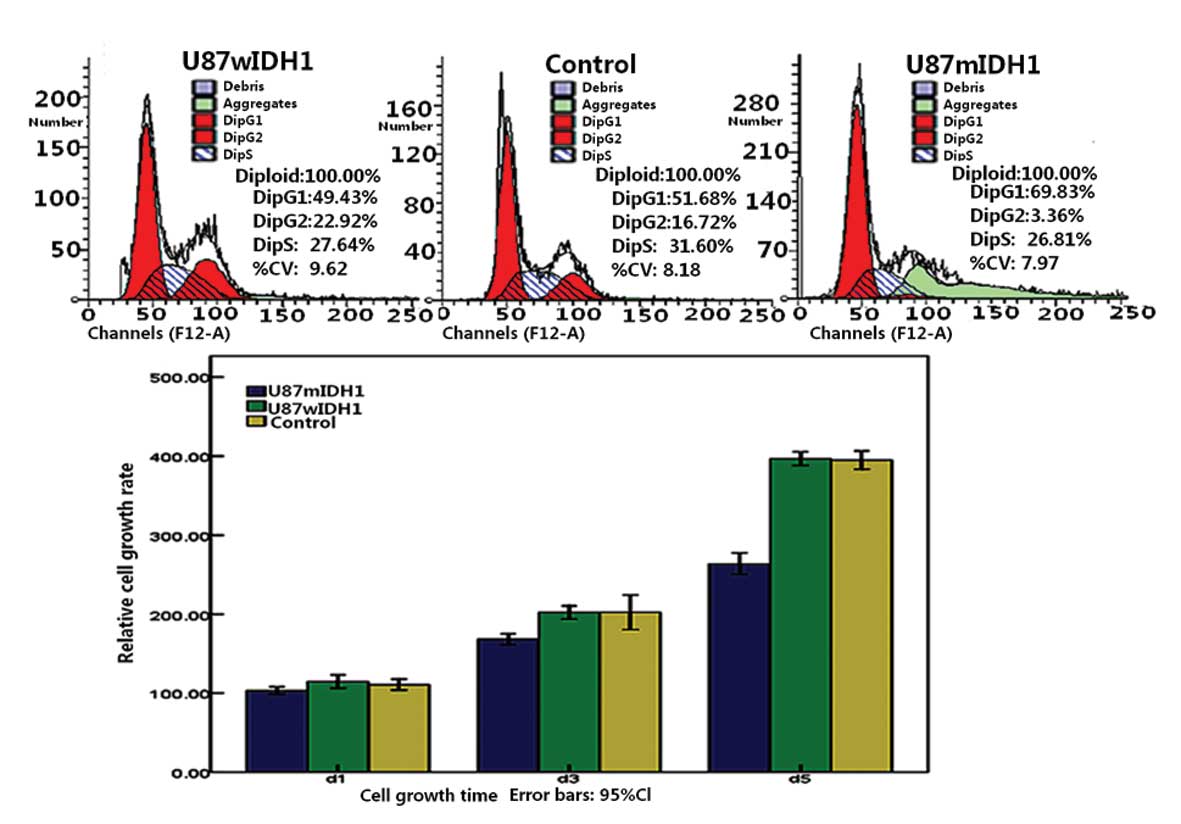
mIDH1 downregulates the proliferation of
U87 cells
The relative growth rates of the three cell lines
are shown in Fig. 3D. It was found
that U87-EGFP-mIDH1 cells had a lower growth rate than the other
two groups (P<0.01). There was no significant difference between
U87-EGFP-wIDH1 and U87-EGFP cells (P>0.05). The CCK-8 assay
exhibited similar results. These results indicate that mIDH1 may
function as an inhibitor of glioma cell growth, while wIDH1 did not
promote further cell growth.
mIDH1 alters the cell cycle of the U87
cell line
Cell cycle analysis showed that U87-EGFP-mIDH1 had a
higher number of cells in the G1 phase following cell culture for
24 h. The number of U87-EGFP-mIDH1 cells arrested in the G1 phase
increased and those in the G2/M stage decreased, compared with
U87-EGFP-wIDH1 and U87-EGFP cells. wIDH1 cells demonstrated a
non-significant difference in the cell cycle from the U87-EGFP
controls (Fig. 3A–C).
mIDH1 does not affect the apoptosis rates
under routine conditions
The Annexin V/RFP method indicated that the
apoptosis rates of all the three groups varied by ~1%. There were
no significant differences between the three groups under routine
culture conditions (P>0.05), indicating that mIDH1 had no effect
on the apoptosis rate of glioma cells.
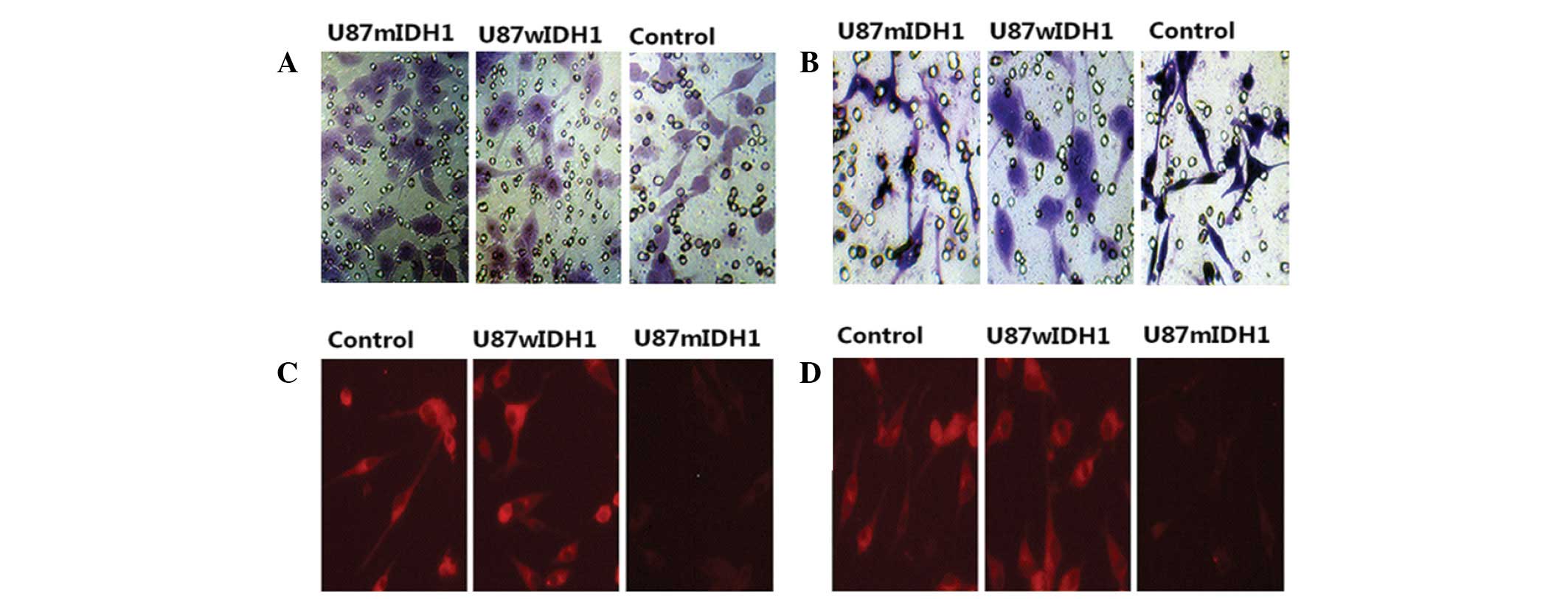
mIDH1 increases cell migration and
reduces the cell invasion
The migration and invasion abilities of cells were
measured by serum chemotaxis. The numbers of U87-EGFP-mIDH1,
U87-EGFP-wIDH1 and U87-EGFP cells that migrated into the membranes
were ~125.5±8.7, ~78.5±7.4 and ~73.8±8.2 cells per microscopic
field (Fig. 4A). The results
indicated that the mutated cells had a higher migration ability
than the other two groups (P<0.01). No significant difference
was detected in the migration rate between U87-EGFP-wIDH1 cells and
the control cells (P>0.05). However, IDH1 mutation reduced the
invasion ability of glioma cells. A small number of U87-EGFP-mIDH1
cells (28.3±4.6) invaded the membrane, compared with the other two
groups of cells (P<0.01; Fig.
4B). Among the three groups, the morphology of the
U87-EGFP-mIDH1 cells changed the most, following invasion of the
membrane. U87-EGFP-wIDH1 cells (65.4±6.8) demonstrated little
difference in invasion ability compared with the control cells
(68.2±7.6) (P>0.05).
mIDH1 induces downregulation of CDC2
levels and upregulation of Brd2 levels
Western blotting and gray-scale scanning analysis
revealed that U87-EGFP-mIDH1 expressed relatively low CDC2 levels
compared with the other two cell lines (P<0.01; Fig. 2C). No significant difference was
found in the CDC2 levels between the U87-EGFP-wIDH1 and U87-EGFP
cells. U87-EGFP-mIDH1 exhibited higher Brd2 protein expression than
the other two cell lines (P<0.01).
mIDH1 reduces downregulation of MMP-2 and
MMP-9 levels
Immunofluorescence (Fig.
4C and D) and western blotting (Fig. 2B) demonstrated that U87-EGFP-mIDH1
exhibited low expression levels of MMP-2 (Fig. 4C) and MMP-9 (Fig. 4D) when compared with the other two
cell lines.
Discussion
IDH has an important role in cell metabolism. Though
different IDH enzymes may have independent functions, they are all
related to the NADPH pool of cells according to the activities of
lipogenesis, antioxidation and immune system response (11,12).
IDH function disorders may cause changes to the NADPH pool and lead
to cell metabolism disorders (10).
IDH1 mutation in glioma has attracted much
attention. Mutation may alter the normal cell metabolism mechanisms
and prevent complete conversion of isocitrate to α-ketoglutarate.
In addition, mutation has been shown to cause 2-hydroxyglutarate
and hypoxia inducible factor-1α accumulation, leading to the added
risk of tumorigenesis (13). This
characteristic may be attributed to oncogene, but this has not been
widely validated. Furthermore, these observations do not correspond
with the high incidence seen with specific congenital and metabolic
disorders. A recent study suggested that the metabolic levels of
2-hydroxyglutarate were not associated with tumorigenesis or tumor
malignancy (14). Therefore,
further studies are needed to elucidate the specific role of IDH1
mutation in tumorigenesis. Due to IDH1 mutation, glioma is
associated with two subtypes which bring about distinctly different
prognoses (7,15,16).
An increasing number of studies have revealed that IDH1 mutation
carriers have an improved prognosis, meaning that IDH1 mutations
may confer a protective effect. A study by Zhu et al was
consistent with these observations (17). The present study focused on the
biological changes induced by IDH1 mutation in glioma cells. The
mechanisms underlying the effect of IDH1 mutation on the enhanced
prognosis of glioma patients was also investigated, in the hope of
providing clues to improve glioma therapy.
Cell cycle control is regulated by checkpoint
control via cell cycle regulatory proteins, including cyclins and
cyclin-dependent kinases. This process is an important means of
inhibiting cancer cell growth and division (18). CDC2 is a fission yeast CDC2 gene.
Cells are unable to enter mitosis without CDC2 activity, which
implies that CDC2 is a key regulator of fission yeast mitosis. A
number of drugs have been designed as antitumor drugs for targeting
CDC2 (19). Qiao et al
previously reported that DAT-230 reduces CDC2 levels and induces
G2/M phase arrest and apoptosis in tumor cells (20). Cell cycle analysis in the present
study showed that IDH1 mutation led to reduced CDC2 levels and
caused increased G1 phase length and reduced S and G2 phases.
Downregulated CDC2 levels may lead to phosphorylation deficiencies
of histone H1 and nuclear lamins, hinder mitotic spindle formation
and further inhibit the cell cycle. CDC2 downregulation markedly
reduced cell proliferation in the present study, which is
consistent with a study by Hogan et al (21). U87-EGFP-wIDH1 and U87-EGFP cells
exhibited a relatively normal distribution within the cell cycle
stages. Compared with U87-EGFP, U87-wIDH1 had a slightly increased
G2 phase, which is likely to be due to the different patterns of
energy metabolism between the two cell lines. This result implies
that the IDH1 gene may play a complementary role in cellular
mitosis.
Cell cycle regulation is a complex process involving
multiple factors. Brd2 belongs to the bromo and extra terminal
(BET) protein family and has been implicated in fundamental
cellular processes, including cell cycle and transcriptional
regulation. Brd2 is closely associated with the cell cycle
transcription factors, E2F1 and E2F2 (22). Ottinger et al have
hypothesized that a BET protein, in combination with the MHV-68
orf73 protein, activates the promoters of G1/S cyclins, while gene
mutation in one binding site inhibits the interaction between Brd2
and E2F and reduces the responses of the BET protein to the
promoters of cyclin D1, D2 and E (23). Ectopic expression of Brd2 has been
found to inhibit S phase progression and induce G1 cell cycle
arrest or exit and Brd2 knockdown promotes S phase entry (24). These observations are consistent
with findings of the present study. In this study, IDHI mutation
resulted in an increased proportion of cells in the G1 phase and
increased Brd2 levels in glioma cells. A previous study has shown
that Brd2-deficient embryonic fibroblast cells proliferate more
slowly than those of wild-type fibroblast cells (25). The present study demonstrated that
glioma cells with mutated IDH1 had higher Brd2 levels and
proliferated more slowly than the other two groups. The U87-mIDH1
cells exhibited a lower proliferation rate than the U87-wIDH1
cells, which is likely to be due to blockage of the U87-mIDH1 cell
cycle. From this, it is possible to conclude that IDH1 mutation
plays an inhibitory role in cell proliferation, due, in part, to
blockage of the cell cycle. This may partly explain why glioma
patients with mutated IDH1 have a longer survival period. Although
IDH1 mutation may not be used as an independent factor for improved
prognosis, it is likely to block the cell cycle and to inhibit cell
proliferation.
MMPs, a family of proteolytic enzymes, help increase
the invasion potential of tumor cells by remodeling the
extracellular matrix. MMP expression has been found to be
associated with tumor progression and survival in several types of
human cancers. In the present study, cells with mutated IDH1
exhibited low expression of MMP-2 and MMP-9 and low invasion
ability, compared with the other two groups. This was shown by a
Transwell invasion experiment. From this perspective, mutated IDH1
cannot be regarded simply as an oncogene. As glioma patients with
mutated IDH1 have a low capacity for degrading various components
of the extracellular matrix, surgery may result in increased
opportunity for invasion of glioma cells. This may partly explain
why the pathological diagnoses of specific patients are promoted to
a higher grade following the initial operation. However, in the
present study, IDH1 mutation induced changes in the biological
behavior of glioma cells and in the mechanism of tumorigenesis.
IDH1 mutation downregulated glioma cell proliferation by blocking
the cell cycle, reduced the cell invasion ability via MMP-2 and
MMP-9 downregulation and promoted the cell migration ability. There
is reason to believe that glioma patients with mutated IDH1 may
have a better prognosis due to blockage of the cell cycle and
inhibition of cell invasion ability.
However, IDH1 mutation cannot be regarded as
exclusively responsible for this enhanced prognosis. The apoptosis
rate did not increase accordingly in the present study; although,
when treated by chemotherapy and radiotherapy, the result may be
different. This is since normal IDH1 enzymes contribute
significantly to the NADPH levels and provide considerable
protection against oxidative stress from chemotherapy and
radiation, which is of great importance in ensuring normal IDH1
regulation. Evidence of this has been identified in several other
tumors (26). Mutated IDH1 may fail
to provide more energy to repair the oxidative stress injury from
chemotherapy and radiation. Mateescu et al (27) indicated that although oxidative
stress promotes tumor growth, it also sensitizes tumors to
treatment.
In conclusion, the present study demonstrated that
IDH1 mutation blocks the cell cycle and inhibits cell proliferation
by downregulating CDC2 levels and increasing Brd2 levels. IDH1
mutation was markedly associated with a significantly reduced
invasion ability, by reducing the levels of MMP-2 and MMP-9. This
study is expected to provide a partial explanation for the improved
prognosis of patients with mutated IDH1 genes. It may be concluded
that IDH1 mutation improves the prognosis of glioma patients by
altering the cell cycle, inhibiting cell proliferation and
downregulating cell invasion ability.
Acknowledgements
The authors thank the Central Laboratory of Xi’an
Jiaotong University for assistance.
References
|
1
|
Wakabayashi T: Clinical trial updates for
malignant brain tumors. Rinsho Shinkeigaku. 51:853–856. 2011.(In
Japanese).
|
|
2
|
Ostrom QT, Gittleman H, Farah P, Ondracek
A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: primary brain and central nervous system
tumors diagnosed in the United States in 2006–2010. Neuro Oncol.
15(Suppl 2): 1–56. 2013.PubMed/NCBI
|
|
3
|
Ohgaki H: Epidemiology of brain tumors.
Methods Mol Biol. 472:323–342. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Combs SE, Gutwein S, Schulz-Ertner D, van
Kampen M, Thilmann C, Edler L, et al: Temozolomide combined with
irradiation as postoperative treatment of primary glioblastoma
multiforme. Phase I/II study. Strahlenther Onkol. 181:372–377.
2005. View Article : Google Scholar
|
|
5
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Onishi M, Ichikawa T, Kurozumi K and Date
I: Angiogenesis and invasion in glioma. Brain Tumor Pathol.
28:13–24. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Parsons DW, Jones S, Zhang X, Lin JC,
Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et
al: An integrated genomic analysis of human glioblastoma
multiforme. Science. 321:1807–1812. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hathaway JA and Atkinson DE: The effect of
adenylic acid on yeast nicotinamide adenine dinucleotide isocitrate
dehydrogenase, a possible control mechanism. J Biol Chem.
238:2875–2881. 1963.
|
|
9
|
Barnes LD, McGuire JJ and Atkinson DE:
Yeast diphosphopyridine nucleotide specific isocitrate
dehydrogenase. Regulation of activity and unidirectional catalysis.
Biochemistry. 11:4322–4329. 1972. View Article : Google Scholar
|
|
10
|
Dang L, White DW, Gross S, Bennett BD,
Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et
al: Cancer-associated IDH1 mutations produce 2-hydroxyglutarate.
Nature. 462:739–744. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Geer BW, Kamiak SN, Kidd KR, Nishimura RA
and Yemm SJ: Regulation of the oxidative NADP-enzyme tissue levels
in Drosophila melanogaster. I. Modulation by dietary
carbohydrate and lipid. J Exp Zool. 195:15–32. 1976. View Article : Google Scholar
|
|
12
|
Wilton AN, Laurie-Ahlberg CC, Emigh TH and
Curtsinger JW: Naturally occurring enzyme activity variation in
Drosophila melanogaster. II. Relationships among enzymes.
Genetics. 102:207–221. 1982.PubMed/NCBI
|
|
13
|
Aghili M, Zahedi F and Rafiee E:
Hydroxyglutaric aciduria and malignant brain tumor: a case report
and literature review. J Neurooncol. 91:233–236. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Capper D, Simon M, Langhans CD, Okun JG,
Tonn JC, Weller M, von Deimling A and Hartmann C; German Glioma
Network. 2-Hydroxyglutarate concentration in serum from patients
with gliomas does not correlate with IDH1/2 mutation status or
tumor size. Int J Cancer. 131:766–768. 2012. View Article : Google Scholar
|
|
15
|
Nobusawa S, Watanabe T, Kleihues P and
Ohgaki H: IDH1 mutations as molecular signature and predictive
factor of secondary glioblastomas. Clin Cancer Res. 15:6002–6007.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yan H, Parsons DW, Jin G, McLendon R,
Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ,
et al: IDH1 and IDH2 mutations in gliomas. N Engl J Med.
360:765–773. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu J, Zuo J, Xu Q, Wang X, Wang Z and
Zhou D: Isocitrate dehydrogenase mutations may be a protective
mechanism in glioma patients. Med Hypotheses. 76:602–603. 2011.
View Article : Google Scholar
|
|
18
|
Chen YN, Chen JC, Yin SC, Wang GS, Tsauer
W and Hsu SL: Effector mechanisms of norcantharidin-induced mitotic
arrest and apoptosis in human hepatoma cells. Int J Cancer.
100:158–165. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim DI, Lee SJ, Lee SB, Park K, Kim WJ and
Moon SK: Requirement for Ras/Raf/ERK pathway in naringin-induced
G1-cell-cycle arrest via p21WAF1 expression. Carcinogenesis.
29:1701–1709. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qiao F, Zuo D, Shen X, Qi H, Wang H, Zhang
W and Wu Y: DAT-230, a novel microtubule inhibitor, exhibits potent
anti-tumor activity by inducing G2/M phase arrest, apoptosis in
vitro and perfusion decrease in vivo to HT-1080. Cancer Chemoth
Pharmacol. 70:259–270. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hogan FS, Krishnegowda NK, Mikhailova M
and Kahlenberg MS: Flavonoid, silibinin, inhibits proliferation and
promotes cell-cycle arrest of human colon cancer. J Surg Res.
143:58–65. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Denis GV, Vaziri C, Guo N and Faller DV:
RING3 kinase transactivates promoters of cell cycle regulatory
genes through E2F. Cell Growth Differ. 11:417–424. 2000.
|
|
23
|
Ottinger M, Pliquet D, Christalla T, Frank
R, Stewart JP and Schulz TF: The interaction of the
gammaherpesvirus 68 orf73 protein with cellular BET proteins
affects the activation of cell cycle promoters. J Virol.
83:4423–4434. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang F, Liu H, Blanton WP, Belkina A,
Lebrasseur NK and Denis GV: Brd2 disruption in mice causes severe
obesity without type 2 diabetes. Biochem J. 425:71–83. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shang E, Wang X, Wen D, Greenberg DA and
Wolgemuth DJ: Double bromodomain-containing gene Brd2 is essential
for embryonic development in mouse. Dev Dyn. 238:908–917. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Woolston CM, Zhang L, Storr SJ, Al-Attar
A, Shehata M, Ellis IO, Chan SY and Martin SG: The prognostic and
predictive power of redox protein expression for
anthracycline-based chemotherapy response in locally advanced
breast cancer. Modern Pathol. 25:1106–1116. 2012. View Article : Google Scholar
|
|
27
|
Mateescu B, Batista L, Cardon M, Gruosso
T, de Feraudy Y, Mariani O, Nicolas A, Meyniel JP, Cottu P,
Sastre-Garau X and Mechta-Grigoriou F: miR-141 and miR-200a act on
ovarian tumorigenesis by controlling oxidative stress response.
Nature Med. 17:1627–1635. 2011. View
Article : Google Scholar : PubMed/NCBI
|