Introduction
Osteosarcoma is the most common type of primary bone
cancer (1). It has been documented
that osteosarcoma has a bimodal age-associated pattern of
distribution with the first peak occurring during the period of
puberty and the second peak at >65 years old (2–4).
Although the incidence of osteosarcoma in childhood and adolescence
is relatively consistent worldwide (5,6),
geographic- and ethnicity-associated variations are evident in the
incidence rates of osteosarcoma in other age groups, particularly
in the elderly (≥60 years), ranging from 0 cases/million
individuals in Kuwait to 11.8 cases/million individuals in the
Philippines.
Significant advances have been made recently in
diagnostic and palliative local treatments (such as isolated limb
perfusion, radiation therapy, embolization, chemoembolization,
thermal ablation and cryoablation) of osteosarcoma (7). As a result, the outcome for patients
with non-metastatic osteosarcoma has been markedly improved
(8). However, patients with a
recurrent metastatic disease at diagnosis or with a recurrent
disease have a poor prognosis, with the long-term survival rate
being only 20% (9). It is therefore
crucial to explore innovative strategies in order to effectively
manage metastatic and recurrent osteosarcoma.
Therapies targeting key molecules in tumorigenesis
have produced exciting and promising developments for cancer
treatment in recent years. Recent evidence has indicated that the
interferon-inducible p200 (IFI-200) family of proteins may serve as
potential therapeutic targets and/or diagnostic biomarkers
(10). Consisting of six members
(Ifi202a, Ifi202b, Ifi203, Ifi204, Mndal and Aim2) in mice and four
homologues, (IFI16, MNDA, IFIX and AIM2) in humans, the IFI-200
family proteins share a high homology each with a conserved domain
of 200 amino acids (11,12) and possess antimicrobial, cell growth
regulatory, differential regulatory and immunomodulatory properties
(13,14). Human and animal studies have
demonstrated that the expression of MNDA is downregulated in
myelodysplastic syndrome (15), and
is markedly (1,000-fold) increased in a mouse strain resistant to
B-cell plasmacytoma but completely absent in another mouse strain
susceptible to B-cell plasmacytoma (16). The above observations suggest that
MNDA is a tumor suppressor. However, the role of MNDA in bone
tumors has not been established. The objective of this study was to
determine the effect of MNDA overexpression on osteosarcoma cell
proliferation and migration/invasiveness in an in vitro cell
culture system.
Materials and methods
Cell culture
An osteosarcoma cell line, sarcoma osteogenic
(Saos-2), was purchased from the Shanghai Academy of Life Sciences
(Shanghai, China). For maintenance, the cells were cultured in DMEM
(Invitrogen, Grand Island, NY, USA) and supplemented with 10% fetal
bovine serum (FBS), 1% l-glutamine and 1% penicillin/streptomycin
at 37°C in a humidified 5% CO2 atmosphere.
Determination of endogenous MNDA mRNA
abundance in wild-type Saos-2 cells
In order to analyze the expression level of
endogenous MNDA in Saos-2 cells, MNDA mRNA abundance was determined
by reverse transcription-polymerase chain reaction (RT-PCR) with
the human monocyte-like U937 cell line included as a positive
control. Total RNA was extracted from wild-type Saos-2 cells and
U937 cells using TRIzol reagent (Life Technologies, Grand Island,
NY, USA). First-strand cDNAs were synthesized with the PrimeScript
1st strand cDNA Synthesis kit from Takara (Tokyo, Japan) according
to the manufacturer’s instructions. PCR amplification of the test
gene MNDA and the internal control gene 3-glyceraldehyde-phosphate
dehydrogenase (GAPDH) were performed with the Premix Taq version
2.0 kit (Takara), using the following reaction conditions: initial
denaturation at 95°C for 5 min, followed by 32 main cycles (94°C
for 45 sec, 60°C for 45 sec, 72°C for 45 sec), and one final
extension cycle at 72°C for 10 min. PCR products were separated by
electrophoresis on a 1.5% agarose gel. MNDA and GAPDH bands were
visualized by ethidium bromide staining. The MNDA primer sequences
used were: sense, 5′-CCACCGCAAGAAACAAAACTGACATCGG-3′ and antisense,
5′-TAAATGGCGCTGTTGCTTTCAGTAC CAC-3′, and the GAPDH primer sequences
were: sense, 5′-TGTTGCCATCAATGCCCCTT-3′ and antisense, 5′-CTCC
ACGACGTACTCAGCG-3′.
Expression vector construction
Total cellular RNA was extracted from the U937 cell
line (ATCC, Manassas, VA, USA) with TRIzol reagent according to the
manufacturer’s instructions. Full-length MNDA cDNA was synthesized
by RT and amplified by PCR using a pair of primers designed based
upon the human MNDA gene sequence (GenBank accession NM_002432.1)
and cloned into pIRES2-enhanced green fluorescent protein (EGFP)
(Clontech Laboratories, Inc., Mountain View, CA, USA). Positive
pIRES2-EGFP-MNDA clones were selected and confirmed by restriction
enzyme mapping and DNA sequencing.
MTT assay
Saos-2 cells were seeded in triplicate in a 96-well
plate at 1×103 cells/well in FBS-containing medium. The
next day the cells were transfected with the vehicle (control),
pIRES2-EGFP and pIRES2-EGFP-MNDA, respectively, using Lipofectamine
2000 (Life Technologies) according to the manufacturer’s
instructions. At 12, 24, 48 and 72 h following transfection, the
DNA-lipofectamine-containing medium was removed and 20 μl (5 mg/ml)
3-(4,5-dimethylthiazol-2-yl-2,5-diphenyltetrazolium bromide (MTT)
(Sigma-Aldrich, St. Louis, MO, USA) solution was added to each
well. Following incubation in the dark for 4 h, the MTT-containing
medium was removed and 150 μl DMSO (Sigma-Aldrich) was added.
Following 15 min of agitation, the plate was read on an automated
plate reader (Perkin-Elmer, Waltham, MA, USA) and the optical
density (OD) at 490 nm (OD490) was obtained for each
well. The percentage growth inhibition in Saos-2 cells transiently
transfected with pIRES2-EGFP or pIRES2-EGFP-MNDA was calculated as
follows: percentage growth inhibition = (control OD490 -
test OD490)/control OD490 × 100. Where the
control OD490 was the average OD490 of the
triplicate wells of control Saos-2 cells, while the test
OD490 was the average OD490 of the triplicate
wells of Saos-2 cells transfected with pIRES2-EGFP or
pIRES2-EGFP-MNDA. Experiments were repeated three times.
Flow cytometric analysis of apoptosis and
cell cycle
Saos-2 cells were seeded in 6-cm dishes at
1×105/well and transfected with the vehicle (control),
pIRES2-EGFP and pIRES2-EGFP-MNDA, respectively, as described
previously (10). At 48 h following
transfection, the transfectants were collected and suspended in PBS
in a Falcon® 12 × 75 mm tube. Following washing with
PBS, the cells were fixed in ethanol and stained with 50 μg/ml
Annexin V-fluorescein isothiocyanate (FITC) (BD Biosciences, San
Jose, CA, USA) and 20 μl of 500 μg/ml propidium iodide (PI)
(Sigma-Aldrich) at 4°C overnight. Stained cells were analyzed by
flow cytometry (Ex, 488 nm; Em, 530 nm). Experiments were performed
three times.
Transwell invasion assay
Saos-2 cells were seeded onto the Matrigel-coated
upper chambers (inserts) of a Corning 24-well Transwell plate
(Sigma-Aldrich) at 5×104 cells/well and transfected with
the vehicle and expression vectors pIRES2-EGFP and pIRES2-EGFP-MNDA
in serum-free DMEM, as described previously (15). The lower chambers of the plate were
filled with DMEM with 10% FBS as a chemoattractant. After being
cultured at 37°C in 5% CO2 for 24 h, the insert was
carefully removed. Cells that did not migrate through the pores and
remained on the upper side of the filter membrane were gently
removed with a cotton swab. Cells on the lower side of the filter
insert were quickly fixed in 5% glutaraldehyde for 10 min and
stained with crystal violet (Sigma-Aldrich). The number of cells on
the lower side of the filter insert were counted under a light
microscope (Nikon, Melville, NY, USA). Triplicate wells were used
for the vehicle transfection and each of the expression vectors
respectively. The experiments were each repeated three times.
Statistical analysis
Data were expressed as the means ± SD and analyzed
using the Student’s t-test using statistical analysis software SPSS
12.0 (SPSS Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Endogenous MNDA mRNA abundance in
wild-type Saos-2 cells
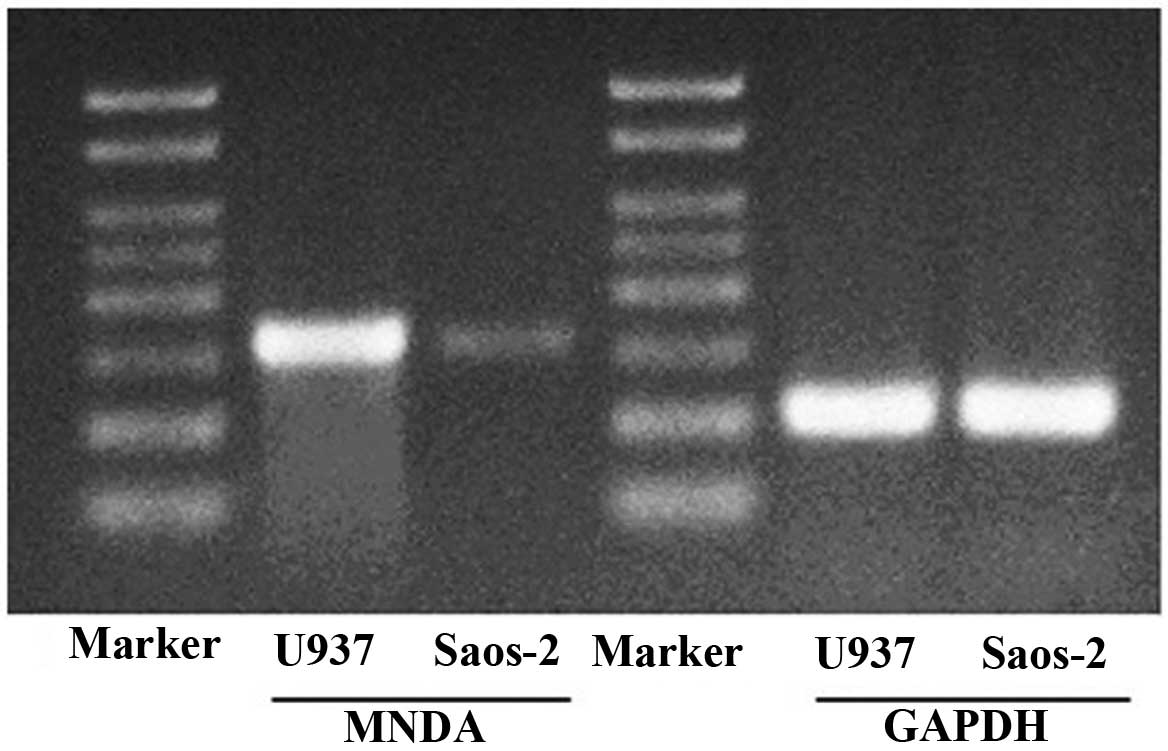
Abundant MNDA mRNA was detected in U937 cells.
Compared with the U937 cells, Saos-2 cells had a lower abundance of
MNDA mRNA (Fig. 1).
Effect of MNDA overexpression on the
proliferation of osteosarcoma cells
In order to determine the effect of MNDA on the
proliferation of osteosarcoma cells and cell viability in Saos-2
cells transiently transfected with the vehicle, pIRES2-EGFP-MNDA
and pIRES2-EGFP, respectively, cells were assessed by MTT assay.
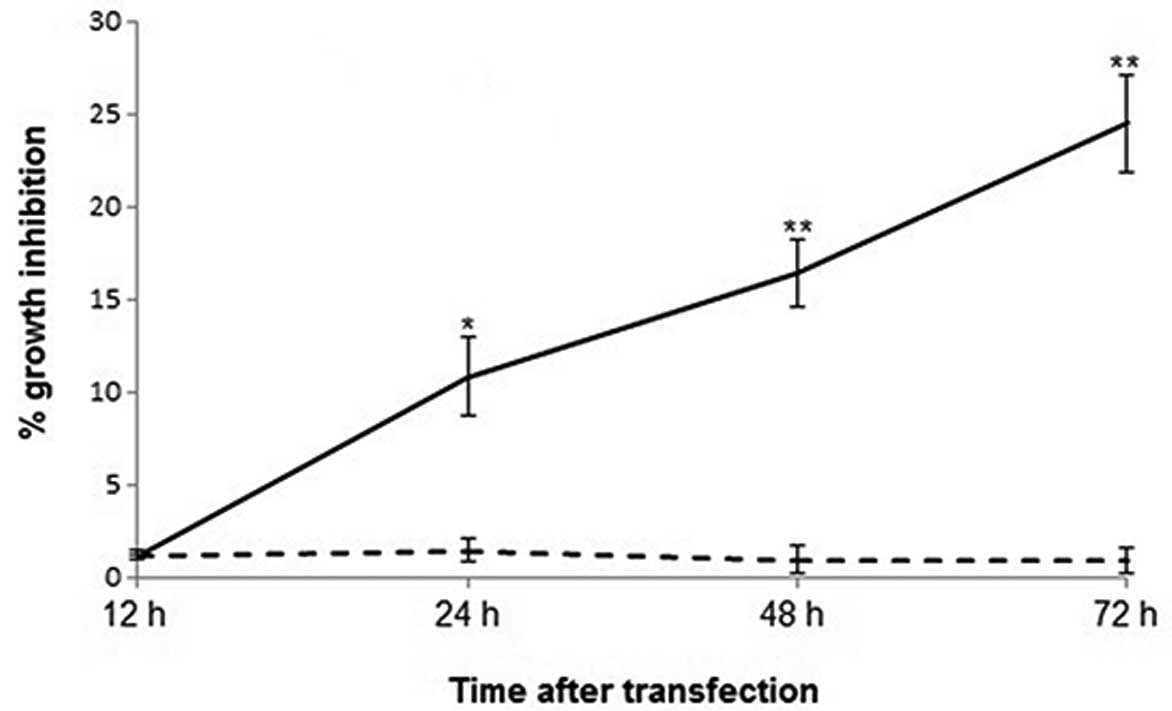
Overexpression of MNDA resulted in a time-dependent increase in the
growth inhibition of Saos-2 cells (Fig.
2). At 12 h following transfection, there was no significant
difference in cell viability between pIRES2-EGFP-MNDA- and
pIRES2-EGFP-transfected Saos-2 cells (P>0.05). However, the
percentage growth inhibition calculated against wild-type Saos-2
cells became significant 24 h after transfection. For
pIRES2-EGFP-MNDA- and pIRES2-EGFP-overexpressing Saos-2 cells
(10.87 vs. 1.51%, P<0.05), the difference became significant 48
h (16.43 vs. 1.01%, P<0.01) and 72 h (24.56 vs. 0.96%,
P<0.01) after transfection, respectively.
Effect of MNDA overexpression on
osteosarcoma cell apoptosis
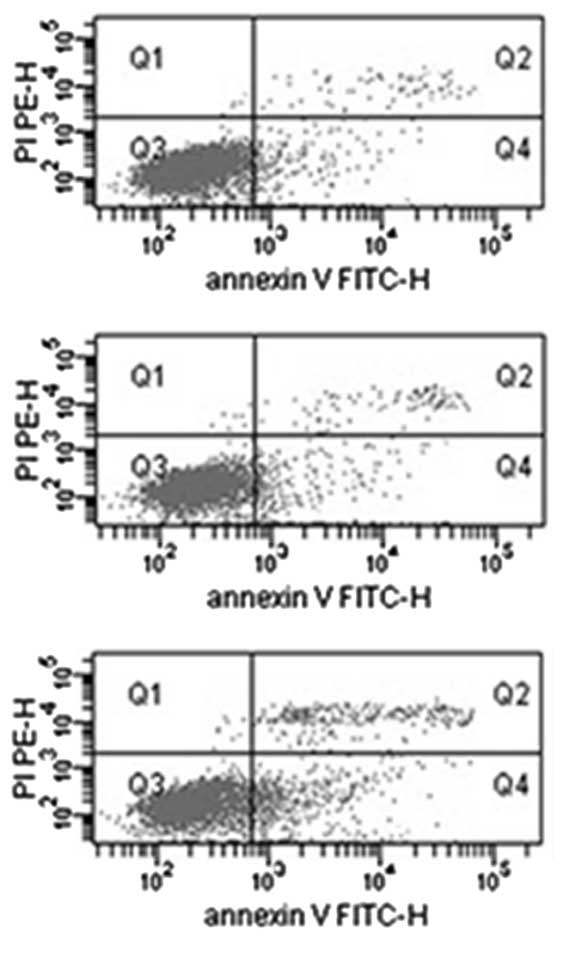
Fig. 3 shows
representative flow cytometric scatter plots of wild-type,
pIRES2-EGFP-MNDA- and pIRES2-EGFP-transfected Saos-2 cells
double-stained with Annexin V-FITC and PI, showing the number of
apoptotic cells 48 h after transfection. When the data from three
replicated experiments were analyzed, the average percentage of
apoptotic cells was significantly higher (P<0.05) in Saos-2
cells transfected with pIRES2-EGFP-MNDA (17.0%) compared with the
wild-type Saos-2 cells (5.7%) and Saos-2 cells transfected with
pIRES2-EGFP (5.8%).
Effect of the MNDA overexpression
osteosarcoma cell migration/invasion
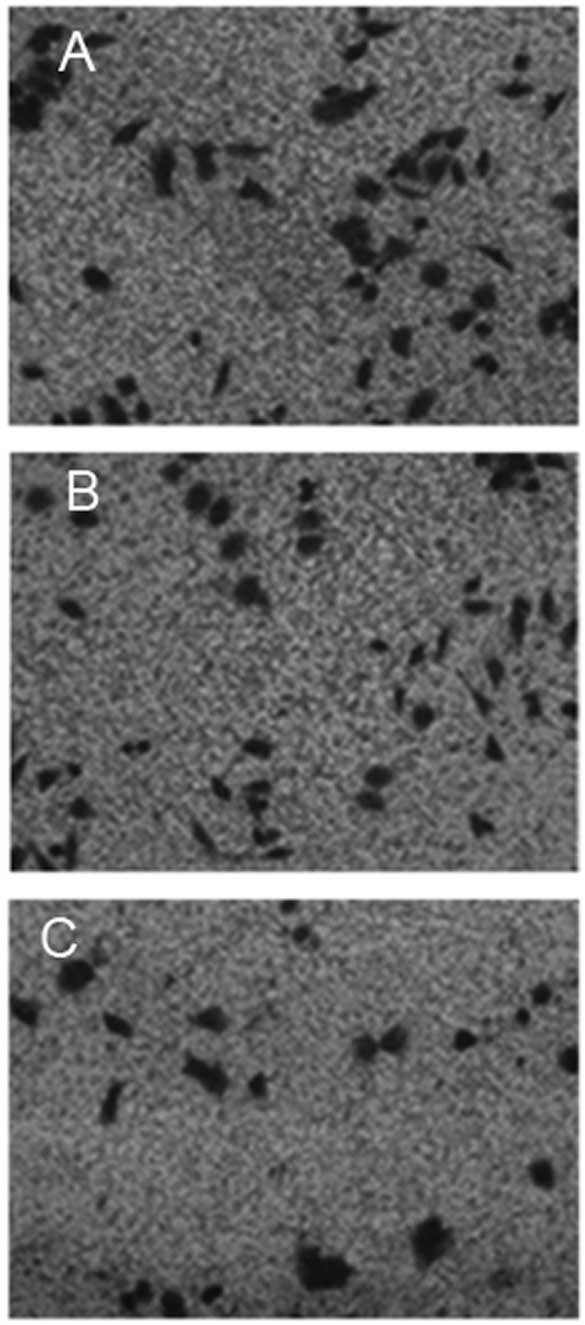
Fig. 4 shows
representative images of wild-type Saos-2 cells, pIRES2-EGFP-MNDA-
and pIRES2-EGFP-transfected Saos-2 cells that migrated out of the
Transwell membrane through the Matrigel matrix. When the average
over the three experiments was calculated, the number of
pIRES2-EGFP-MNDA-overexpressing Saos-2 cells that migrated through
the Matrigel and Transwell membrane was 28±3, which was
significantly lower (P<0.01) than that of the wild-type control
cells (77±5) and cells overexpressing pIRES2-EGFP (80±3).
Discussion
Targeted cancer therapies, also known as
‘molecularly targeted drugs’ or ‘molecularly targeted therapies’,
have been used for various malignancies in a clinical setting
(17). Since interfering with
specific molecules involved in tumorigenesis and the subsequent
blocking of tumor growth and progression are the fundamental bases
of targeted cancer therapies, the identification of appropriate
targets (molecules known to play a key role in cancer cell growth
and survival) is the key initial step in the development of
targeted therapies.
MNDA, a member of the human IFI-200 family, may
serve as a novel molecule for targeted therapies. Although MNDA was
originally identified in, and thought to be restricted to, myeloid
cells (11) there is evidence that
MNDA and other IFI-200 family proteins are also expressed in a
variety of non-myeloid cell types (18). Somatic inactivation of the genes of
the IFI-200 family have been observed in several solid tumors
including prostate cancer, melanoma and colon cancer (19–21),
suggesting a tumor suppressor role for these family gene products.
In a previous study, we demonstrated that MNDA was highly expressed
in normal bone tissue but was expressed at markedly low levels in
osteosarcoma cells (22). In this
study, we observed that MNDA mRNA was significantly less abundant
in Saos-2 cells compared to U937 cells (Fig. 1), confirmation of the downregulation
of MNDA expression in human osteosarcoma.
In order to evaluate the tumor suppressor role of
MNDA in osteosarcoma and explore the possibility of developing an
MNDA-based targeted therapy for bone cancers, we also determined
the effect of MNDA overexpression on the proliferation, apoptosis
and migration/invasion of osteosarcoma cells. Our results
demonstrated that the overexpression of MNDA affected osteosarcoma
cells by effectively inhibiting proliferation (Fig. 2), inducing apoptosis (Fig. 3) and reducing their migration
(Fig. 4) in vitro. To the
best of our knowledge, we are the first group to have observed MNDA
overexpression-mediated cell growth inhibition, cell cycle
arrest/apoptosis and reduced invasiveness in osteosarcoma cells.
However, the overexpression of other IFI-200 family members has
been previously reported to inhibit growth and induce cell cycle
arrest, apoptosis or senescence in several other cell models
(11,23).
At present, the mechanisms underlying MNDA mediation
of growth inhibition, apoptosis and a reduction in the invasiveness
of osteosarcoma cells are not completely understood. The majority
of IFI-200 family proteins possess a conserved interferon response
motif that is associated with protein-protein interactions in the
regulation of apoptotic and inflammatory signaling pathways.
Furthermore, IFI-200 family proteins may also regulate cell cycle
progression/apoptosis and differentiation through interacting with,
and modulating the activities of, multiple transcriptional factors
such as Rb and p53 (11).
Nevertheless, whether similar mechanisms are involved in
MNDA-mediated osteosarcoma cell growth inhibition, apoptosis and
reduced metastasis requires further investigation.
In conclusion, the present study further confirmed
the downregulation of MNDA in osteosarcoma cells as demonstrated in
our previous study and has demonstrated that the overexpression of
MNDA in osteosarcoma cells results in growth inhibition, cell cycle
arrest, apoptosis and reduced invasiveness. These observations
suggest that MNDA is a potential novel therapeutic target for
osteosarcoma.
Acknowledgements
This study was supported by grants from the Natural
Science Foundation of Shandong Province, China (no.
ZR2009CM042).
References
|
1
|
Longhi A, Errani C, De Paolis M, Mercuri M
and Bacci G: Primary bone osteosarcoma in the pediatric age: state
of the art. Cancer Treat Rev. 32:423–436. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar
|
|
3
|
Mirabello L, Troisi RJ and Savage SA:
International osteosarcoma incidence patterns in children and
adolescents, middle ages and elderly persons. Int J Cancer.
125:229–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: data
from the Surveillance, Epidemiology, and End Results Program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stiller CA and Parkin DM: Geographic and
ethnic variations in the incidence of childhood cancer. Brit Med
Bull. 52:682–703. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stiller CA: International patterns of
cancer incidence in adolescents. Cancer Treat Rev. 33:631–645.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mavrogenis AF, Rossi G, Palmerini E, et
al: Palliative treatments for advanced osteosarcoma. J BUON.
17:436–445. 2012.
|
|
8
|
Bacci G, Ferrari S, Bertoni F, et al:
Long-term outcome for patients with nonmetastatic osteosarcoma of
the extremity treated at the istituto ortopedico rizzoli according
to the istituto ortopedico rizzoli/osteosarcoma-2 protocol: an
updated report. J Clin Oncol. 18:4016–4027. 2000.
|
|
9
|
Bielack S, Carrle D and Casali PG; the
ESMO Guidelines Working Group. Osteosarcoma: ESMO clinical
recommendations for diagnosis, treatment and follow-up. Ann Oncol.
20:137–139. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bellos F, Alpermann T, Gouberman E, et al:
Evaluation of flow cytometric assessment of myeloid nuclear
differentiation antigen expression as a diagnostic marker for
myelodysplastic syndromes in a series of 269 patients. Cytometry B
Clin Cytom. 82:295–304. 2012. View Article : Google Scholar
|
|
11
|
Asefa B, Klarmann KD, Copeland NG, Gilbert
DJ, Jenkins NA and Keller JR: The interferon-inducible p200 family
of proteins: a perspective on their roles in cell cycle regulation
and differentiation. Blood Cells Mol Dis. 32:155–167. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Choubey D, Duan X, Dickerson E, et al:
Interferon-inducible p200-family proteins as novel sensors of
cytoplasmic DNA: role in inflammation and autoimmunity. J
Interferon Cytokine Res. 30:371–380. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Parmar S and Platanias LC: Interferons:
mechanisms of action and clinical applications. Curr Opin Oncol.
15:431–439. 2003. View Article : Google Scholar
|
|
14
|
Zhang K, Kagan D, DuBois W, et al: Mndal,
a new interferon-inducible family member, is highly polymorphic,
suppresses cell growth, and may modify plasmacytoma susceptibility.
Blood. 114:2952–2960. 2009. View Article : Google Scholar
|
|
15
|
Briggs RC, Shults KE, Flye LA, et al:
Dysregulated human myeloid nuclear differentiation antigen
expression in myelodysplastic syndromes: evidence for a role in
apoptosis. Cancer Res. 66:4645–4651. 2006. View Article : Google Scholar
|
|
16
|
Rozzo SJ, Allard JD, Choubey D, et al:
Evidence for an interferon-inducible gene, Ifi202, in the
susceptibility to systemic lupus. Immunity. 15:435–443. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Keller U, von Bubnoff N, Peschel C and
Duyster J: Oncologist’s/haematologist’s view on the roles of
pathologists for molecular targeted cancer therapy. J Cell Mol Med.
14:805–817. 2010.
|
|
18
|
Choubey D, Deka R and Ho SM:
Interferon-inducible IFI16 protein in human cancers and autoimmune
diseases. Front Biosci. 13:598–608. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Alimirah F, Chen J, Davis FJ and Choubey
D: IFI16 in human prostate cancer. Mol Cancer Res. 5:251–259. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
DeYoung KL, Ray ME, Su YA, et al: Cloning
a novel member of the human interferon-inducible gene family
associated with control of tumorigenicity in a model of human
melanoma. Oncogene. 15:453–457. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Woerner SM, Kloor M, Schwitalle Y, et al:
The putative tumor suppressor AIM2 is frequently affected by
different genetic alterations in microsatellite unstable colon
cancers. Genes Chromosomes Cancer. 46:1080–1089. 2007. View Article : Google Scholar
|
|
22
|
Li X, Wu WK, Sun B, et al:
Dihydroptychantol A, a macrocyclic bisbibenzyl derivative, induces
autophagy and following apoptosis associated with p53 pathway in
human osteosarcoma U2OS cells. Toxicol Appl Pharm. 251:146–154.
2011. View Article : Google Scholar
|
|
23
|
Ouchi M and Ouchi T: Role of IFI16 in DNA
damage and checkpoint. Front Biosci. 13:236–239. 2008. View Article : Google Scholar : PubMed/NCBI
|