Introduction
Furin is the best-characterized representative of
the mammalian subtilisin-like family of proprotein convertases. It
is synthesized as an inactive proenzyme and is rapidly matured by
autocatalytic cleavage between the prodomain and the catalytic
domain in the endoplasmic reticulum (ER) (1,2).
Following this initial cleavage, the propeptide-furin complex
leaves the ER and enters the trans-Golgi network (TGN) for its
second cleavage (3,4). Thus, furin becomes active to process
substrate molecules in multiple compartments in the TGN/endosomal
system (5). Numerous protein
precursors, including matrix metalloproteinases (MMPs), hormones,
growth factors, serum proteins, receptors and adhesion molecules,
have been identified as furin substrates (6–8).
Membrane type I (MT1)-MMP proenzyme cleavage by furin is considered
to be a principal event in the activation of this substrate and may
play a vital role in tumor cell migration (9).
Furin activation plays a vital role in tumor
development (10). The furin
inhibitor α1-antitrypsin Portland (α1-PDX) has been used to block
furin activity and to prevent cancer metastasis in biochemical,
cellular and animal studies (11).
The Wnt signaling pathway plays a vital role in
normal development, but also in tumorigenesis (12,13).
Inappropriate activation of the Wnt signaling pathway results in
the onset of several types of cancer (14). Based on the different interactions
between Wnt receptors or co-receptors, the Wnt signaling pathway is
divided into three signaling pathways, namely the canonical
Wnt/β-catenin signaling pathway, and the non-canonical (or
heretical) Wnt/ Ca2+ and planar cell polarity (PCP)
signaling pathways (15). Previous
studies have suggested that the interaction of the Wnt/PCP
signaling pathway with certain key molecules is associated with
cancer cell migration and invasion (16,17).
The canonical Wnt signaling pathway involves a key mediator,
β-catenin, which is able to enter the cell nucleus and associate
with the transcription factors lymphoid enhancer-binding factor 1
and T-cell factor, leading to the transcription of Wnt target genes
(18,19). The stabilization of β-catenin is
regulated by phosphorylation modification by glycogen synthase
kinase 3β, followed by degradation via the proteasome. Abnormal
activation of the Wnt/β-catenin signaling pathway has been detected
in a number of types of human tumor, including lung, breast,
cervical and liver, and is due to lack of degradation and
ultimately the nuclear accumulation of β-catenin. In patients with
hepatocellular carcinoma, β-catenin accumulation has been linked to
poor differentiation and high proliferative activity of cells, and
a poor prognosis (20,21). The levels of β-catenin are regulated
by numerous types of protein, which may lead to the onset of cancer
if not regulated or expressed appropriately. By forming a complex
with transcription factor 4, β-catenin activates the transcription
of target genes, including MT1-MMP, whose expression levels
correlate with the levels of cell migration and invasion (22). Abnormal expression of MT1-MMP has
been detected in numerous types of cancer. Such induction of the
expression of MT1-MMP could be regulated by the Wnt/β-catenin
signaling pathway; this is based on the observation that depletion
of β-catenin in SW480 colorectal carcinoma cells results in the
downregulation of the expression levels of MT1-MMP (23). In a previous study, it was
demonstrated that the migration of MG-63 and Saos-2 osteosarcoma
cells was inhibited significantly by a certain range of
concentrations of α1-PDX treatment (10), but the exact molecular mechanism of
this effect remains unknown.
Materials and methods
Cell culture and experimental
reagents
MG-63 and Saos-2 osteosarcoma cells were purchased
from the American Type Culture Collection (Manassas, VA, USA)
cultured in RPMI-1640 (Invitrogen Life Technologies, Carlsbad, CA,
USA) supplemented with 10% fetal bovine serum (FBS), 100 U/ml
penicillin and 100 μg/ml streptomycin, in a 5% CO2
humidified atmosphere at 37°C. α1-PDX (126850-2.5MG;
Calbiochem-Merck KGaA, Darmstadt, Germany) was added to the medium
at a concentration of 480 nM where indicated. Primary antibodies
against Wnt, β-catenin, MT1-MMP and β-actin were purchased from
Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Other
reagents were used, including anti-mouse-IgG-HRP and
anti-rabbit-IgG-HRP (BD Biosciences, CA, USA) and Transwell
invasion chambers (Promega, Madison, WI, USA).
Monolayer cell migration assay
A monolayer wound-healing model was performed as a
cell migration assay. MG-63 and Saos-2 osteosarcoma cells were
seeded in a six-well-plate for 48 h in complete RPMI-1640 medium. A
confluent monolayer of MG-63 and Saos-2 osteosarcoma cells was then
scraped with a sterile 200-μl pipette tip into another
six-well-plate and washed with phosphate-buffered saline (PBS).
Following incubation with complete RPMI-1640 or α1-PDX (480 nM) for
48 h, cell migration images were captured using an inverted phase
contrast microscope at ×100 magnification.
Transwell invasion assay
The Matrigel invasion chambers were hydrated for 4 h
prior to starting the invasion assay. Log-phase cells
(4×104) were plated in 200 μl complete RPMI-1640
containing 10% FBS in the upper chamber of the Transwell, and the
lower chamber was filled with 500 μl complete RPMI-1640 containing
10% FBS. Following incubation for 2 h, the cells were treated with
α1-PDX as decribed previously and allowed to migrate for 10 h at
37°C and 5% CO2. The cells were fixed for 15 min at room
temperature by replacing the culture medium in the bottom and top
of the chamber with 4% formaldehyde buffer. Subsequently, the
chambers were rinsed in PBS and stained with 0.1% crystal violet
for 10 min, then the migrated cells were photographed under an
optical microscope. The cell number was counted at 12 different
areas. Data were averaged from three parallel experiments, which
were normalized to those of the controls.
Reverse transcription-polymerase chain
reaction (RT-PCR) analysis
The cells were incubated with α1-PDX for 48 h prior
to RT-PCR. Total RNA was extracted from the MG-63 and Saos-2
osteosarcoma cells using the TRIzol method (Invitrogen Life
Technologies). RT was performed with 1 μg total RNA and 10 μM of
the specific primers. The cDNAs were amplified by PCR for MT1-MMP
(sense, 5′-AGCCCCGAAGCCTGGCTACA-3′; and antisense,
5′-GCCGCCCTCACCATCGAAGG-3′; 492-bp product), or glyceraldehyde
3-phosphate dehydrogenase was used as the endogenous reference
housekeeping gene (sense, 5′-ACCACAGTCCATGCCATCAC-3′; and
antisense, 5′-TCCACCACCCTGTTGCTGTA-3′; 556-bp product). The PCR
conditions were as follows: 95°C for 5 min, followed by 30 cycles
of 95°C for 15 sec, 60°C for 30 sec and 72°C for 45 sec.
Western blot analysis
The cells were incubated with α1-PDX for 48 h prior
to western blotting. MG-63 and Saos-2 osteosarcoma cells were lysed
in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris pH 7.4,
150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, 5
mM EDTA, 100 mM NaF, and 1 mM Na3VO4)
containing a protease inhibitor cocktail (product no., 04693116001;
Roche, Madison, WI, USA)for 30 min on ice, followed by
centrifugation for 30 min at 35,800 × g. The protein concentrations
were determined by the bicinchoninic acid assay method (Pierce BCA
Protein Assay kit; Pierce Biotechnology, Inc., Rockford, IL, USA).
Equal quantities of total proteins were electrophoresed by 12%
SDS-PAGE gel, followed by transfer to polyvinylindene difluoride
membranes using a wet transblot system (Bio-Rad, Hercules, CA,
USA). The membranes were blocked for 1 h at room temperature with
5% nonfat dry milk and incubated overnight at 4°C with antibodies
[rabbit anti-Wnt, β-catenin, MT1-MMP and mouse anti-β-actin
(1:1,000)]. After washing, the membranes were incubated for 1 h
with HRP-conjugated goat anti-rabbit or anti-mouse-IgG-HRP
secondary antibodies diluted to 1:5,000 in PBS Tween-20. After
further washing and processing using SuperSignal West Pico
Chemiluminescent substrate (Pierce Biotechnology, Inc.), the
membranes were exposed to a Fujifilm LAS-3000 Imager (Fuji, Tokyo,
Japan). The band densities of the western blots were normalized
relative to the relevant β-actin band density with ImageJ Analysis
software (National Institutes of Health, Bethesda, MD, USA).
Chromatin immunoprecipitation (ChIP)
assay
The cells were incubated with α1-PDX for 48 h prior
to performing the ChIP assay. A ChIP assay was performed using a
ChIP kit (Sigma-Aldrich, St. Louis, MO, USA) with slight
modifications. MG-63 and Saos-2 osteosarcoma cells
(2×107) were cross-linked with 1% formaldehyde for 10
min at room temperature, followed by the addition of 1 ml of 125 mM
glycine to inactivate the formaldehyde. The cells were washed twice
with ice-cold PBS and then scraped and centrifuged at 1,000 × g at
4°C for 5 min. The pelleted cells were lysed with 1 ml
modified-RIPA lysis buffer (0.1% SDS, 10 mM EDTA, 1% Triton X-100
and 50 mM Tris-HCl pH 8.1) containing a protease inhibitor cocktail
and incubated on ice for 10 min. Following sonication to produce
genomic DNA with lengths of 0.2–0.5 kb, the samples were
centrifuged at 13,000 × g for 10 min to remove insoluble cell
debris. The lysates were diluted in ChIP dilution buffer (0.01%
SDS; 1.1% Triton X-100; 2 mM EDTA; 20 mM Tris-HCl, pH 8.1; and 500
mM NaCl) and protease inhibitor cocktail. Dilutions of the
chromatin preparations were stored at −20°C. The chromatin solution
was precleared with 20 μl of 3% bovine serum albumin/protein A
agarose beads for 2 h at 4°C with rotation. Anti-β-catenin
polyclonal antibody (Santa Cruz Biotechnology, Inc.) was added to
the precleared supernatant and incubated overnight at 4°C with
rotation. The negative controls included a sample incubated without
antibody and one incubated with rabbit IgG (Santa Cruz
Biotechnology, Inc.) to determine whether the interactions were due
to nonspecific IgG interactions. The bead complexes were washed
with low-salt immune complex wash buffer (Sigma-Aldrich), followed
by high-salt immune complex wash buffer (Sigma-Aldrich) and a final
LiCl immune complex wash buffer (Sigma-Aldrich) for 5 min each on a
rotating platform followed by brief centrifugation at 35,800 × g
for 10 min. Two final washes in 1X Tris EDTA buffer were performed
for 5 min each. Following the final wash, the DNA was extracted by
incubating the beads twice for 15 min with 200 μl freshly prepared
elution buffer (1% SDS and 50 mM NaHCO3). The samples
were then uncrosslinked in a 65°C water bath overnight and the DNA
was purified using a QIAquick Nucleotide Removal kit (Qiagen Inc.,
Valencia, CA, USA). The purified DNA was analyzed by PCR. The PCR
primers used to amplify the MT1-MMP promoter region were as
follows: GTCTCCCGCCCCAAGACCCT (forward) and GGAACACCACATCGGGGGCG
(reverse).
Statistical analysis
All experiments were performed three times and the
data were expressed as the mean ± standard error of the mean.
Statistical analysis was performed by SPSS software, version 11.0
(SPSS, Inc., Chicago, IL, USA). Differences between the groups were
statistically evaluated using the t-test or one-way analysis of
variance with post-hoc analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
Inhibitory effect of α1-PDX on MG-63 and
Saos-2 osteosarcoma cell migration
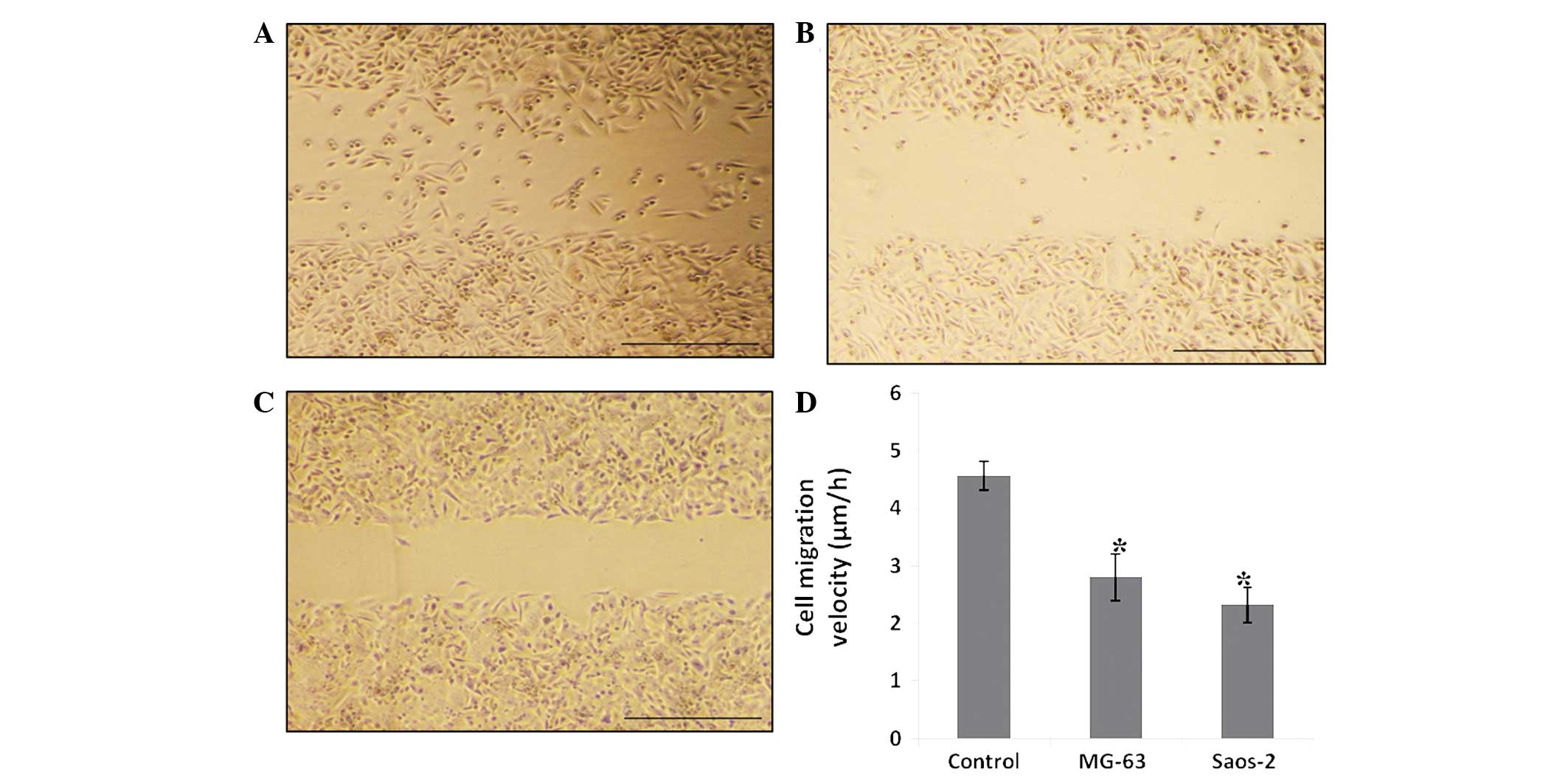
The effect of α1-PDX on MG-63 and Saos-2
osteosarcoma cell migration was monitored by a monolayer
wound-healing assay. Log-phase cells were seeded on six-well plates
and incubated with complete cell medium or 480 nM α1-PDX for 24 h.
Following wounding by a sterile 200-μl pipette tip, the cells
treated with normal cell medium migrated clearly. The cells that
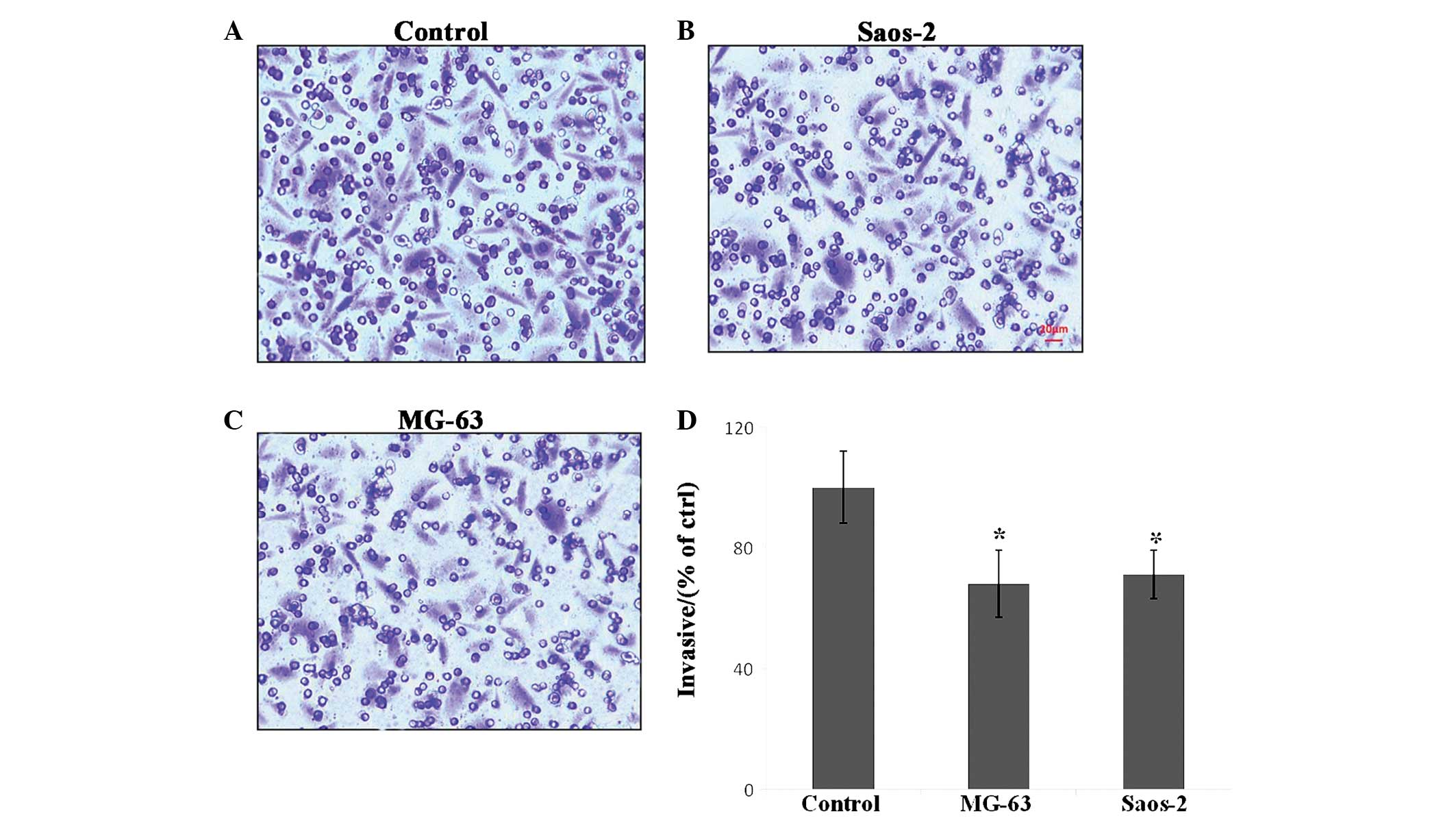
were treated with α1-PDX showed no evident migration (Fig. 1). In the three-dimensional cell
migration assay with the Transwell system, the cells treated with
α1-PDX were found to migrate less than the control cells (Fig. 2). This data indicates that MG-63 and
Saos-2 osteosarcoma cell migration was inhibited upon α1-PDX
treatment. However, these assays did not reveal the mechanism of
the inhibitory effect.
Downregulation effect of α1-PDX on the
expression levels of MT1-MMP
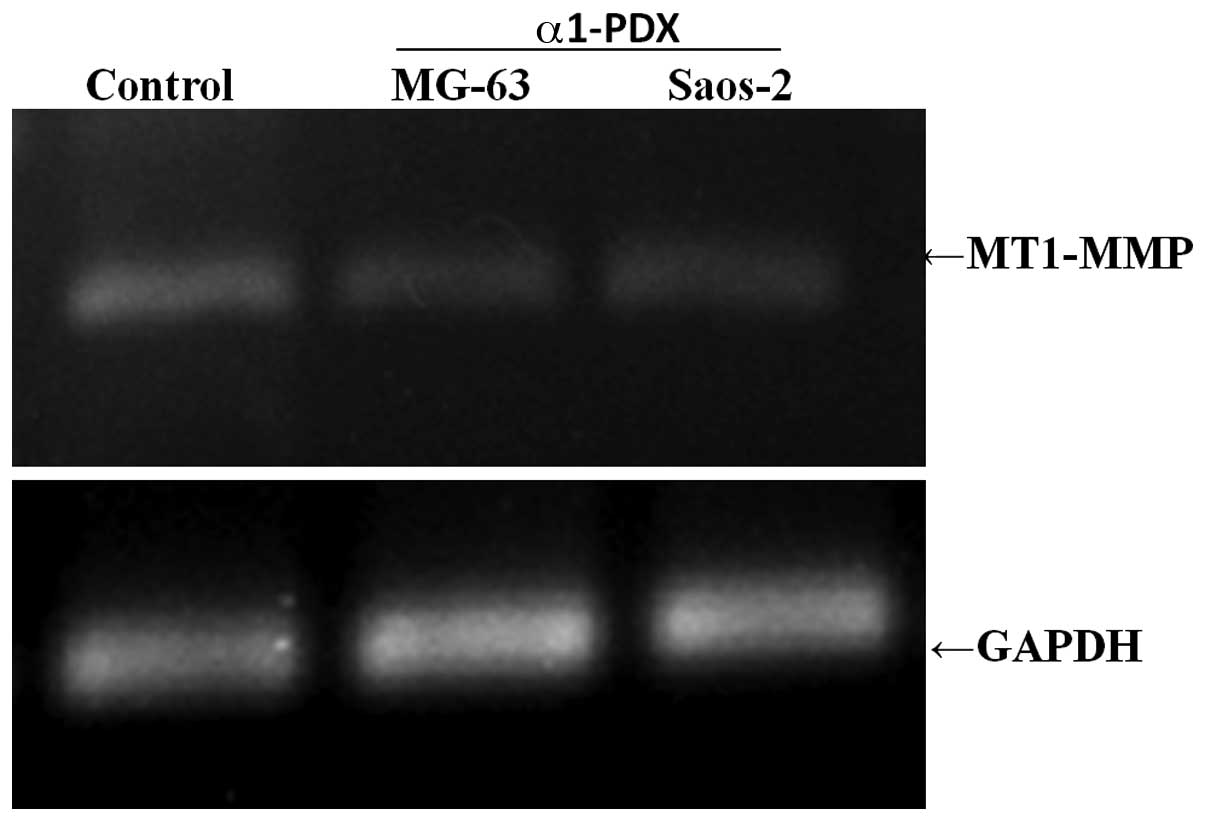
To explore the possible mechanism of the inhibitory
effect of α1-PDX on MG-63 and Saos-2 osteosarcoma cell migration,
the expression levels of MT1-MMP, which is the key mediator of cell
migration and invasion, were detected. RT-PCR was used for
detection of the levels of gene expression of MT1-MMP in MG-63 and
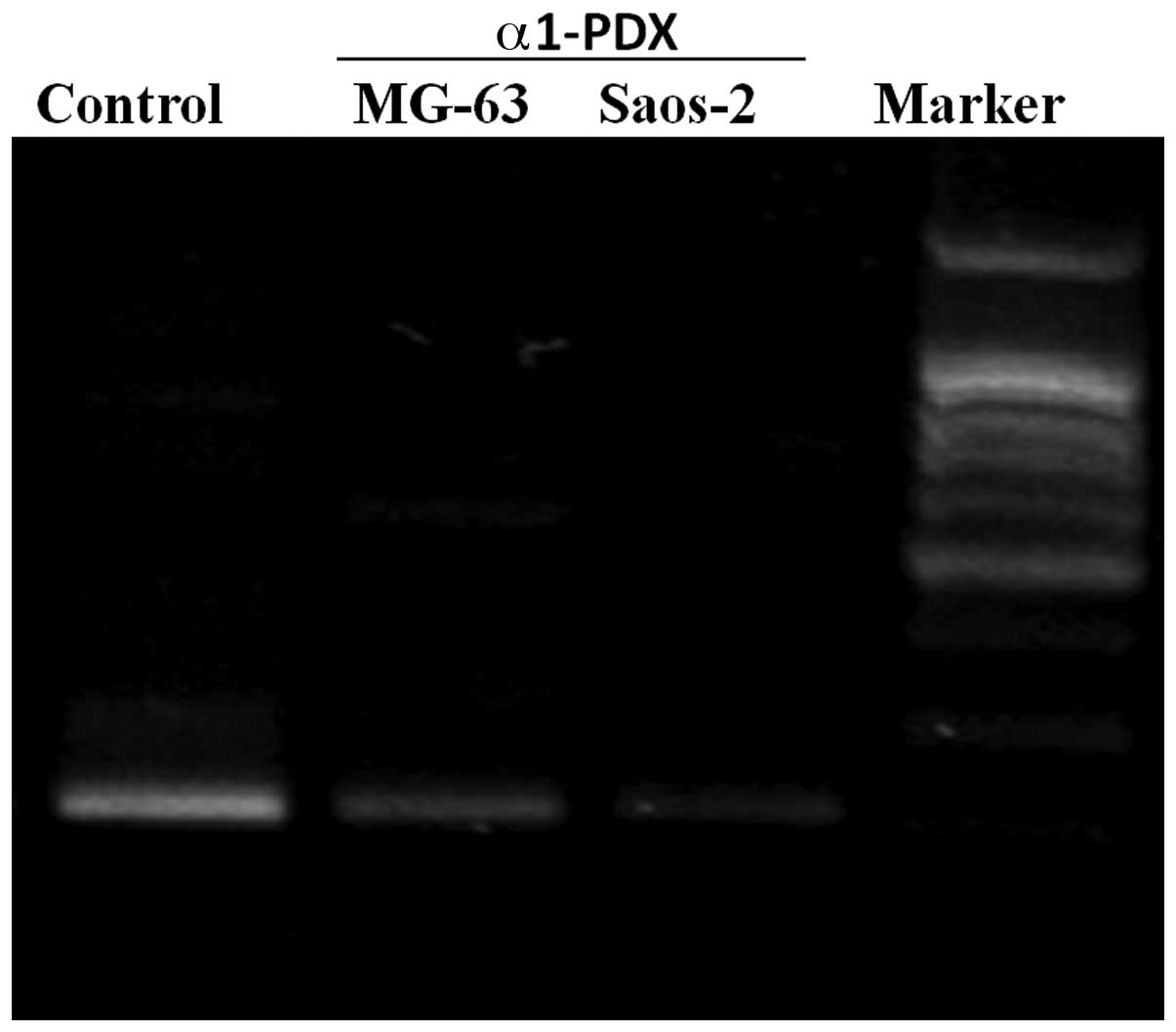
Saos-2 osteosarcoma cells upon α1-PDX treatment. From the results
(Fig. 3), the gene expression
levels of MT1-MMP were reduced significantly in the α1-PDX
treatment cells, compared with those of the control group. The
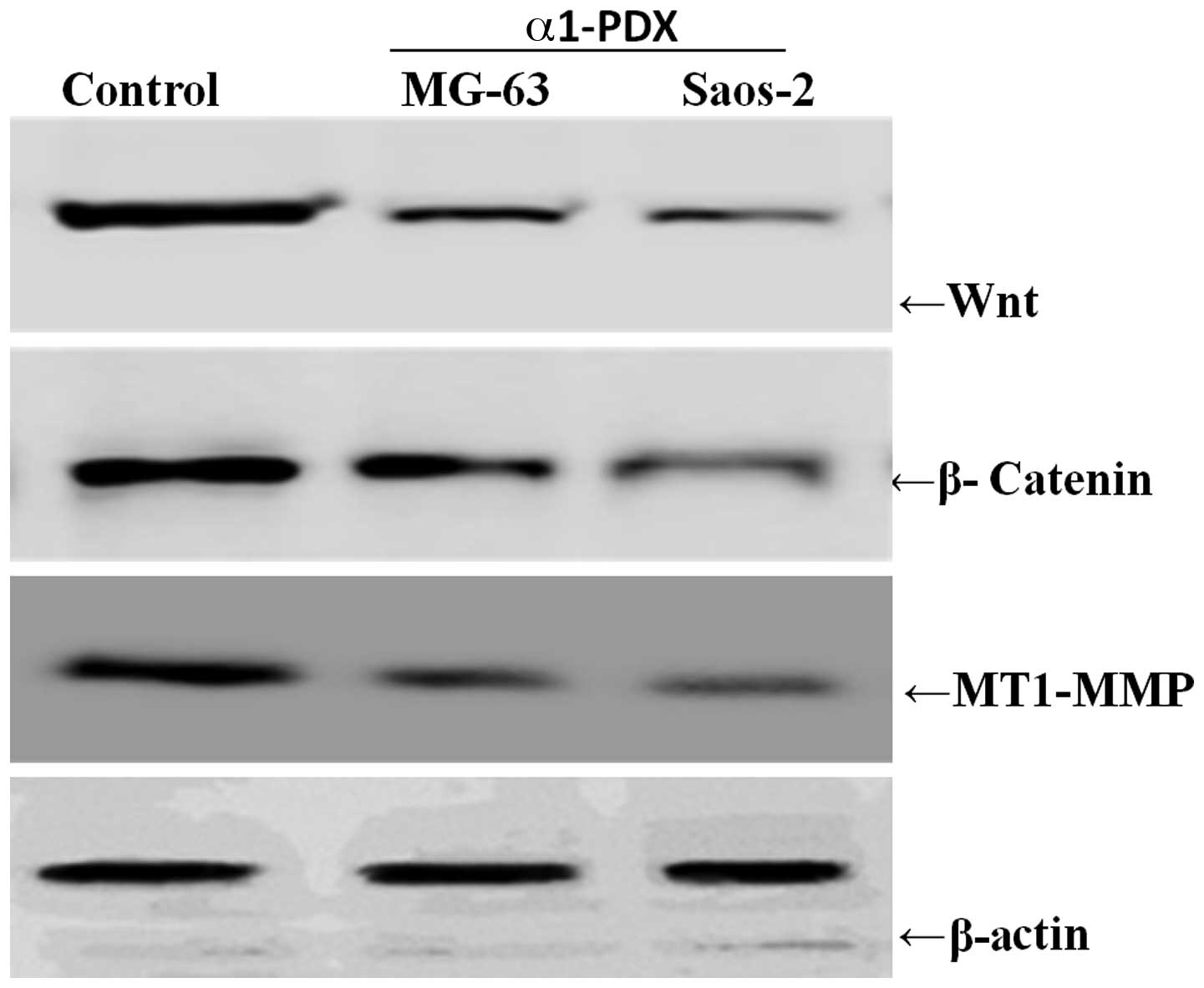
protein levels of MT1-MMP were also detected upon α1-PDX treatment
and, as expected, the protein levels also decreased evidently
compared with those of the control cells (Fig. 4). These results suggest that α1-PDX
suppresses MG-63 and Saos-2 osteosarcoma cell migration, possibly
via downregulation of the expression of MT1-MMP gene and protein
levels.
Wnt signaling pathway may be involved in
the downregulation of the levels of MT1-MMP by α1-PDX
treatment
To further explore the exact mechanism by which
α1-PDX downregulates the MT1-MMP expression levels, the effect of
α1-PDX on Wnt signaling pathway-related properties was
investigated. When the MG-63 and Saos-2 osteosarcoma cells were
treated with α1-PDX for the indicated times, the expression levels
of Wnt and β-catenin decreased significantly, paralleled with the
reduction in the MT1-MMP expression levels (Fig. 4). These results suggested that the
downregulating effect of α1-PDX on the levels of MT-MMP may be via
the Wnt signaling pathway.
Effect of α1-PDX on the levels of MT1-MMP
transcriptional activity via the Wnt signaling pathway
To determine whether the levels of MT1-MMP
transcriptional activity correlated with the levels of Wnt
signaling pathway activity, a ChIP assay was performed. The MG-63
and Saos-2 osteosarcoma cells were treated with α1-PDX for 24 h and
β-catenin antibody was used for immunoprecipitation with target
genes. The results showed that the levels of MT1-MMP
transcriptional activity decreased evidently compared with those of
the control (Fig. 5), which
demonstrated that the effect of α1-PDX on MT1-MMP expression levels
was mediated through Wnt signaling.
Discussion
Previous studies have indicated that α1-PDX has the
potential role of inhibiting cancer cell migration, but the exact
mechanism is unknown. In the present study, it has been
demonstrated that α1-PDX inhibited the migration and invasion of
MG-63 and Saos-2 osteosarcoma cells through downregulation of the
expression levels of MT1-MMP via the Wnt/β-catenin signaling
pathway. The invasion ability of osteosarcoma MG-63 and Saos-2
cells upon α1-PDX treatment was detected through two- and
three-dimensional migration assays (wound-healing and Transwell
assays). It was observed that the migration ability was reduced
significantly upon α1-PDX treatment for 24 h. Tumor invasion and
metastasis is a multistage and multifactorial process, which is
regulated by complicated mechanisms and multiple signaling pathways
(24).
Numerous protein molecules are involved in the
regulation of cell adhesion, migration and invasion in tumor
biology behaviors. MMPs are a family of zinc-binding proteases that
have been shown to contribute to cancer cell invasion through the
ability to degrade the extracellular matrix (25,26).
MT1-MMP (also known as MMP-14) is the first identified and also the
most common member of the MT-MMP subfamily involved in pericellular
proteolysis associated with cell migration (27,28).
Harada et al and Arii et al have
demonstrated that MT1-MMP is involved in hepatocarcinoma cell
migration (29,30); however, the molecular mechanism is
unknown. In searching for the underlying mechanism of α1-PDX
inhibition of the migration and invasion of MG-63 and Saos-2
osteosarcoma cells in the present study, the expression levels of
MT1-MMP mRNA and protein in MG-63 and Saos-2 osteosarcoma cells
upon α1-PDX treatment were detected. It was identified that the
mRNA and protein expression levels of MT1-MMP were decreased
evidently upon α1-PDX treatment.
The activity levels of the majority of MMPs are very
low in normal steady state tissues; however, their expression
levels are regulated by various inflammatory cytokines, growth
factors and hormones, as well as by cell-cell interactions
(31). Furthermore, the proteolytic
activity of MMPs is strictly controlled at several levels,
including transcriptional, post-transcriptional and
post-translational, as well as via their endogenous inhibitors
(31,32). The transcription of MT1-MMP is
strictly regulated by the Wnt signaling pathway (33,34);
therefore, we hypothesized that inhibition of MG-63 and Saos-2
osteosarcoma cell migration and invasion by α1-PDX may be through
downregulation of the Wnt signaling pathway. Based on this
assumption, the expression levels of Wnt and β-catenin were
detected in the present study. Western blotting indicated that the
expression levels of Wnt and β-catenin decreased markedly in the
α1-PDX-treated cells compared with those of the control; however,
the expression levels of the positive control, the
docetaxel-treated group, decreased weakly. These results suggested
that α1-PDX has a potential role of downregulating the expression
levels of Wnt and β-catenin. To investigate whether the effect of
α1-PDX on the transcriptional activity of MT1-MMP is mediated
through the Wnt signaling pathway, the levels of MT1-MMP
transcriptional activity were detected through a ChIP assay. As
expected, upon α1-PDX treatment a small level of MT1-MMP was
detected, which was decreased significantly compared with that of
the control.
In summary, the data of the present study
demonstrated that α1-PDX has the potential role of inhibiting the
migration and invasion of MG-63 and Saos-2 osteosarcoma cells,
which may be through downregulating the expression levels of
MT1-MMP via the canonical Wnt signaling pathway. It is of note that
α1-PDX downregulates the expression levels of transcripts and
protein of MT1-MMP, an activator of proMMP-2 (pro-gelatinase A/72
kDa type IV collagenase) (35),
which is a major and specific basement membrane matrix protein.
Since the degradation of the basement membrane by MMP-2 is likely
an essential step for cancer invasion (36–38),
it is necessary to study whether α1-PDX mediates the activity of
other MMPs. Therefore, numerous aspects of the mechanism of α1-PDX
remain to be resolved.
References
|
1
|
Gawlik K, Shiryaev SA and Zhu W:
Autocatalytic activation of the furin zymogen requires removal of
the emerging enzyme’s N-terminus from the active site. PLoS One.
4:e50312009.PubMed/NCBI
|
|
2
|
Vey M, Schäfer W, Berghöfer S, Klenk HD
and Garten W: Maturation of the trans-Golgi network protease furin:
compartmentalization of propeptide removal, substrate cleavage, and
COOH-terminal truncation. J Cell Biol. 127:1829–1842. 1994.
View Article : Google Scholar
|
|
3
|
Creemers JW, Vey M, Schäfer W, Ayoubi TA,
Roebroek AJ, Klenk HD, Garten W and Van de Ven WJ: Endoproteolytic
cleavage of its propeptide is a prerequisite for efficient
transport of furin out of the endoplasmic reticulum. J Biol Chem.
270:2695–2702. 1995. View Article : Google Scholar
|
|
4
|
Anderson ED, VanSlyke JK, Thulin CD, Jean
F and Thomas G: Activation of the furin endoprotease is a
multiple-step process: requirements for acidification and internal
propeptide cleavage. EMBO J. 16:1508–1518. 1997. View Article : Google Scholar
|
|
5
|
Molloy SS, Thomas L, VanSlyke JK, et al:
Intracellular trafficking and activation of the furin proprotein
convertase: localization to the TGN and recycling from the cell
surface. EMBO J. 13:18–33. 1994.PubMed/NCBI
|
|
6
|
Fujisawa T, Kamimura H, Hosaka M, et al:
Functional localization of proprotein-convertase furin and its
substrate TGFbeta in EGF receptor-expressing gastric chief cells.
Growth Factors. 22:51–59. 2004. View Article : Google Scholar
|
|
7
|
Louagie E, Taylor NA, Flamez D, et al:
Role of furin in granular acidification in the endocrine pancreas:
identification of the V-ATPase subunit Ac45 as a candidate
substrate. Proc Natl Acad Sci USA. 105:12319–12324. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yana I and Weiss SJ: Regulation of
membrane type-1 matrix metalloproteinase activation by proprotein
convertases. Mol Biol Cell. 11:2387–2401. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dangi-Garimella S, Krantz SB, Barron MR,
et al: Three-dimensional collagen I promotes gemcitabine resistance
in pancreatic cancer through MT1-MMP-mediated expression of HMGA2.
Cancer Res. 71:1019–1028. 2011. View Article : Google Scholar
|
|
10
|
López de Cicco R, Bassi DE, Zucker S, et
al: Human carcinoma cell growth and invasiveness is impaired by the
propeptide of the ubiquitous proprotein convertase furin. Cancer
Res. 65:4162–4171. 2005.PubMed/NCBI
|
|
11
|
Thomas G: Furin at the cutting edge: from
protein traffic to embryogenesis and disease. Nat Rev Mol Cell
Biol. 3:753–766. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nusse R and Varmus HE: Wnt genes. Cell.
69:1073–1087. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
McMahon AP, Gavin BJ, Parr B, et al: The
Wnt family of cell signalling molecules in postimplantation
development of the mouse. Ciba Found Symp. 165:199–218.
1992.PubMed/NCBI
|
|
14
|
Polakis P: The many ways of Wnt in cancer.
Curr Opin Genet Dev. 17:45–51. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Smalley MJ and Dale TC: Wnt signalling in
mammalian development and cancer. Cancer Metastasis Rev.
18:215–230. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y: Wnt/Planar cell polarity
signaling: a new paradigm for cancer therapy. Mol Cancer Ther.
8:2103–2109. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Katoh M: WNT/PCP signaling pathway and
human cancer (review). Oncol Rep. 14:1583–1588. 2005.PubMed/NCBI
|
|
18
|
Hecht A, Vleminckx K, Stemmler MP, et al:
The p300/CBP acetyltransferases function as transcriptional
coactivators of beta-catenin in vertebrates. EMBO J. 19:1839–1850.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takemaru KI and Moon RT: The
transcriptional coactivator CBP interacts with beta-catenin to
activate gene expression. J Cell Biol. 149:249–254. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Inagawa S, Itabashi M, Adachi S, et al:
Expression and prognostic roles of beta-catenin in hepatocellular
carcinoma: correlation with tumor progression and postoperative
survival. Clin Cancer Res. 8:450–456. 2002.
|
|
21
|
Wong CM, Fan ST and Ng IO: beta-Catenin
mutation and overexpression in hepatocellular carcinoma:
clinicopathologic and prognostic significance. Cancer. 92:136–145.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pulyaeva H, Bueno J, Polette M, Birembaut
P, Sato H, Seiki M and Thompson EW: MT1-MMP correlates with MMP-2
activation potential seen after epithelial to mesenchymal
transition in human breast carcinoma cells. Clin Exp Metastasis.
15:111–120. 1997. View Article : Google Scholar
|
|
23
|
Takahashi M, Tsunoda T, Seiki M, Nakamura
Y and Furukawa Y: Identification of membrane-type matrix
metalloproteinase-1 as a target of the beta-catenin/Tcf4 complex in
human colorectal cancers. Oncogene. 21:5861–5867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kessenbrock K, Plaks V and Werb Z: Matrix
metalloproteinases: regulators of the tumor microenvironment. Cell.
141:52–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Itoh Y and Seiki M: MT1-MMP: a potent
modifier of pericellular microenvironment. J Cell Physiol. 206:1–8.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kajita M, Itoh Y, Chiba T, Mori H, Okada
A, Kinoh H and Seiki M: Membrane-type 1 matrix metalloproteinase
cleaves CD44 and promotes cell migration. J Cell Biol. 153:893–904.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Deryugina EI, Ratnikov BI, Postnova TI,
Rozanov DV and Strongin AY: Processing of integrin alpha(v) subunit
by membrane type 1 matrix metalloproteinase stimulates migration of
breast carcinoma cells on vitronectin and enhances tyrosine
phosphorylation of focal adhesion kinase. J Biol Chem.
277:9749–9756. 2002. View Article : Google Scholar
|
|
28
|
Overall CM: Molecular determinants of
metalloproteinase substrate specificity: matrix metalloproteinase
substrate binding domains, modules, and exosites. Mol Biotechnol.
22:51–86. 2002. View Article : Google Scholar
|
|
29
|
Harada T, Arii S, Mise M, Imamura T,
Higashitsuji H, et al: Membrane-type matrix
metalloproteinase-1(MT1-MMP) gene is overexpressed in highly
invasive hepatocellular carcinomas. J Hepatol. 28:231–239. 1998.
View Article : Google Scholar
|
|
30
|
Arii S, Mise M, Harada T, Furutani M,
Ishigami S, Niwano M, et al: Overexpression of matrix
metalloproteinase-9 gene in hepatocellular carcinoma with invasive
potential. Hepatology. 24:316–322. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nagase H, Visse R and Murphy G: Structure
and function of matrix metalloproteinases and TIMPs. Cardiovasc
Res. 69:562–573. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Egeblad M and Werb Z: New functions for
the matrix metalloproteinases in cancer progression. Nat Rev
Cancer. 2:161–174. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kähäri VM and Saarialho-Kere U: Matrix
metalloproteinases and their inhibitors in tumour growth and
invasion. Ann Med. 31:34–45. 1999.PubMed/NCBI
|
|
34
|
Sato H, Takino T, Okada Y, Cao J, Shinagaw
A, Yamamoto E and Seiki M: A matrix metalloproteinase expressed on
the surface of invasive tumour cells. Nature. 370:61–65. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sounni NE and Noel A: Membrane type-matrix
metalloproteinases and tumor progression. Biochimie. 87:329–342.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sato H, Takino T and Miyamori H: Roles of
membrane-type matrix metalloproteinase-1 in tumor invasion and
metastasis. Cancer Sci. 96:212–217. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Seiki M: Membrane-type 1 matrix
metalloproteinase: a key enzyme for tumor invasion. Cancer Lett.
194:1–11. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Holmbeck K, Bianco P, Yamada S and
Birkedal-Hansen H: MT1-MMP: a tethered collagenase. J Cell Physiol.
200:11–19. 2004. View Article : Google Scholar : PubMed/NCBI
|