Introduction
Eradication of early gastric carcinoma (GC) could
contribute to a reduction in the mortality of GC, given that most
early GCs progress to become advanced GCs (1). Therapeutic interventions for
late-stage GC are usually limited to non-curative gastrectomy,
lymphadenectomy and post-operative chemoradiotherapy (2). The five-year relative survival rates
for GC patients are <30% in the majority of countries (3). Hence, the identification of novel
biomarkers is of great clinical importance for early diagnosis,
targeted treatment and prognosis evaluation in GCs.
Gastric tumorigenesis is a heterogeneous process
that occurs following a series of clonal molecular genetic
alterations, including genomic gains and losses, particularly
deletion of tumor-suppressor genes and amplification of oncogenes.
Unveiling abnormalities of specific genes may offer novel insights
into the mechanisms of local growth or the metastatic potential of
early GCs, and allow patients to be stratified into different risk
categories or be treated with novel options for targeted therapy
(4).
Cytogenetic studies have been performed to evaluate
genetic alterations associated with early GCs (1,2,4,5).
Defining the genetic instability aids in the identification of the
tumor-specific signatures involved in the initiation and
progression of GCs, and thus aids in locating genomic biomarkers
for the early detection of GC (5).
However, the molecular mechanism of GC development remains to be
understood, and identification of the predictive markers in the
early stage is crucial. There is a critical requirement for
identifying biomarkers for early detection and novel treatments for
GC. Therefore, in the present study, whole genome array comparative
genomic hybridization (CGH) was conducted to investigate DNA copy
number alterations and new candidate genes that may be indicative
and specific for early GC.
Materials and methods
Study materials
A total of 22 gastric tumor samples were obtained
from patients treated at the Department of General Surgery of
Chungnam National University Hospital in Taejeon, South Korea. None
of these patients had received pre-operative chemotherapy or
radiation. The stage of disease was based on the
tumor-node-metastasis classification using the Union International
Cancer Center staging system. The original diagnostic material of
all patients was reviewed to verify the original histopathological
diagnosis and staging according to the World Health Organization
classification system (6). The
present study was reviewed and approved by the Institutional Review
Board of the Chungnam National University Hospital, and written
informed consent was obtained from each patient according to the
institutional regulations.
Array-CGH experiment
DNA isolation was performed using a DNA isolation
kit according to the manufacturer’s instructions (Promega, Madison,
WI, USA), with certain modifications as previously described
(7,8). Array-CGH was conducted using the
MacArray™ Karyo 1.4 K BAC-chip (Macrogen, Seoul, Korea) (9–11)
according to the manufacturer’s instructions and as described in
our previous studies (12,13). Briefly, all clones were two-end
sequenced using an ABI PRISM 3700 DNA Analyzer (Applied Biosystems,
Foster City, CA, USA), and their sequences were blasted (BLAST;
http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Hybridizations were carried out using a standard
direct method as previously described (14,15).
Briefly, 500 ng normal male DNA (reference) and digested tumor DNA
(test) was labeled with Cy5-dCTP and Cy3-dCTP, respectively, by
random primed labeling (Array CGH Genomic Labeling System;
Invitrogen, Carlsbad, CA, USA). Hybridizations were performed in a
sealed chamber for 48 h at 37°C. Subsequent to hybridization,
slides were washed according to the manufacturer’s instructions and
immediately scanned on a GenePix 4200A two-color fluorescence
scanner (Axon Instruments, Union City, CA, USA). The acquired
images were analyzed using GenePix Pro 4.1 imaging software (Axon
Instruments).
Array CGH data analysis
Breakpoint detection and status assignment of the
genomic regions were performed using a Gaussian model-based
approach (GLAD). The median of the signal ratio (test
signal/reference signal) of each triplicated spot was defined as a
gain or a loss when it was >0.25 or <−0.25, respectively.
High-level amplification of clones was defined when their intensity
ratios were >1.0 in log2 scale, and vice versa for
homozygous deletion. The threshold value was determined empirically
as a value 3-fold that of the standard deviations calculated from
30 normal males and females in hybridization experiments. The R
2.2.1 package of the Bioconductor Project (http://www.bioconductor.org) was used for the
detection of the frequency of gain or loss and for statistical
analysis. The Benjamini-Hochberg false discovery rate was applied
for multiple testing corrections for the high number of
false-positive calls.
Results
Genome wide array analysis in early GC
cases
In total, 22 tumor samples were assessed by
high-resolution array CGH to investigate DNA copy number
alterations and new candidate genes associated with early GCs. The
majority of clones were frequently gained (log2 ratio
>0.25) or lost (log2 ratio <−0.25), with 97.5% of
the clones being gained or lost in 62.7% of the cases. As the first
step of the analysis, chromosome 8q, the most frequently gained
[77.3% (17/22)] and amplified [log2 ratio >1, 18.2%
(4/22)] region in GC cases, was focused upon. There were high-level
amplifications on chromosome 8 at four distinct loci, centered at
106.4, 89.6, 77.6 and 110.5 Mb (containing MYC). The most
common region with copy number increase in GC cases was more
clearly defined and narrowed down to 8q22.1-q24.3, encompassing
BAC177_N09 to BAC145_J12. A list of the delineations of the
8q22.1-q24.3 chromosomal region and possible target genes of GC is
presented in Table I.
 | Table IVarious chromosomal recurrent minimal
regions of genetic alterations on the long arm of chromosome 8 in
22 GCs. |
Table I
Various chromosomal recurrent minimal
regions of genetic alterations on the long arm of chromosome 8 in
22 GCs.
| Regions | Gene contained in
clones | % of gainsa | % of
amplificationsb |
|---|
| 8q21.1-q21.3 | IL7, PMP2, FABP9,
FABP4, MMP16, NBS1, DECR1 | 22.7 (5/22) | |
| 8q22.1-q23.3 | TFCP2L3,
MGC35555 | 18.2 (4/22) | 9.1 (2/22) |
| 8q24.11-q24.3 | EXT1, NOV, LOC392264,
MYC, PVT1, LOC401475, SIAT4A, WISP1, NDRG1, TG, LOC286094, NDRG1,
LOC392271, COL22A1, CYHR1, KIFC2, FOXH1, PPP1R16A, GPT, LOC113655,
RECQL4, ZC3HDC3, LOC389693, GSDMDC1, LOC389694, PP3856, EEF1D | 77.3 (17/22) | 9.1 (2/22) |
Chromosomal alterations of 8q22.1-q24.3
are the most common genetic changes in early GCs
In the 8q22.1-q24.3 region, two common regions of
alterations across the chromosome were further clarified. The first
loci spanned 96.3–116.8 kb, and was mapped at the 8q22.1-q23.3
region. Notably, two amplified loci in these regions were
identified in 9.1% (2/22) of the cases. One locus at 8q22.3
contained amplified clones covering a region of ~106.4 kb, and
comprised the transcription factor CP2-like 3 (TFCP2L3) gene
and the other locus spanning ~89.6 kb on 8q23.1, without associated
genes. The median span of the copy number amplifications was 16.8
kb (range, 89.6–106.4 kb), and all amplifications were located
between BAC157_F12 and BAC169_B05.
The second loci spanned 119.0–144.7 kb on
8q24.11-q24.3, and this region contained 27 possible target genes
according to the information archived from human genome databases
(http://genome.ucsc.edu/). Significantly, a high
frequency of copy number gains (log2 ratio >0.25) and
high-level gains (log2 ratio >0.5) in the
8q24.11-q24.3 region were detected in 77.3 (17/22) and 36.4% (8/22)
of the GC cases, respectively. More specifically, two amplified
(log2 ratio >1) loci in these regions were noted in
9.1% of the GC cases. One locus at 8q24.21 comprised the
representative oncogenes of myelocytomatosis (MYC) and
plasmacytoma variant translocation 1 (PVT1) (4.5%). The
other locus spanning ~110.5 kb on 8q24.3 was found to contain
cysteine/histidine rich 1 (CYHR1), kinesin family member C2
(KIFC2), forkhead box H1 (FOXH1), protein phosphatase
1 regulatory subunit 16A (PPP1R16A), glutamic-pyruvate
transaminase (GPT), LOC113655 and RecQ protein-like 4
(RECQL4) genes with the highest level of amplification. The
median span of the copy number amplifications of the 8q24.11-q24.3
region was 32.9 kb (range, 77.6–110.5 kb), and all amplifications
were located between BAC192_N08 and BAC145_J11. An example of an
individual profile at the 8q22.1-q24.3 region in the 22 GC cases is
shown in Fig. 1. A representative
weighted frequency (%) diagram with high-level amplifications in
the 8q22.1-q24.3 region for all 22 GC cases is displayed in
Fig. 2.
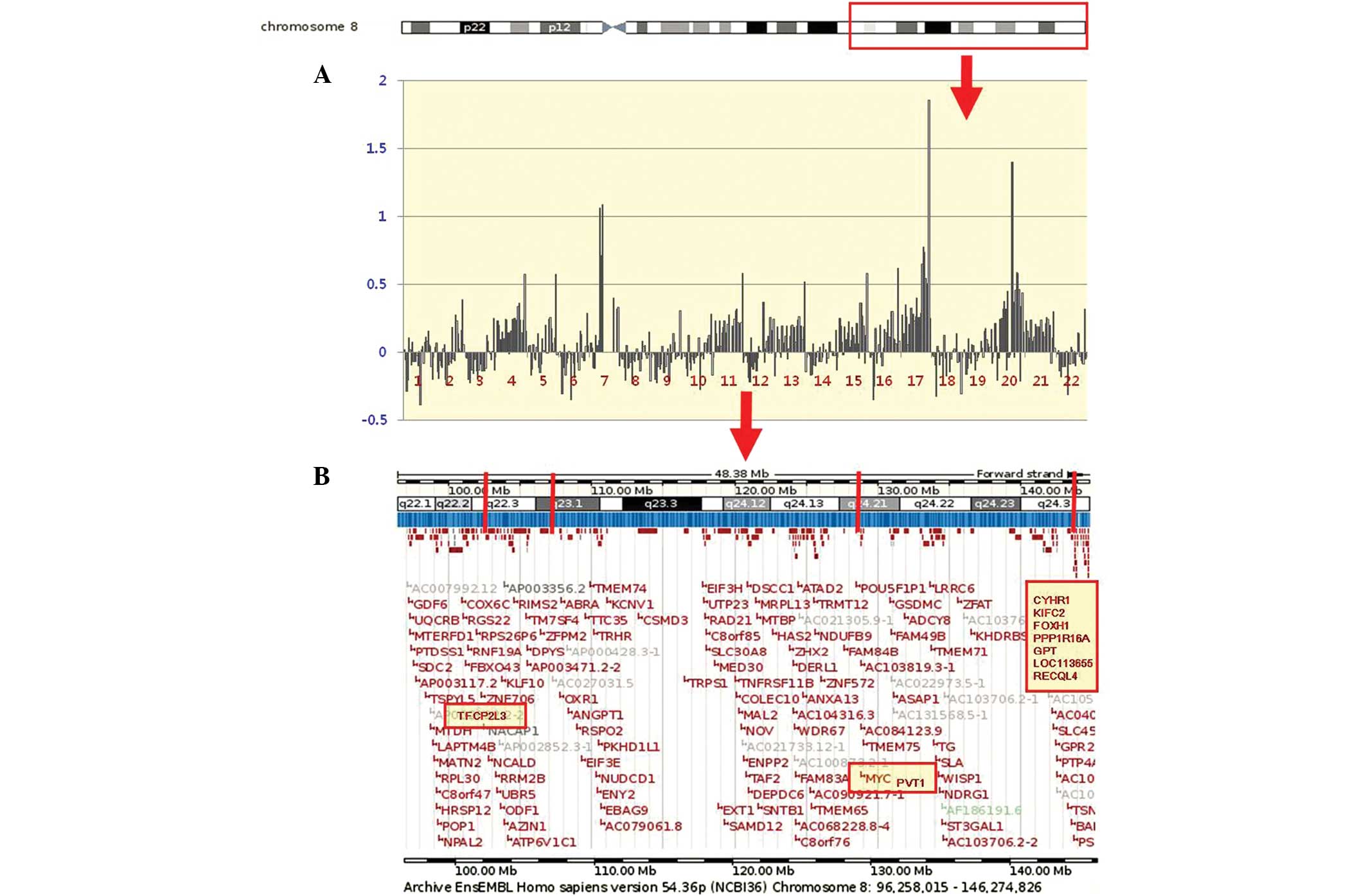 | Figure 1(A) An example of an individual
profile at the 8q22.1-q24.3 region in the 22 GC cases. High-level
amplifications are clearly seen in cases 7, 17 and 20. Cytobands in
the ideogram are shown in the upper image. (B) The schematic
presentation of cytogenetic bands, and a map position from the UCSC
genome browser is shown below the plot. The candidate target genes
(TFCP2L3, MYC, PVT1, CYHR1, KIFC2, FOXH1, PPP1R16A, GPT,
LOC113655 and RECQL4) at the 8q22.1-q24.3 region are
shaded in yellow. GC, gastric carcinoma; UCSC, University of
California Santa Cruz; TFCP2L3, transcription factor
CP2-like 3; MYC, myelocytomatosis; PVT1, plasmacytoma
variant translocation 1; CYHR1, cysteine/histidine rich 1;
KIFC2, kinesin family member C2; FOXH1, forkhead box
H1; PPP1R16A, protein phosphatase 1 regulatory subunit 16A;
GPT, glutamic-pyruvate transaminase; RECQl4, RecQ
protein-like 4. |
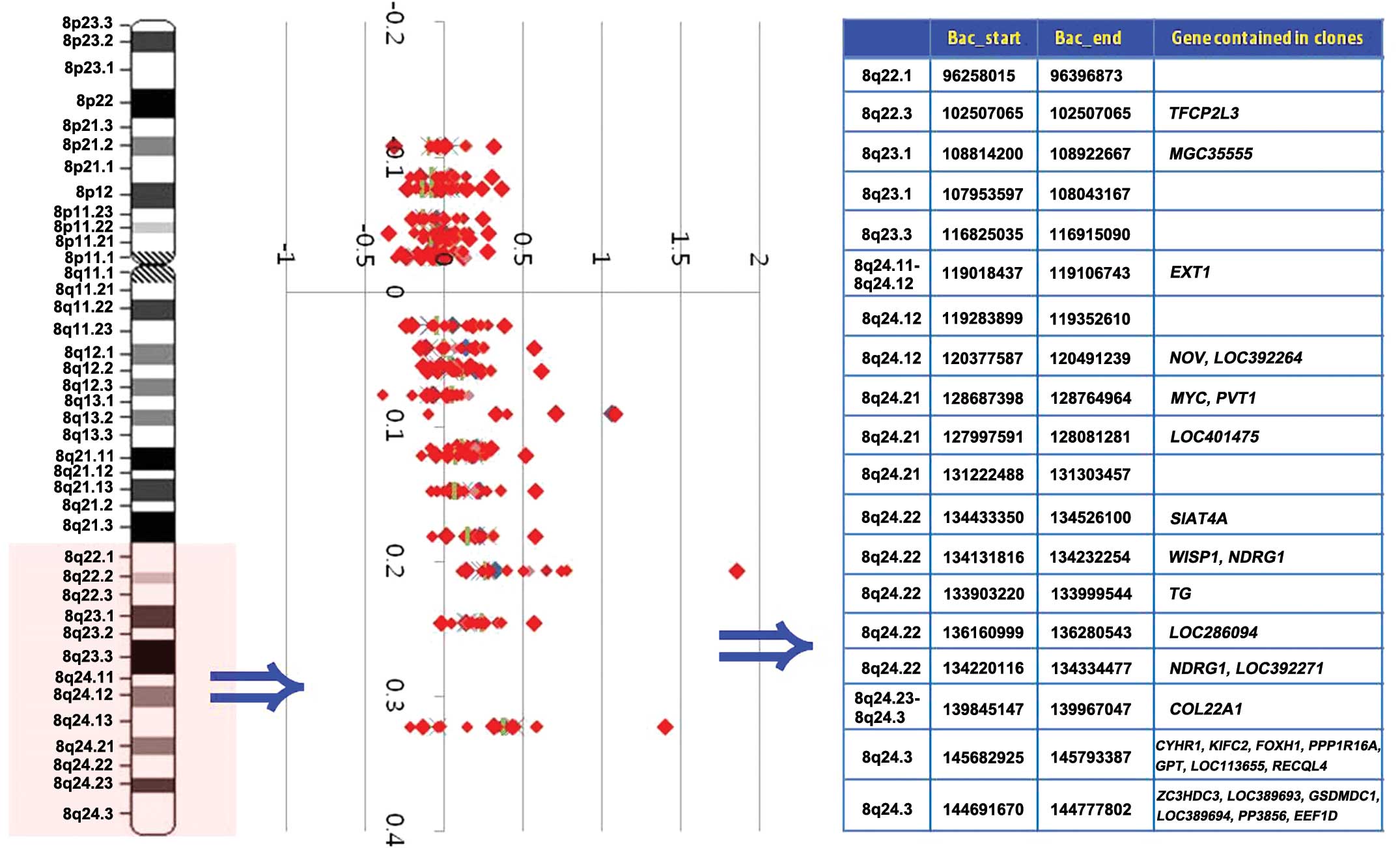 | Figure 2Weighted frequency (%) diagram of the
8q22.1-q24.3 region in GC cases. In the intensity ratio profiles,
the Y-axis represents the map position of the corresponding clone
and the intensity ratios are assigned to the X-axis. Cytobands in
the ideogram are shown on the left. Genes contained in clones are
shown on the right. GC, gastric carcinoma; MYC,
myelocytomatosis; PVT1, plasmacytoma variant translocation
1; CYHR1, cysteine/histidine rich 1; KIFC2, kinesin
family member C2; FOXH1, forkhead box H1; PPP1R16A,
protein phosphatase 1 regulatory subunit 16A; GPT,
glutamic-pyruvate transaminase; RECQl4, RecQ protein-like
4. |
Discussion
In the present study, the most notable finding was
the frequent genetic abnormality of chromosome 8q in early GC
cases. The long arm of chromosome 8 has long been suspected to
contain critical oncogenes that lead to numerous cancer types,
including GC (14–17). Abi-Ayad et al (16) reported that copy number gains of the
8q arm (70%) or the entirety of chromosome 8 were the most frequent
imbalance in melanoma. de Krijger et al (17) also reported that chromosome 8q gains
were the most frequent genetic abnormality in pleuropulmonary
blastoma. Notably, the two surviving patients in the series
exhibited gains of chromosome 8q material as the only genetic
abnormality. Furthermore, Buffart et al (14) documented copy number gains of genes
on chromosome 8q in 9.5–73.0% of the GC cases, with the highest
frequency of gains in MYC (73.0%). More recently, Cheng
et al (2) documented the
highest frequency (70%) of copy number gains at 8q in GCs by array
CGH analysis. By combining the results of the present study with
those in other studies, it is indicated that copy number gains on
chromosome 8q occur as an early event in the multistage development
of various tumors, including GC, which commences in the mildly
abnormal epithelium.
More specifically, the smallest region of overlap,
the 8q22.11-q24.3 region, was determined. A high frequency of
single copy number gains (log2 ratio >0.25) and
high-level gains (log2 ratio >0.5) in this region was
detected in 77.3 and 36.4% of the cases, respectively. Notably, two
amplified (log2 ratio >1) loci in these regions were
detected in 18.2% (4/22) of the GC cases. One locus on 8q24.3 was
found to contain CYHR1, KIFC2, FOXH1, PPP1R16A, GPT,
LOC113655 and RECQL4 genes, placing the highest level of
amplifications in the GC cases. To the best of our knowledge, the
involvement of these genes in the pathogenesis of GC has not been
described previously; however, genetic mutations of these genes are
commonly found in various types of cancers. Katoh et al
(15) reported high-level
amplification and overexpression of the FOXA1 gene in
esophageal and lung cancer. Upregulation of the FOXA1 gene
in pancreatic cancer and basal cell carcinoma, due to the
transcriptional regulation by the Sonic Hedgehog (SHH) pathway, has
also been documented. Furthermore, overexpression of the
RECQL4 gene with chromosomal aberrations and instability in
osteosarcoma has also been reported (18,19).
These findings indicate that chromosomal alterations of these
developmental genes may contribute in playing a role in the
underlying mechanism of cancer development.
The other amplified locus at 8q24.21 comprised the
representative oncogene of MYC. Several experimental studies
have shown that MYC amplification or overexpression is
observed in early GC patients when tumor invasion is confined to
the mucosa or submucosa, regardless of the presence of lymph node
metastasis (2,18–22).
In the study by Onoda et al (20), MYC expression was found to be
more frequent and stronger in early lesions compared with advanced
lesions, and Ishii et al (21) also documented increased MYC
expression in early GCs in comparison with decreased MYC
expression in late-stage GCs. Based on these findings and the data
from the present study, MYC genetic aberrations may be an
early event in the pathogenesis of gastric carcinogenesis, and the
detection of MYC locus amplification could be used as an
auxiliary tool in GC diagnosis and as a predictor of GC
aggressiveness (22).
In addition, one potential oncogene candidate of
PVT1 was identified as a potential target within the 8q24.21
amplicon. PVT1 encodes a non-coding RNA and is a host gene
for several miRNAs, namely hsa-miR-1204, 1205, 1206 and 1207
(23). Although involvement of the
PVT1 gene in the pathogenesis of GC has not been mentioned
thus far, genetic mutations of the PVT1 gene have been
consistently reported in multiple types of tumors (23–26).
Nagoshi et al (24)
indicated that PVT1 rearrangements represent a novel
molecular paradigm underlying the pathology of 8q24.21
rearrangement-positive multiple myeloma. PVT1 also showed
increased expression in prostate cell lines compared to normal
prostate tissue (25). In addition,
Meyer et al (23) reported
that the risk locus can interact with two downstream genes,
MYC and PVT1, and that PVT1, a novel target
gene candidate, regulates the 8q24 risk region. Additionally, an
association between genetic variants within PVT1 and
Hodgkin’s lymphoma has also been postulated (26). Taken together, the results of the
present study and the findings of other studies also present
evidence that PVT1 is a new target gene candidate, regulated
by the 8q24 risk region, and that it could be defined as an
independent target region for chromosome 8q amplifications in
various tumors, including GCs. Further functional and biological
studies are required to validate and evaluate the role of the
PVT1 gene as a novel candidate oncogene in GC in larger
series and on multiple samples.
In summary, the present study confirms and expands
upon previous observations that 8q genetic mutations accumulate
early during the multistage pathogenesis of GC. The confirmation of
previously reported 8q gains and the identification of novel target
genes at 8q22.1-q24.3 amplified chromosomal sites should provide
important clues with regard to the genetic mechanisms of initiation
and progression, and provide new insights into the clinical
behavior and management of GC. Additional genome-wide studies with
a larger number of patients are warranted to confirm the results of
the present study and to improve our understanding of GC.
Acknowledgements
This study was financially supported by the research
fund of Korea Nazarene University in 2013.
References
|
1
|
Nakayama T, Ling ZQ, Mukaisho K, Hattori T
and Sugihara H: Lineage analysis of early and advanced tubular
adenocarcinomas of the stomach: continuous or discontinuous? BMC
Cancer. 10:3112010. View Article : Google Scholar
|
|
2
|
Cheng L, Wang P, Yang S, et al:
Identification of genes with a correlation between copy number and
expression in gastric cancer. BMC Med Genomics. 5:142012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brenner H, Rothenbacher D and Arndt V:
Epidemiology of stomach cancer. Methods Mol Biol. 472:467–477.
2009. View Article : Google Scholar
|
|
4
|
Gümüs-Akay G, Unal AE, Elhan AH, et al:
DNA copy number changes in gastric adenocarcinomas: high
resolution-comparative genomic hybridization study in Turkey. Arch
Med Res. 40:551–560. 2009.
|
|
5
|
Cheng L and Zhang Q, Yang S, Yang Y, Zhang
W, Gao H, Deng X and Zhang Q: A 4-gene panel as a marker at
chromosome 8q in Asian gastric cancer patients. Genomics.
102:323–330. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mihailovici MS, Danciu M, Teleman S,
Stanciu C, Stan M, Bălan G and Potoroacă A: Diagnosis of gastric
cancer on endobiopsies using the WHO classification. Rev Med Chir
Soc Med Nat Iasi. 106:725–729. 2002.(In Romanian).
|
|
7
|
Sun YN and Li Y: Expression of mRNA for
membrane-type 1, 2, and 3 matrix metalloproteinases in human
laryngeal cancer. Chin Med Sci J. 19:170–173. 2004.PubMed/NCBI
|
|
8
|
Lowy AM, Clements WM, Bishop J, et al:
beta-Catenin/Wnt signaling regulates expression of the membrane
type 3 matrix metalloproteinase in gastric cancer. Cancer Res.
66:4734–4741. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nanjundan M, Nakayama Y, Cheng KW, Lahad
J, Liu J, Lu K, Kuo WL, Smith-McCune K, Fishman D, Gray JW and
Mills GB: Amplification of MDS1/EVI1 and EVI1, located in the
3q26.2 amplicon, is associated with favorable patient prognosis in
ovarian cancer. Cancer Res. 67:3074–3084. 2007. View Article : Google Scholar
|
|
10
|
Kim KR, Oh SY, Park UC, et al: Gene
expression profiling using oligonucleotide microarray in atrophic
gastritis and intestinal metaplasia. Korean J Gastroenterol.
49:209–224. 2007.(In Korean).
|
|
11
|
Sanjmyatav J, Steiner T, Wunderlich H,
Diegmann J, Gajda M and Junker K: A specific gene expression
signature characterizes metastatic potential in clear cell renal
cell carcinoma. J Urol. 186:289–294. 2011. View Article : Google Scholar
|
|
12
|
Hashimoto T, Kusakabe T, Watanabe K, et
al: Liver-type fatty acid-binding protein is highly expressed in
intestinal metaplasia and in a subset of carcinomas of the stomach
without association with the fatty acid synthase status in the
carcinoma. Pathobiology. 71:115–122. 2004. View Article : Google Scholar
|
|
13
|
Hashimoto T, Kusakabe T, Sugino T, et al:
Expression of heart-type fatty acid-binding protein in human
gastric carcinoma and its association with tumor aggressiveness,
metastasis and poor prognosis. Pathobiology. 71:267–273. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Buffart TE, van Grieken NC, Tijssen M,
Coffa J, Ylstra B, Grabsch HI, van de Velde CJ, Carvalho B and
Meijer GA: High resolution analysis of DNA copy-number aberrations
of chromosomes 8, 13, and 20 in gastric cancers. Virchows Arch.
455:213–223. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Katoh M and Katoh M: Human FOX gene family
(Review). Int J Oncol. 25:1495–1500. 2004.PubMed/NCBI
|
|
16
|
Abi-Ayad N, Couturier J,
Devouassoux-Shisheboran M, Grange JD, Kodjikian L and Calender A:
Genomic profiling by comparative genomic hybridization: analysis of
ten enucleated uveal melanoma cases. J Fr Ophtalmol. 34:17–23.
2011.(In French).
|
|
17
|
de Krijger RR, Claessen SM, van der Ham F,
van Unnik AJ, Hulsbergen-van de Kaa CA, van Leuven L, van Noesel M
and Speel EJ: Gain of chromosome 8q is a frequent finding in
pleuropulmonary blastoma. Mod Pathol. 20:1191–1199. 2007.PubMed/NCBI
|
|
18
|
Costa Raiol LC, Figueira Silva EC, Mendes
da Fonseca D, et al: Interrelationship between MYC gene numerical
aberrations and protein expression in individuals from northern
Brazil with early gastric adenocarcinoma. Cancer Genet Cytogenet.
181:31–35. 2008.
|
|
19
|
Calcagno DQ, Leal MF, Assumpcao PP, Smith
MA and Burbano RR: MYC and gastric adenocarcinoma carcinogenesis.
World J Gastroenterol. 14:5962–5968. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Onoda N, Maeda K, Chung YS, Yano Y,
Matsui-Yuasa I, Otani S and Sowa M: Overexpression of c-myc
messenger RNA in primary and metastatic lesions of carcinoma of the
stomach. J Am Coll Surg. 182:55–59. 1996.PubMed/NCBI
|
|
21
|
Ishii H, Gobé G, Kawakubo Y, Sato Y and
Ebihara Y: Interrelationship between Epstein-Barr virus infection
in gastric carcinomas and the expression of apoptosis-associated
proteins. Histopathology. 38:111–119. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Suzuki S, Tenjin T, Watanabe H, Matsushima
S, Shibuya T and Tanaka S: Low level c-myc gene amplification in
gastric cancer detected by dual color fluorescence in situ
hybridization analysis. J Surg Oncol. 66:173–178. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Meyer KB, Maia AT, O’Reilly M, et al: A
functional variant at a prostate cancer predisposition locus at
8q24 is associated with PVT1 expression. PLoS Genet.
7:e10021652011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nagoshi H, Taki T, Hanamura I, et al:
Frequent PVT1 rearrangement and novel chimeric genes PVT1-NBEA and
PVT1-WWOX occur in multiple myeloma with 8q24 abnormality. Cancer
Res. 72:4954–4962. 2012. View Article : Google Scholar
|
|
25
|
Jia L, Landan G, Pomerantz M, Jaschek R,
Herman P, Reich D, Yan C, Khalid O, Kantoff P, Oh W, Manak JR,
Berman BP, Henderson BE, Frenkel B, Haiman CA, Freedman M, Tanay A
and Coetzee GA: Functional enhancers at the gene-poor 8q24
cancer-linked locus. PLoS Genet. 5:e10005972009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Enciso-Mora V, Broderick P, Ma Y, et al: A
genome-wide association study of Hodgkin’s lymphoma identifies new
susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3). Nat
Genet. 42:1126–1130. 2010.
|
















