Introduction
Adult T-cell leukemia (ATL) is a highly aggressive
malignant disease of CD4+ T cells, caused by human
T-cell lymphotropic virus type I (HTLV-I) (1). Infection with HTLV-1, the first
oncogenic human retrovirus to be identified, also causes various
chronic inflammatory disorders, including HTLV-I-associated
myelopathy/tropical spastic paraparesis (2). The majority of infected individuals
remain clinically asymptomatic, although 2–5% of HTLV-I-infected
carriers develop ATL through genetic and epigenetic changes in the
cell following a latent period of 40–60 years (3). Tax, the viral oncoprotein, plays a
central role in tumorigenesis and contributes to the pathogenesis
of ATL and inflammatory diseases by inducing persistent activation
of numerous cellular transcription factors, including nuclear
factor-κB, cyclic adenosine 3′,5′-monophosphate response
element-binding protein and activator protein 1, leading to
transactivation of cellular gene promoters (4). However, based on the absence of Tax
protein expression in numerous late-stage ATL cells, this viral
protein appears to be required only for the initiation of
transformation (3,4).
High mobility group box 1 (HMGB1) is an abundant and
ubiquitous nuclear protein that binds to DNA and nucleosomes and
induces structural changes in the chromatin fiber. It regulates
numerous cellular activities, including transcription, replication
and repair (5). In addition, HMGB1
is released from damaged, necrotic or activated immune cells and
functions as an extracellular signaling molecule (6,7). Thus,
intranuclear and extranuclear HMGB1 are therapeutic targets for
inflammation and infection (8).
Previous studies have identified the roles of HMGB1 in cancer
(9), with high protein expression
in colon, breast, lung, prostate, cervical and gastric cancer,
hepatocellular carcinoma and leukemia compared with normal tissues
and healthy controls (10).
More recently, Zhang et al (11) have demonstrated that HMGB1
expression is upregulated by HTLV-I Tax in T cells. However, the
expression levels of HMGB1 in HTLV-I-infected T-cell lines have not
been determined. In the present study, the levels HMGB1 in several
T-cell lines and in the plasma of patients with ATL were
analyzed.
Materials and methods
Cell lines
The HTLV-I-infected T-cell lines, MT-2 (12), MT-4 (13), C5/MJ (14), SLB-1 (15), HuT-102 (16), MT-1 (17), TL-OmI (18) and ED-40515(−) (19), were cultured in RPMI-1640 medium
supplemented with 10% heat-inactivated fetal bovine serum. MT-2,
MT-4, C5/MJ and SLB-1 are HTLV-I-transformed T-cell lines
established by an in vitro coculture protocol. MT-1, TL-OmI
and ED-40515(−) are T-cell lines of leukemic cell origin
established from patients with ATL. HuT-102 was also established
from a patient with ATL and constitutively expresses viral genes,
but its clonal origin is not clear. In JPX-9 cells, Tax is under
the transcriptional control of the metallothionein gene promoter
and can be induced by the addition of CdCl2 to the
medium (20). Experiments using
JPX-9 cells were carried out after a 24-h cultivation period in the
absence and presence of 20 μM CdCl2.
Patients and plasma samples
The diagnosis of acute ATL was based on clinical
features, hematological findings and the presence of anti-HTLV-I
antibodies in the serum. Plasma samples were collected at the time
of admission to the Naha City Hospital (Naha, Japan) prior to
chemotherapy and stored at −80°C until use. Informed consent was
obtained from all blood donors.
Polymerase chain reaction (PCR)
Total RNA was prepared from various cell cultures
using TRIzol (Invitrogen Life Technologies, Carlsbad, CA, USA)
according to the instructions provided by the manufacturer.
First-strand cDNA was synthesized from 1 μg total cellular
RNA using a PrimeScript RT-PCR kit (Takara Bio Inc., Otsu, Japan)
with random primers. The primer sequences for HMGB1, receptor for
advanced glycation end products (RAGE), Tax and GAPDH are listed in
Table I. PCR was halted during the
exponential phase of DNA amplification and the reaction products
were fractionated on 2% agarose gels and visualized by ethidium
bromide staining. The obtained bands of amplified DNA were
quantified using Image J (National Institutes of Health, Bethesda,
MD, USA).
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Gene name | Forward sequence | Reverse sequence |
|---|
| HMGB1 |
5′-ATGGGCAAAGGAGATCCTAAGAA-3′ |
5′-TTATTCATCATCATCATCTTCTT-3′ |
| RAGE |
5′-ATGGAAACTGAACACAGGCC-3′ |
5′-CACACATGTCCCCACCTTAT-3′ |
| Tax |
5′-CCGGCGCTGCTCTCATCCCGGT-3′ |
5′-GGCCGAACATAGTCCCCCAGAG-3′ |
| GAPDH |
5′-GCCAAGGTCATCCATGACAACTTTGG-3′ |
5′-GCCTGCTTCACCACCTTCTTGATGTC-3′ |
Assay for HMGB1
The concentrations of HMGB1 were measured in
cultured supernatants from JPX-9 cells and plasma samples from
patients and healthy donors using a commercially available ELISA
kit II (Shino-Test Corporation, Tokyo, Japan) according to the
manufacturer’s instructions. The minimum detection level for HMGB1
was 1 ng/ml.
Statistical analysis
Differences between groups were examined for
statistical significance using the unpaired Student’s t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of HMGB1 and RAGE mRNA in
HTLV-I-infected T-cell lines
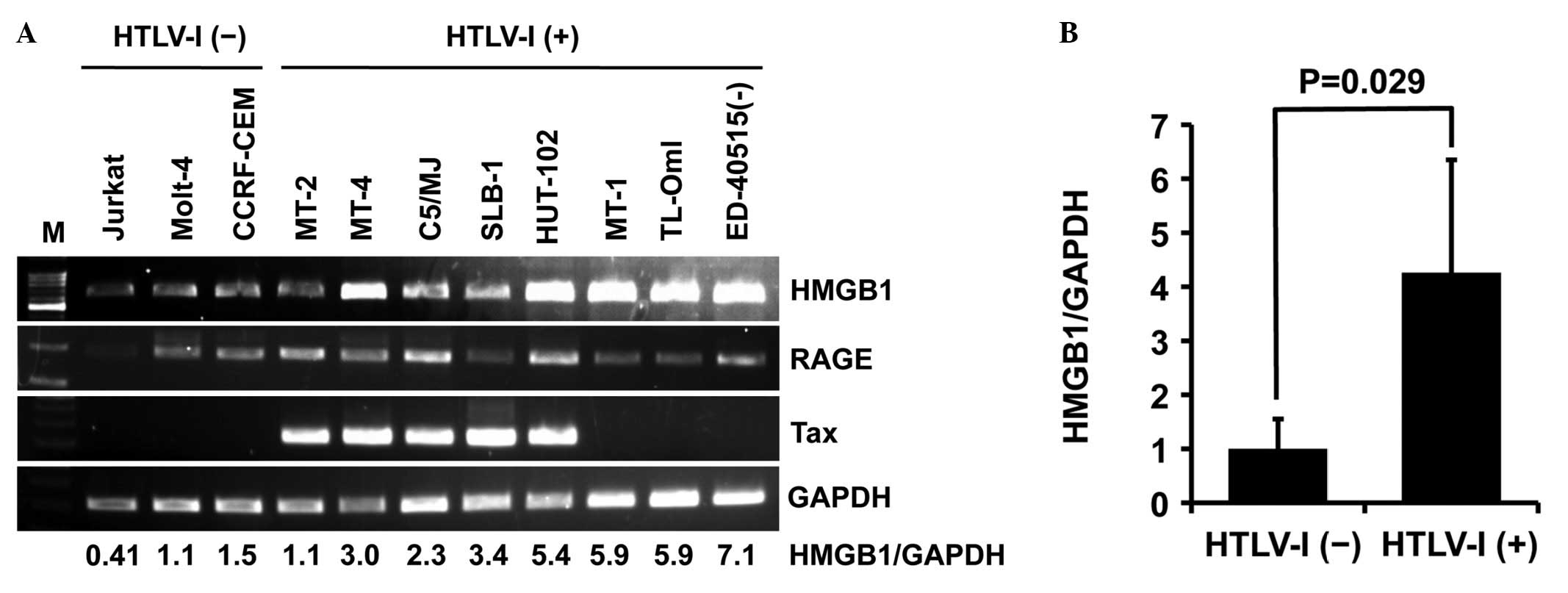
First, the expression levels of HMGB1, RAGE (HMGB1
receptor) and Tax were analyzed by PCR in eight HTLV-I-infected
T-cell lines [MT-2, MT-4, C5/MJ, SLB-1, HuT-102, MT-1, TL-OmI and
ED-40515(−)]. HTLV-I-transformed T-cell lines (MT-2, MT-4, C5/MJ
and SLB-1) and HuT-102 constitutively expressed Tax mRNA, but
ATL-derived T-cell lines [MT-1, TL-OmI and ED-40515(−)] did not
(Fig. 1A). PCR also revealed high
expression of HMGB1 mRNA in all HTLV-I-infected T-cell lines except
MT-2 compared with uninfected T-cell lines (Jurkat, Molt-4 and
CCRF-CEM), regardless of Tax expression (Fig. 1A). The relative expression of HMGB1
in HTLV-I-infected T-cell lines was four-fold higher than in
uninfected T-cell lines (Fig. 1B).
By contrast, RAGE was expressed in all T-cell lines except Jurkat,
regardless of HTLV-I infection, although its expression levels
varied widely (Fig. 1A).
Tax protein enhances HMGB1 release in T
cells
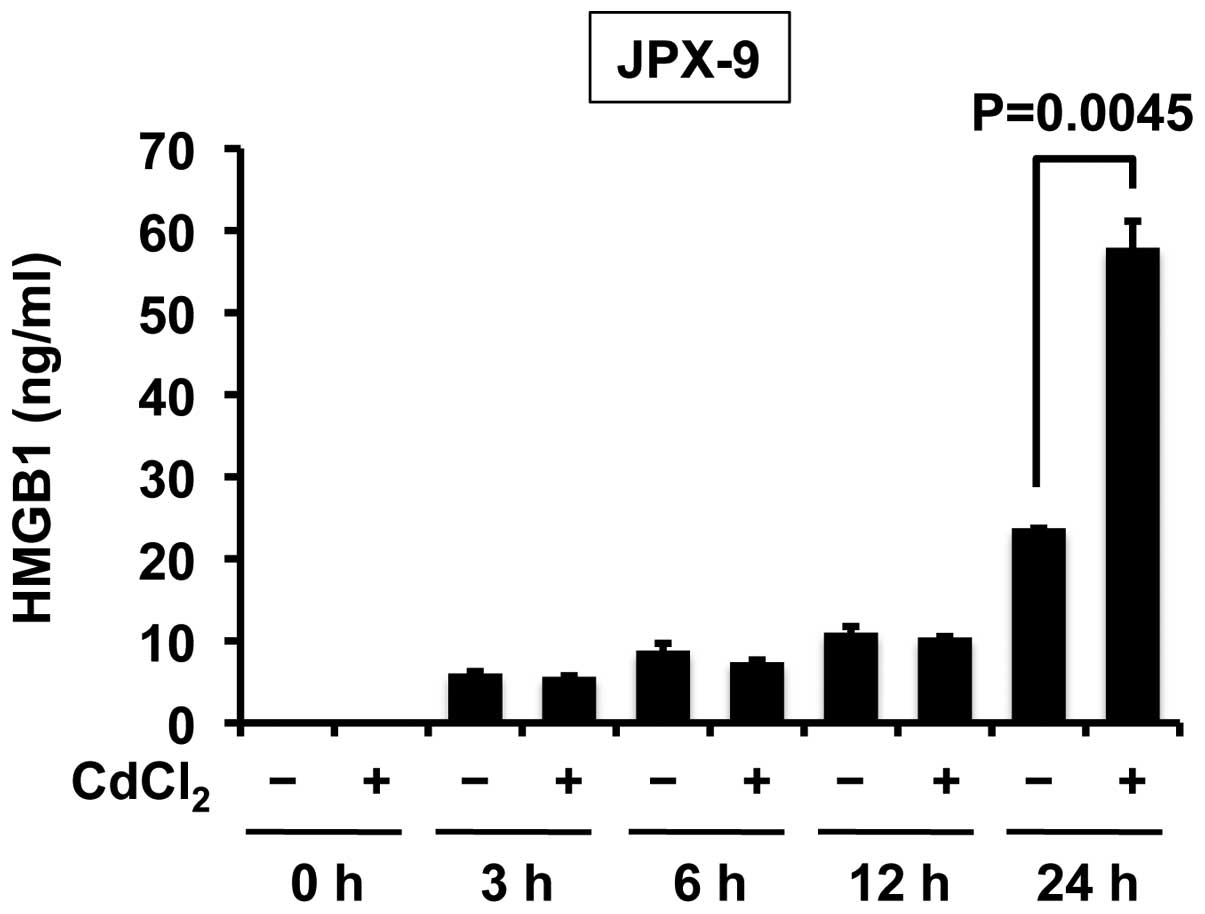
Next, the study sought to discern whether Tax
induces the release of HMGB1 in T cells. Treatment of JPX-9 cells
(a Jurkat subline that carries the Tax gene under the control of
the metallothionein gene promoter) with CdCl2 rapidly
induced Tax expression (20). ELISA
revealed that Tax enhanced the release of HMGB1 in T cells in the
24 h after treatment with CdCl2 (Fig. 2).
Plasma levels of HMGB1 in patients with
ATL
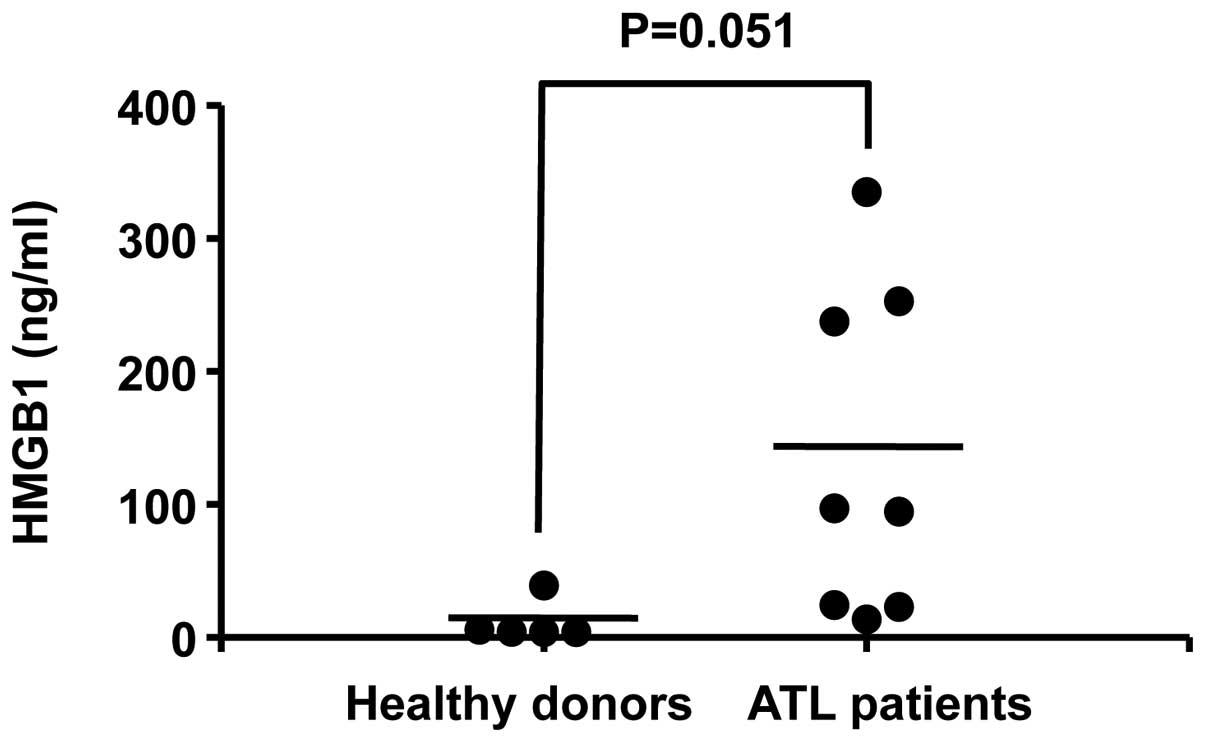
Finally, HMGB1 levels in the plasma of ATL patients
were investigated. The plasma levels of HMGB1 in patients with
acute ATL (n=8) tended to be higher than those in healthy donors
(n=5), albeit without statistical significance (P=0.051) (Fig. 3). The mean plasma HMGB1 levels in
ATL patients (134 ng/ml) were ten-fold higher than those of healthy
donors (11 ng/ml). These data demonstrate that HMGB1 is markedly
released in the plasma of ATL patients.
Discussion
The HTLV-I transactivator protein, Tax, is a protein
of significant interest in HTLV-I pathogenesis, as it is a potent
activator of a variety of transcription pathways and is in itself
sufficient to immortalize T cells in vitro. Thus, it plays
an important role in cellular transformation (3,4).
Recently, it has been reported that Tax is involved in upregulation
of HMGB1 expression by interaction with CCAAT/enhancer binding
protein (11). The present study
found that HMGB1 mRNA was abundantly expressed in HTLV-I-infected
T-cell lines. In addition, Tax increased HMGB1 secretion in T
cells. These results are consistent with those reported previously
(11). Tax expression did not
correlate with the upregulation of HMGB1 mRNA in HTLV-I-infected
T-cell lines. However, significantly higher plasma levels of HMGB1
were noted in ATL patients with peripheral leukemic cells negative
for Tax protein than in healthy donors. Thus, it may be that
another factor is essential for the induction of HMGB1 expression
and secretion in ATL.
While the exact function of HMGB1 protein is not
clear at present, it is reported to play important roles in
sustained angiogenesis, evasion of apoptosis, growth signal
self-sufficiency, insensitivity to antigrowth signals, the
inflammatory microenvironment, immortalization, tissue invasion and
metastasis in cancer cells (10).
In addition, HMGB1 is reported to promote drug resistance in
osteosarcoma (21). Previous
studies have also shown that endogenous HMGB1 regulates autophagy
(22) and that HMGB1-induced
autophagy promotes resistance to chemotherapy in leukemia cells
(23). Therefore, HMGB1 appears to
play important roles in malignant progression, inflammation, organ
infiltration and drug resistance in HTLV-I-associated diseases,
including ATL. These findings suggest that HMGB1 is a potential
biomarker and therapeutic target for ATL.
In conclusion, the present study demonstrated that
there are elevated plasma HMGB1 levels in patients with ATL and an
abundant HMGB1 expression in HTLV-I-infected T-cell lines.
Acknowledgements
The authors thank Dr Masataka Nakamura for providing
JPX-9 cells, Dr Michiyuki Maeda for providing ED-40515(−) cells and
the Fujisaki Cell Center, Hayashibara Biomedical Laboratories
(Okayama, Japan) for providing HuT-102, C5/MJ and MT-1 cells. The
authors would also like to thank Naha City Hospital (Naha, Japan)
for providing plasma samples from patients.
References
|
1
|
Yoshida M, Miyoshi I and Hinuma Y:
Isolation and characterization of retrovirus from cell lines of
human adult T-cell leukemia and its implication in the disease.
Proc Natl Acad Sci USA. 79:2031–2035. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Watanabe T: HTLV-1-associated diseases.
Int J Hematol. 66:257–278. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Matsuoka M and Jeang KT: Human T-cell
leukaemia virus type 1 (HTLV-1) infectivity and cellular
transformation. Nat Rev Cancer. 7:270–280. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Grassmann R, Aboud M and Jeang KT:
Molecular mechanisms of cellular transformation by HTLV-1 Tax.
Oncogene. 24:5976–5985. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hock R, Furusawa T, Ueda T and Bustin M:
HMG chromosomal proteins in development and disease. Trends Cell
Biol. 17:72–79. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lotze MT and Tracey KJ: High-mobility
group box 1 protein (HMGB1): nuclear weapon in the immune arsenal.
Nat Rev Immunol. 5:331–342. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dumitriu IE, Baruah P, Manfredi AA,
Bianchi ME and Rovere-Querini P: HMGB1: guiding immunity from
within. Trends Immunol. 26:381–387. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Andersson U and Tracey KJ: HMGB1 is a
therapeutic target for sterile inflammation and infection. Annu Rev
Immunol. 29:139–162. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sims GP, Rowe DC, Rietdijk ST, Herbst R
and Coyle AJ: HMGB1 and RAGE in inflammation and cancer. Annu Rev
Immunol. 28:367–388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tang D, Kang R, Zeh HJ III and Lotze MT:
High-mobility group box 1 and cancer. Biochim Biophys Acta.
1799:131–140. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang CG, Wang H, Niu ZG, Zhang JJ, Yin
MM, Gao ZT and Hu LH: Tax is involved in up-regulation of HMGB1
expression levels by interaction with C/EBP. Asian Pac J Cancer
Prev. 14:359–365. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Miyoshi I, Kubonishi I, Yoshimoto S, Akagi
T, Ohtsuki Y, Shiraishi Y, Nagata K and Hinuma Y: Type C virus
particles in a cord T-cell line derived by co-cultivating normal
human cord leukocytes and human leukaemic T cells. Nature.
294:770–771. 1981. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yamamoto N, Okada M, Koyanagi Y, Kannagi M
and Hinuma Y: Transformation of human leukocytes by cocultivation
with an adult T cell leukemia virus producer cell line. Science.
217:737–739. 1982. View Article : Google Scholar
|
|
14
|
Popovic M, Sarin PS, Robert-Gurroff M,
Kalyanaraman VS, Mann D, Minowada J and Gallo RC: Isolation and
transmission of human retrovirus (human t-cell leukemia virus).
Science. 219:856–859. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Koeffler HP, Chen IS and Golde DW:
Characterization of a novel HTLV-infected cell line. Blood.
64:482–490. 1984.PubMed/NCBI
|
|
16
|
Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn
PA, Minna JD and Gallo RC: Detection and isolation of type C
retrovirus particles from fresh and cultured lymphocytes of a
patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA.
77:7415–7419. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Miyoshi I, Kubonishi I, Sumida M, Hiraki
S, Tsubota T, Kimura I, Miyamoto K and Sato J: A novel T-cell line
derived from adult T-cell leukemia. Gann. 71:155–156.
1980.PubMed/NCBI
|
|
18
|
Sugamura K, Fujii M, Kannagi M, Sakitani
M, Takeuchi M and Hinuma Y: Cell surface phenotypes and expression
of viral antigens of various human cell lines carrying human T-cell
leukemia virus. Int J Cancer. 34:221–228. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Maeda M, Shimizu A, Ikuta K, Okamoto H,
Kashihara M, Uchiyama T, Honjo T and Yodoi J: Origin of human
T-lymphotrophic virus I-positive T cell lines in adult T cell
leukemia. Analysis of T cell receptor gene rearrangement. J Exp
Med. 162:2169–2174. 1985. View Article : Google Scholar
|
|
20
|
Ohtani K, Nakamura M, Saito S, Nagata K,
Sugamura K and Hinuma Y: Electroporation: application to human
lymphoid cell lines for stable introduction of a transactivator
gene of human T-cell leukemia virus type I. Nucleic Acids Res.
17:1589–1604. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang J, Ni J, Liu K, Yu Y, Xie M, Kang R,
Vernon P, Cao L and Tang D: HMGB1 promotes drug resistance in
osteosarcoma. Cancer Res. 72:230–238. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tang D, Kang R, Livesey KM, Cheh CW,
Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ III
and Lotze MT: Endogenous HMGB1 regulates autophagy. J Cell Biol.
190:881–892. 2010. View Article : Google Scholar
|
|
23
|
Liu L, Yang M, Kang R, Wang Z, Zhao Y, Yu
Y, Xie M, Yin X, Livesey KM, Lotze MT, Tang D and Cao L:
HMGB1-induced autophagy promotes chemotherapy resistance in
leukemia cells. Leukemia. 25:23–31. 2011. View Article : Google Scholar : PubMed/NCBI
|