In 2011, breast cancer ranked first in the incidence
and mortality of tumor diseases (1). Currently, it is the leading cause of
cancer and continues to be the second most common cause of cancer
mortality in females worldwide (2).
In developed countries, the lower mortality rate is attributed to
mammographic screening and advances in adjuvant therapy (3). The most commonly diagnosed breast
cancer subtypes are luminal A and B tumors (4,5),
together defined as the ER-positive (ER+) and
progesterone receptor-positive (PgR+) tumors (6–8). In
total, >70% of breast cancers are ER+ (9,10) and,
thus, ERs remain the most informative biomarkers in specific
subtypes of breast tumors (3,11). The
ERs (subtypes α and β) are members of the nuclear receptor family
of proteins modulating the expression of genes in response to
ligand binding (12–14). ERα expression occurs in bones, the
uterus, mammary gland, liver and adipose tissue, whereas ERβ is
predominantly expressed in the ovary, mammary gland and intestinal
tract. There is also expression of the two subtypes in the brain
and cardiovascular system (15).
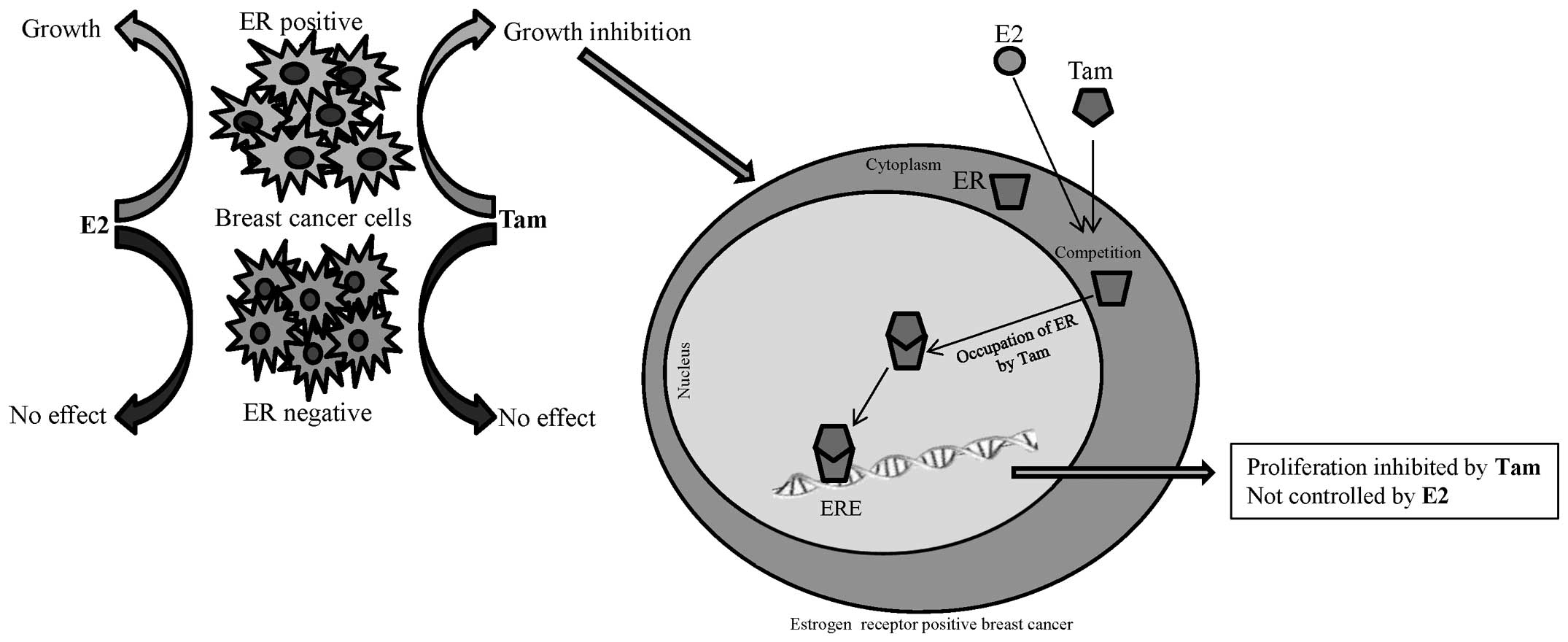
ERs are located in the cell cytoplasm in a complex with the heat
shock protein 90 chaperone, which dissociates following ligand
binding (16). The ER-ligand
complex is translocated into the nucleus, where it interacts with
coregulators of transcription and binds to the estrogen response
element (ERE) promoter region of a target gene and thereby
activates mRNA transcription (17–19).
Identification of biomarkers using matrix-assisted laser
desorption/ionization (MALDI) is currently of increasing
significance and has contributed to rapid advances in metabolomics
(20). MALDI may also be a powerful
tool for investigation of biomarkers in biological systems, through
the direct analysis of thin tissue sections (21), for example ERs in breast tissue. The
present review aimed to summarize the evidence for the use of MALDI
time of flight mass spectrometry (TOF MS) for the identification of
ER proteins in breast cancer tissues.
Molecules acting as ER agonists generally exert a
stimulatory effect on the proliferation of estrogen-sensitive
breast carcinoma cells (12). In
human breast cancer, ER+ tumors exhibit an
overexpression of ERα as a result of transcription from a promoter
inactive in normal breast epithelium. In addition, behavior of ERα
depends on the structure of the bound ligand [e.g. estradiol, the
most active estrogen (22)]
modulating the transcriptional activity of the estrogen responsive
genes (18,23). ERα, as a main target in breast
cancer, is influenced by a number of types of coregulator following
ligand binding, including coactivators and corepressors (24). The balance between coregulators is
crucial for regulation of gene transcription by ERα (25). Overexpression of coactivators, for
example coactivator-associated arginine methyltransferase 1, may
also increase the expression of ERα target genes involved in breast
tumor cell differentiation and proliferation (26), including breast cancer (BRCA) 1 and
BRCA2 genes (27). In addition,
reduction of ERα spliced variant 46 (46 kDa) and 36 (36 kDa) mRNA
levels have been observed in colon tumors (28) and overexpression of ERα36 has been
observed in gastric (29) and
endometrial cancers (30).
It is well known that ER levels and emplacement of
breast tumor metastasis are the fundamental and critical
determinants of clinical outcome, with high prognostic values
having the greatest impact on patient survival chances (31,32).
The importance of ERs as breast carcinoma biomarkers is also due to
the ability of the hormone receptor protein to provide detailed
information about breast tumor subtype. ER+ breast
cancer types exhibit favorable responses to hormone therapy
(33–35), for example tamoxifen (36), or to aromatase inhibitors (37), designed to block aberrant signaling
within oncogenic pathways (Fig. 1).
The use of neoadjuvant chemotherapy for the treatment of
ER+ tumors is associated with a major obstacle;
chemoresistance (38,39). Hence, the identification of cancer
subtypes using protein analysis is likely to enable the treatment
effects of chemotherapy to be maximized (40). At present, the most utilized method
for ER protein analysis in practice is immunohistochemistry
(41–45). Great potential has also been
attributed to MALDI TOF MS offering reliable, robust and efficient
analysis, renowned for its ease of operation and inexpensive
matrixes required for sample preparation, as well as its
derivative, surface-enhanced laser desorption/ionization
spectrometry (20).
MALDI TOF has been hypothesized to represent one of
the most comprehensive and versatile tools for investigation of new
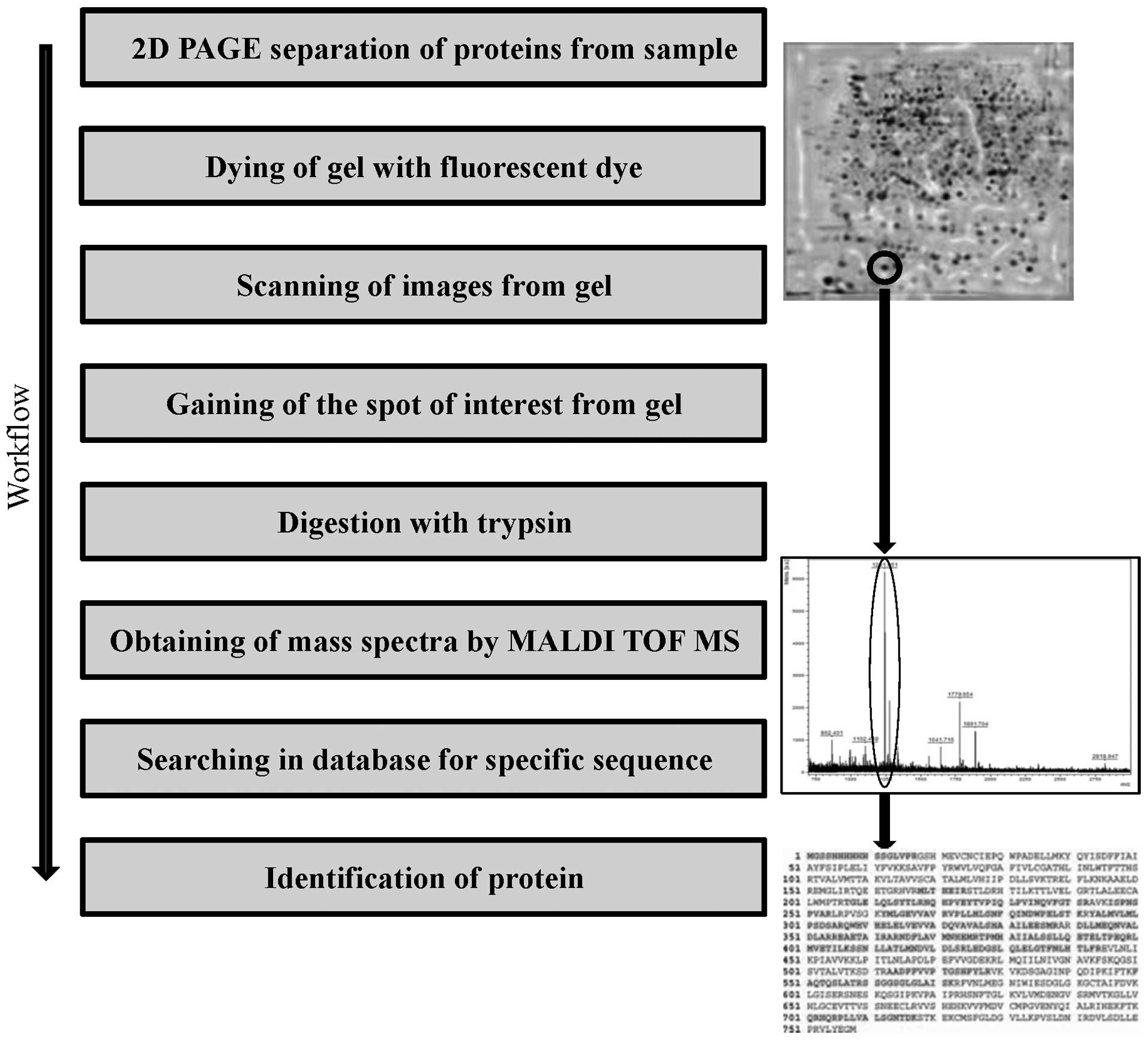
biomarkers and protein analysis (46). A key element of the proteomic
application of MALDI TOF is the separation of proteins from a
sample using two-dimensional gel electrophoresis, prior to
subsequent analysis by MS (Fig. 2)
(47,48). The MALDI TOF result, exhibited as
protein peak spectra, may be quantitatively and statistically
evaluated for determination of differential protein expression in
response to a particular biological state (49). Nalvarte et al reported an
approach for the isolation of ERα from MCF-7 cells based on the
natural affinity of ER proteins towards the estrogen response
element immobilized on a Sepharose column with subsequent
two-dimensional electrophoresis, and identification using MALDI TOF
mass spectrometry (50). This
method provided a rapid method to identify ERα cofactor and
transcription factor recruitment under various conditions. However,
it has been hypothesized that the equivalent analysis of ER
proteins in clinical samples is likely to be subject to extensive
chemical noise that may invalidate results (51). The quantity and identity of
biomarkers observed in tissue profiles are also influenced by a
number of factors, including the volume of matrix solution used and
the sites of laser shots application, used for ionization, which
provides charge to molecules and thus enables proper mass
detection, which facilitates rendering of the data into spatial
distribution maps, or images for the many hundreds of ions measured
in the mass spectra (52). A
potential problem may be found also in the variability in sample
preparation, leading to crystal heterogeneity, and thus to
discrimination and suppression of certain signals. There are
various approaches to minimize MALDI analysis, including the
production of thin films by rapid drying of volatile solvents, or
the use of electrospray with the ability to produce thin
homogeneous films (51). The lack
of further evidence associated with the diagnosis of breast cancer
by MALDI TOF analysis of ERs highlights the issues associated with
ER protein analysis in real biological samples. However, this
method demonstrates excellent results for the visualization of
protein expression (46,53), DNA methylation status (54), monitoring of ER interactions
(55,56) and in searching for new biomarkers
for breast cancer diagnosis (57,58).
At present, the most commonly used method for
differentiation of breast cancer subtypes is immunohistochemical
classification, based on the level of expression of ERs and
progesterone receptors (41,42).
This method provides relatively accurate results (false negativity,
15.1%), however, it can be time-consuming when analysis of a large
number of samples is necessary, requiring sample staining,
incubation, application of antibodies and visualization. By
contrast, MALDI TOF MS may be useful for the analysis of large
amounts of tissue samples. The greatest disadvantage of MALDI MS is
the acquisition costs, however, this is balanced by reduced
operating costs, reliability, robustness and efficiency.
Additionally, ER isolation must be performed using two-dimensional
gel electrophoresis (47) or a
chromatographic system, in which several issues limit the isolation
and proteomic analysis of ER complexes. The greatest of these is
the low amount of endogenous ERs complexed with EREs, increasing
the requirement for sensitivity of analytical methods used for
isolation, and therefore it is necessary to find compromise between
protein isolation efficiency and accuracy of the method utilized
for its detection (50). However,
MALDI may be useful for other applications, for example the
monitoring of cancer gene expression (53,59).
ER proteins are important for diagnostics and
classification of breast tumors subtypes. In particular, the need
for identification of the cancer subtype is vital for selection of
the appropriate treatment, and to predict the chemoresistance which
is commonly noted in ER+ tumors. At present,
immunohistochemistry provides good results, however, this technique
is laborious. Large diagnostic potential has been attributed to
MALDI TOF MS but, due to the relatively recent development and high
cost, the use of this application in clinical practice remains
uncommon.
This study was supported by grants from the Internal
Grant Agency of the University of Veterinary and Pharmaceutical
Sciences Brno (project 4/2013/FVHE) and MSMT (no. 6215712402).
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar
|
|
3
|
Patani N, Martin LA and Dowsett M:
Biomarkers for the clinical management of breast cancer:
international perspective. Int J Cancer. 133:1–13. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carey LA, Perou CM, Livasy CA, et al:
Race, breast cancer subtypes, and survival in the Carolina Breast
Cancer Study. JAMA. 295:2492–2502. 2006. View Article : Google Scholar
|
|
5
|
Yang XR, Chang-Claude J, Goode EL, et al:
Associations of breast cancer risk factors with tumor subtypes: a
pooled analysis from the Breast Cancer Association Consortium
studies. J Natl Cancer Inst. 103:250–263. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Muss HB: Coming of age: breast cancer in
seniors. Oncologist. 16(Suppl 1): S79–S87. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ma H, Wang Y, Sullivan-Halley J, et al:
Use of four biomarkers to evaluate the risk of breast cancer
subtypes in the women’s contraceptive and reproductive experiences
study. Cancer Res. 70:575–587. 2010.
|
|
8
|
Shubbar E, Helou K, Kovács A, et al: High
levels of γ-glutamyl hydrolase (GGH) are associated with poor
prognosis and unfavorable clinical outcomes in invasive breast
cancer. BMC Cancer. 13:472013.
|
|
9
|
Tian W, Chen J, He H and Deng Y: MicroRNAs
and drug resistance of breast cancer: basic evidence and clinical
applications. Clin Transl Oncol. 15:335–342. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ombra MN, Di Santi A, Abbondanza C,
Migliaccio A, Avvedimento EV and Perillo B: Retinoic acid impairs
estrogen signaling in breast cancer cells by interfering with
activation of LSD1 via PKA. Biochim Biophys Acta. 1829:480–486.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fasching PA, Heusinger K, Haeberle L, et
al: Ki67, chemotherapy response, and prognosis in breast cancer
patients receiving neoadjuvant treatment. BMC Cancer. 11:4862011.
View Article : Google Scholar
|
|
12
|
Sharan S, Nikhil K and Roy P: Effects of
low dose treatment of tributyltin on the regulation of estrogen
receptor functions in MCF-7 cells. Toxicol Appl Pharmacol.
269:176–186. 2013. View Article : Google Scholar
|
|
13
|
Yan Y, Liu H, Wen H, et al: The novel
estrogen receptor GPER regulates the migration and invasion of
ovarian cancer cells. Mol Cell Biochem. 378:1–7. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Oh Y and Chung KC: Zinc finger protein 131
inhibits estrogen signaling by suppressing estrogen receptor α
homo-dimerization. Biochem Biophys Res Commun. 430:400–405.
2013.PubMed/NCBI
|
|
15
|
Komm BS and Mirkin S: Evolution of the
tissue selective estrogen complex (TSEC). J Cell Physiol.
228:1423–1427. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng Q, Chang JT, Geradts J, et al:
Amplification and high-level expression of heat shock protein 90
marks aggressive phenotypes of human epidermal growth factor
receptor 2 negative breast cancer. Breast Cancer Res. 14:R622012.
View Article : Google Scholar
|
|
17
|
Coughlan N, Thillainadesan G, Andrews J,
Isovic M and Torchia J: β-Estradiol-dependent activation of the
JAK/STAT pathway requires p/CIP and CARM1. Biochim Biophys Acta-Mol
Cell Res. 1833:1463–1475. 2013.
|
|
18
|
Sengupta S, Obiorah I, Maximov PY, Curpan
R and Jordan VC: Molecular mechanism of action of bisphenol and
bisphenol A mediated by oestrogen receptor alpha in growth and
apoptosis of breast cancer cells. Br J Pharmacol. 169:167–178.
2013. View Article : Google Scholar
|
|
19
|
Levin ER: Integration of the extranuclear
and nuclear actions of estrogen. Mol Endocrinol. 19:1951–1959.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pirman DA, Efuet E, Ding XP, et al:
Changes in cancer cell metabolism revealed by direct sample
analysis with MALDI mass spectrometry. PLoS One. 8:e613792013.
View Article : Google Scholar
|
|
21
|
Cornett DS, Reyzer ML, Chaurand P and
Caprioli RM: MALDI imaging mass spectrometry: molecular snapshots
of biochemical systems. Nat Methods. 4:828–833. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang HS, Wu HM, Cheng BH, et al:
Functional analyses of endometriosis-related polymorphisms in the
estrogen synthesis and metabolism-related genes. PLoS One.
7:e473742012. View Article : Google Scholar
|
|
23
|
Srinivasan S, Nwachukwu JC, Parent AA, et
al: Ligand-binding dynamics rewire cellular signaling via estrogen
receptor-α. Nat Chem Biol. 9:326–332. 2013.PubMed/NCBI
|
|
24
|
Aust S, Horak P, Pils D, et al: The
prognostic value of estrogen receptor beta and proline-, glutamic
acid- and leucine-rich protein 1 (PELP1) expression in ovarian
cancer. BMC Cancer. 13:1152013. View Article : Google Scholar
|
|
25
|
Borjesson AE, Farman HH, Engdahl C, et al:
The role of activation functions 1 and 2 of estrogen receptor-α for
the effects of estradiol and selective estrogen receptor modulators
in male mice. J Bone Miner Res. 28:1117–1126. 2013.
|
|
26
|
Zeng H, Wu JC, Bedford MT, et al: A
TR-FRET-based functional assay for screening activators of CARM1.
Chembiochem. 14:827–835. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Meric-Bernstam F, Gutierrez-Barrera AM,
Litton J, et al: Genotype in BRCA-associated breast cancers. Breast
J. 19:87–91. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang H, Teng R, Wang Q, et al:
Transcriptional analysis of estrogen receptor alpha variant mRNAs
in colorectal cancers and their matched normal colorectal tissues.
J Steroid Biochem Mol Biol. 112:20–24. 2008. View Article : Google Scholar
|
|
29
|
Wang J, Li J, Fang R, Xie S, Wang L and Xu
C: Expression of ERα36 in gastric cancer samples and their matched
normal tissues. Oncol Lett. 3:172–175. 2012.
|
|
30
|
Tu BB, Lin SL, Yan LY, Wang ZY, Sun QY and
Qiao J: ER-α36, a novel variant of estrogen receptor α, is involved
in EGFR-related carcinogenesis in endometrial cancer. Am J Obstet
Gynecol. 205:227.e1–e6. 2011.
|
|
31
|
Kammerer M, Gutzwiller S, Stauffer D,
Delhon I, Seltenmeyer Y and Fournier B: Estrogen receptor α (ERα)
and estrogen related receptor α (ERRα) are both transcriptional
regulators of the Runx2-I isoform. Mol Cell Endocrinol.
369:150–160. 2013.
|
|
32
|
Gamucci T, Vaccaro A, Ciancola F, et al:
Recurrence risk in small, node-negative, early breast cancer: a
multicenter retrospective analysis. J Cancer Res Clin Oncol.
139:853–860. 2013. View Article : Google Scholar
|
|
33
|
Althuis MD, Fergenbaum JH, Garcia-Closas
M, Brinton LA, Madigan MP and Sherman ME: Etiology of hormone
receptor-defined breast cancer: a systematic review of the
literature. Cancer Epidemiol Biomarkers Prev. 13:1558–1568.
2004.PubMed/NCBI
|
|
34
|
Thrane S, Lykkesfeldt AE, Larsen MS,
Sorensen BS and Yde CW: Estrogen receptor α is the major driving
factor for growth in tamoxifen-resistant breast cancer and
supported by HER/ERK signaling. Breast Cancer Res Treat. 139:71–80.
2013.
|
|
35
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ramirez-Ardila DE, Helmijr JC, Look MP, et
al: Hotspot mutations in PIK3CA associate with first-line treatment
outcome for aromatase inhibitors but not for tamoxifen. Breast
Cancer Res Treat. 139:39–49. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hiscox S, Davies EL and Barrett-Lee P:
Aromatase inhibitors in breast cancer. Maturitas. 63:275–279. 2009.
View Article : Google Scholar
|
|
38
|
Hodgkinson VC, Agarwal V, El Fadl D, et
al: Pilot and feasibility study: comparative proteomic analysis by
2-DE MALDI TOF/TOF MS reveals 14-3-3 proteins as putative
biomarkers of response to neoadjuvant chemotherapy in ER-positive
breast cancer. J Proteomics. 75:2745–2752. 2012. View Article : Google Scholar
|
|
39
|
Kim SI, Sohn J, Koo JS, Park SH, Park HS
and Park BW: Molecular subtypes and tumor response to neoadjuvant
chemotherapy in patients with locally advanced breast cancer.
Oncology. 79:324–330. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Haddock CL, Holtz B, Senzer N and
Nemunaitis J: Applications of HPLC-MALDI-TOF MS/MS phosphoproteomic
analysis in oncological clinical diagnostics. Curr Proteomics.
8:153–167. 2011. View Article : Google Scholar
|
|
41
|
Tangjitgamol S, Tanvanich S,
Srijaipracharoen S and Manusirivithaya S: Expression of estrogen
receptor, progesterone receptor, and Her-2/neu in primary and
extra-corporeal endometrial cancer. Histol Histopathol. 28:787–794.
2013.PubMed/NCBI
|
|
42
|
Alkner S, Bendahl PO, Grabau D, et al: The
role of AIB1 and PAX2 in primary breast cancer: validation of AIB1
as a negative prognostic factor. Ann Oncol. 24:1244–1252. 2013.
View Article : Google Scholar
|
|
43
|
Seferina SC, Nap M, van den Berkmortel F,
Wals J, Voogd AC and Tjan-Heijnen VC: Reliability of receptor
assessment on core needle biopsy in breast cancer patients. Tumour
Biol. 34:987–994. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wik E, Ræder MB, Krakstad C, et al: Lack
of estrogen receptor-α is associated with epithelial-mesenchymal
transition and PI3K alterations in endometrial carcinoma. Clin
Cancer Res. 19:1094–1105. 2013.
|
|
45
|
Kinsella MD, Birdsong GG, Siddiqui MT,
Cohen C and Hanley KZ: Immunohistochemical detection of estrogen
receptor, progesterone receptor and human epidermal growth factor
receptor 2 in formalin-fixed breast carcinoma cell block
preparations: correlation of results to corresponding tissue block
(needle core and excision) samples. Diagn Cytopathol. 41:192–198.
2013.
|
|
46
|
Sang QX, Man YG, Sung YM, et al:
Non-receptor tyrosine kinase 2 reaches its lowest expression levels
in human breast cancer during regional nodal metastasis. Clin Exp
Metastasis. 29:143–153. 2012. View Article : Google Scholar
|
|
47
|
Jin Y, Zhang X, Lu D, et al:
Histopathological and proteomic analysis of hepatic tissue from
adult male zebrafish exposed to 17β-estradiol. Environ Toxicol
Pharmacol. 29:91–95. 2010.PubMed/NCBI
|
|
48
|
Pietrowska M, Marczak L, Polanska J, et
al: Mass spectrometry-based serum proteome pattern analysis in
molecular diagnostics of early stage breast cancer. J Transl Med.
7:602009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bouchal P, Dvorakova M, Scherl A, Garbis
SD, Nenutil R and Vojtesek B: Intact protein profiling in breast
cancer biomarker discovery: protein identification issue and the
solutions based on 3D protein separation, bottom-up and top-down
mass spectrometry. Proteomics. 13:1053–1058. 2013. View Article : Google Scholar
|
|
50
|
Nalvarte I, Schwend T and Gustafsson JA:
Proteomics analysis of the estrogen receptor alpha receptosome. Mol
Cell Proteomics. 9:1411–1422. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Atsriku C, Benz CC, Scott GK, Gibson BW
and Baldwin MA: Quantification of cysteine oxidation in human
estrogen receptor by mass spectrometry. Anal Chem. 79:3083–3090.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Cornett DS, Mobley JA, Dias EC, et al: A
novel histology-directed strategy for MALDI-MS tissue profiling
that improves throughput and cellular specificity in human breast
cancer. Mol Cell Proteomics. 5:1975–1983. 2006. View Article : Google Scholar
|
|
53
|
Kabbage M, Trimeche M, Bergaoui S, et al:
Calreticulin expression in infiltrating ductal breast carcinomas:
relationships with disease progression and humoral immune
responses. Tumour Biol. 34:1177–1188. 2013. View Article : Google Scholar
|
|
54
|
Castilla MÁ, Diaz-Martin J, Sarrió D, et
al: MicroRNA-200 family modulation in distinct breast cancer
phenotypes. PLoS One. 7:e477092012.PubMed/NCBI
|
|
55
|
Paramanik V and Thakur MK: Estrogen
receptor β and its domains interact with casein kinase 2,
phosphokinase C, and N-myristoylation sites of mitochondrial and
nuclear proteins in mouse brain. J Biol Chem. 287:22305–22316.
2012.
|
|
56
|
Bovet C, Plet B, Ruff M, et al: Towards
high-throughput identification of endocrine disrupting compounds
with mass spectrometry. Toxicol In Vitro. 23:704–709. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Collodoro M, Lemaire P, Eppe G, et al:
Identification and quantification of concentration-dependent
biomarkers in MCF-7/BOS cells exposed to 17β-estradiol by 2-D DIGE
and label-free proteomics. J Proteomics. 75:4555–4569.
2012.PubMed/NCBI
|
|
58
|
Lai TC, Chou HC, Chen YW, et al:
Secretomic and proteomic analysis of potential breast cancer
markers by two-dimensional differential gel electrophoresis. J
Proteome Res. 9:1302–1322. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Obazee O, Justenhoven C, Winter S, et al:
Confirmation of the reduction of hormone replacement
therapy-related breast cancer risk for carriers of the
HSD17B1_937_G variant. Breast Cancer Res Treat. 138:543–548. 2013.
View Article : Google Scholar : PubMed/NCBI
|