Introduction
Breast cancer is one of the most common types of
cancer among females worldwide, as well as a leading cause of
mortality. However, the survival of patients has increased over the
past decades due to earlier diagnosis and effective therapies
(1). In addition, cancer biomarkers
provide useful information for the prognosis and assessment of
cancer treatment.
The identification of cancer biomarkers is important
for cancer biology and clinical applications. With the development
and improvement of high-throughput biotechnologies, cancer
biomarkers can be identified by comparing normal cells with cancer
cells through genomic, transcriptomic and proteomic analyses. At
present, the most promising biomarkers are proteins (2,3) and
proteomic analysis provides an opportunity to identify altered
protein groups and the complex pathways in breast cancer. The
mapping of proteomic profiles and differential proteomics has been
widely performed in breast cancer to identify potential biomarkers
(4). Some of these proteins have
been reported to have potential clinical significance and key
proteins, such as the receptor tyrosine-protein kinase erbB-2
(ERBB2) and breast cancer 1 and 2 early onset, may be used as
potential diagnostic, prognostic or predictive biomarkers (5,6).
Although cancer markers may indicate the status of cancer
development, they alone are not sufficient to determine the cancer
biology. In addition, due to the heterogeneity of experimental
methods and specimen preparation (7), the proteomic results lack good
reproducibility and require further validation prior to their use
in clinical detection and to explain the underlying mechanisms of
breast cancer. The transformation of normal cells to cancer cells
requires the complex regulation of networks and altered molecules.
In addition, the networks associated with cancer cause abnormal
cell proliferation and invasion. The identification of these
intricate pathways is essential to understanding the biological
mechanisms of cancer and may aid in predicting or monitoring cancer
progression, as well in developing a therapeutic strategy by
focusing on the pathways instead of individual proteins. The
enriched pathways or functions are the most probable causes of
cancer (8,9), and the enriched proteins involved in
these processes may in turn serve as target agents in the diagnosis
or treatment of cancer.
The aim of cancer proteomics is to identify altered
proteins and to correlate them with the tumorigenesis and
progression of cancer. The development and application of proteomic
technologies has resulted in a surplus of potential breast cancer
biomarkers; however, these results require validation by
immunohistochemistry or western blot analysis for clinical
diagnostics. Immunohistochemistry is being increasingly used in the
pathology of breast cancer to provide a definitive histological
diagnosis and information for treatment and prognosis. A panel of
immunohistochemical markers can be used for estimating prognosis
and predicting therapy response (10). Conversely, immunohistochemistry may
also be useful for identifying additional cancer markers.
Currently, the Human Protein Atlas (www.proteinatlas.org) is used to generate a global
immunohistochemistry map of protein expression profiles in normal
and cancer tissues, and it provides a reliable resource for the
identification of biomarkers. The present study performed a direct
comparison of the protein expression levels in breast cancer with
those in normal breast tissues to identify differentially expressed
proteins in breast cancer. In addition, a functional enrichment
analysis was performed to identify new functional modules in breast
cancer. The results identified additional potential marker proteins
that could be used as background to reveal the altered pathways in
human breast cancer. The combinational protein profiles are likely
to present a more sensitive and specific evaluation of the
heterogeneity of cancer and could be applied to investigate the
mechanisms of cancer formation at the functional pathway level.
Materials and methods
Patient characteristics and serum
collection
Blood samples were collected from 30 breast cancer
patients and 30 healthy volunteers at the Yu-Huang-Ding Hospital
(Yantai, China) who had provided written informed consent. Ethical
approval for the study was obtained from the Yu-Huang-Ding Hospital
research and ethics committee. Venous blood was drawn from each
subject into 10-ml fasting blood tubes, which were allowed to clot
at room temperature for 1 h. The serum was then separated by
centrifugation at 2,000 × g for 15 min at 4°C.
Data collection
Staining profiles for proteins in normal breast and
breast cancer tissues were downloaded from the Human Protein Atlas.
The expression level of each protein was then graded into four
levels: Strong, >75%; moderate, 25–75%; weak, <25% and
negative, 0% for use as retrieval parameters. The differentially
expressed proteins were defined as those which exhibited a change
in expression of more than two levels between the previously
described groups. Finally, the selected proteins were grouped into
upregulated and downregulated proteins in human breast cancer.
Broad functional analysis
All differentially expressed proteins were
classified broadly into several catalogs according to the Gene
Ontology (GO) annotation (www.geneontology.org), PANTHER classification
(www.pantherdb.org) and functions annotated in UniProt
(www.uniprot.org).
Over-representation analysis
Ontological analysis
The over-representation analyses of GO terms,
including biological process and molecular function, were performed
using the ConsensusPathDB-human database system (http://cpdb.molgen.mpg.de/CPDB), which is a
molecular functional interaction database. The GO level 2 and 3
categories and a P-value cut-off of 0.01 were selected.
Pathway analysis
The enriched pathway analysis was performed using
the DAVID (http://david.abcc.ncifcrf.gov/) and PANTHER tools. For
the pathway analysis, the Kyoto Encyclopedia of Genes and Genomes
(KEGG) and Reactome databases were selected. The minimum overlap
with the input list was set at two proteins, with P<0.01.
Analysis of the membrane
organization
The secreted and membrane proteins were screened
through tools in LOCATE (http://locate.imb.uq.edu.au/), which is a curated
database for describing the membrane organization. The membrane
proteins included type I, II and III proteins.
ELISA assay
The serum samples were collected from breast cancer
patients and healthy age-matched volunteers. The ELISA kits for
peroxiredoxin (PRDX)2, PRDX6, cathepsin (CTS)B and CTSD (Abnova
Corporation, Taibei, Taiwan) were used and the assays were run
according to the manufacturer’s instructions. The absorbance was
measured at 450 nm using a 680 microplate reader (Bio-Rad,
Hercules, CA, USA).
Statistical analysis
The ELISA data were statistically analyzed and the
differences between the two groups were assessed by the independent
samples t-test. P<0.01 was considered to indicate a
statistically significant difference.
Results
Differentially expressed proteins in
human breast cancer
The human breast cancer protein profile was
constructed by screening the Human Protein Atlas quantitative
dataset stained using immunohistochemistry. A total of 1,688
proteins were found to be differentially expressed between breast
cancer and normal breast tissues, including 773 upregulated and 915
downregulated proteins in human breast cancer.
Broad functional analysis
All the proteins were placed into broad functional
categories on the basis of the GO and PANTHER databases. As shown
in Fig. 1, a total of 1,688
proteins were grouped into several classes according to their major
functions. The major protein class was the nucleic acid binding
proteins (13.1%), followed by the cytoskeletal proteins (9.5%),
receptors (7.8%), signaling molecules (7.6%) and transporters
(7.5%). These molecules exhibited different functions, of which the
leading function was metabolism (19.2%), followed by binding
(17.4%), transport (8.5%) and cell motility (8.2%).
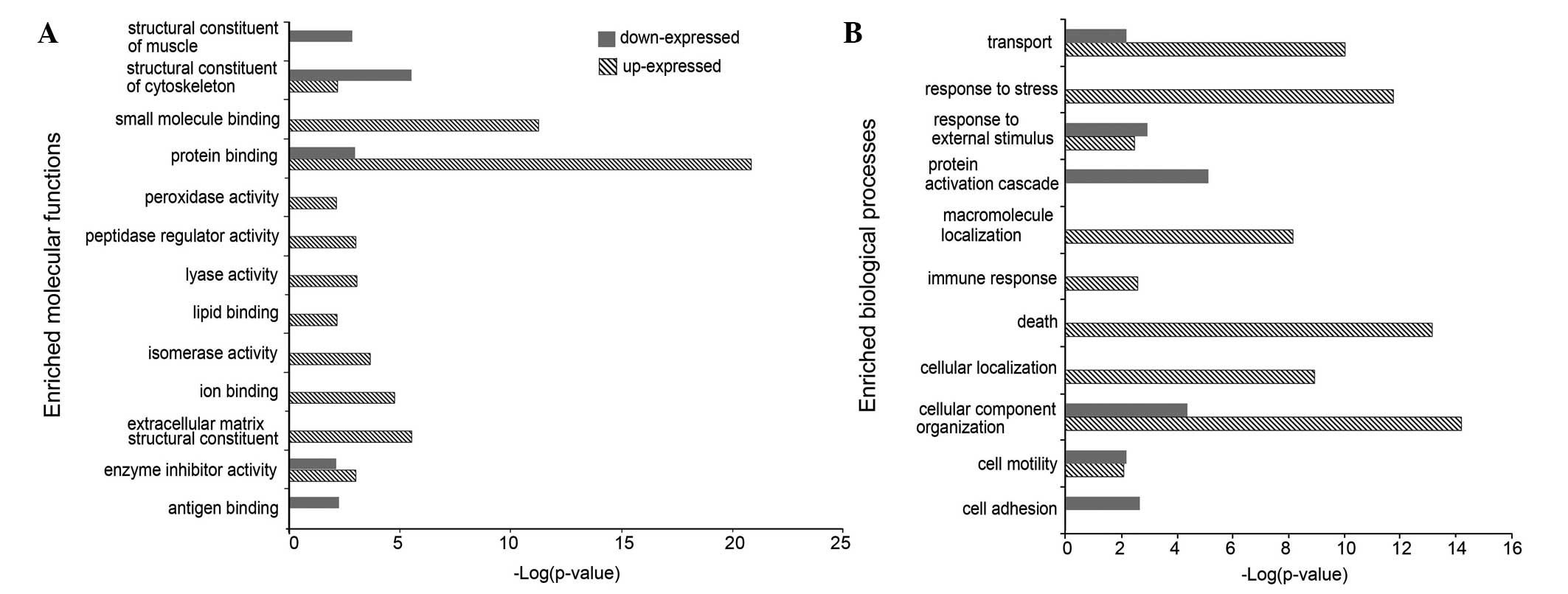
Enriched ontological analysis
The main enriched GO terms were categorized with
respect to upregulated and downregulated proteins, to determine
which molecular functions or biological processes were enriched in
the protein groups differentially expressed in human breast cancer
(Fig. 2). As to be expected, more
important molecular functions and biological processes were
enriched with upregulated proteins as opposed to downregulated
proteins. The important enriched molecular functions were enzyme
activity and binding functions (including peroxidase, lyase,
isomerase and peptidase regulator activity), as well as small
molecule, lipid and ion binding functions. These enriched molecular
functions were identified to be important in the processes of
response to stress, cell death and localization.
Enriched pathway analysis
An enriched pathway analysis of upregulated and
downregulated proteins in breast cancer was performed using the
KEGG and Reactome pathway databases; in total, 42 pathways were
obtained. As predicted, the more important pathways were enriched
with upregulated proteins than downregulated proteins. As listed in
Table I, 25 pathways were uniquely
enriched with upregulated proteins and four pathways were uniquely
enriched with downregulated proteins.
 | Table IEnriched pathways in upregulated and
downregulated breast cancer proteins. |
Table I
Enriched pathways in upregulated and
downregulated breast cancer proteins.
| | Upregulated | Downregulated | |
|---|
| |
|
| |
|---|
| Pathway name | Set size | n (%) | P-value | n (%) | P-value | Data source |
|---|
| Glycolysis | 29 | 11 (40.7) | 6.82E-08 | - | - | Reactome |
| Glucose
metabolism | 67 | 16 (24.6) | 2.68E-07 | - | - | Reactome |
| Proteasome | 44 | 13 (29.5) | 3.61E-07 | - | - | KEGG |
| Folding of actin by
CCT/TriC | 9 | 6 (66.7) | 1.88E-06 | - | - | Reactome |
| Focal adhesion | 206 | 26 (12.7) | 4.96E-05 | 24 (11.7) | 1.29E-04 | KEGG |
| SHC-mediated
signaling | 15 | 6 (40.0) | 8.44E-05 | - | - | Reactome |
| RAF/MAP kinase
cascade | 10 | 5 (50.0) | 9.54E-05 | - | - | Reactome |
| Citrate cycle (TCA
cycle) | 30 | 8 (26.7) | 1.51E-04 | 7 (23.3) | 6.49E-04 | KEGG |
| Calnexin/calreticulin
cycle | 11 | 5 (45.5) | 1.67E-04 | - | - | Reactome |
| Smooth muscle
contraction | 24 | 7 (29.2) | 2.13E-04 | 6 (25.0) | 1.08E-03 | Reactome |
| Metabolism of
proteins | 572 | 48 (9.1) | 3.04E-04 | - | - | Reactome |
| Mitotic prophase | 35 | 8 (23.5) | 3.85E-04 | - | - | Reactome |
| Membrane
trafficking | 154 | 19 (12.7) | 5.10E-04 | - | - | Reactome |
| Collagen
formation | 88 | 13 (14.9) | 8.21E-04 | 12 (13.8) | 1.53E-03 | Reactome |
| ARMS-mediated
activation | 16 | 5 (31.2) | 1.26E-03 | - | - | Reactome |
| Metabolism of
nucleotides | 81 | 12 (14.8) | 1.39E-03 | - | - | Reactome |
| RAF activation | 5 | 3 (60.0) | 1.49E-03 | - | - | Reactome |
| Axon guidance | 260 | 26 (10.1) | 1.84E-03 | - | - | Reactome |
| Signaling to
RAS | 25 | 6 (24.0) | 1.87E-03 | - | - | Reactome |
| Frs2-mediated
activation | 18 | 5 (27.8) | 2.25E-03 | - | - | Reactome |
| Hemostasis | 472 | 40 (8.6) | 2.98E-03 | - | - | Reactome |
| Cell cycle | 124 | 15 (12.1) | 3.03E-03 | - | - | KEGG |
| Peroxisome | 81 | 11 (13.9) | 3.58E-03 | - | - | KEGG |
| FRS2-mediated
cascade | 39 | 7 (18.4) | 4.00E-03 | - | - | Reactome |
| MEK activation | 7 | 3 (42.9) | 4.80E-03 | - | - | Reactome |
| Pentose phosphate
pathway (hexose monophosphate shunt) | 8 | 3 (42.9) | 4.80E-03 | - | - | Reactome |
| Proteoglycans in
cancer | 226 | 22 (9.8) | 5.71E-03 | - | - | KEGG |
| Integrin cell
surface interactions | 84 | 11 (13.1) | 5.76E-03 | - | - | Reactome |
| Regulation of actin
cytoskeleton | 215 | 21 (9.9) | 6.23E-03 | - | - | KEGG |
| PI3K-Akt signaling
pathway | 346 | 30 (8.7) | 7.13E-03 | 30 (8.7) | 7.13E-03 | KEGG |
| Vitamin C
(ascorbate) metabolism | 8 | 3 (37.5) | 7.37E-03 | 3 (37.5) | 7.37E-03 | Reactome |
| Serine
biosynthesis | 3 | 2 (66.7) | 7.60E-03 | 2 (66.7) | 7.60E-03 | Reactome |
| Glyoxylate and
dicarboxylate metabolism | 24 | 5 (20.8) | 8.52E-03 | 6 (25.0) | 1.08E-03 | KEGG |
| ERK1
activation | 3 | 2 (66.7) | 8.60E-03 | 2 (66.7) | 8.60E-03 | Reactome |
| PERK regulated gene
expression | 3 | 2 (66.7) | 8.60E-03 | 2 (66.7) | 8.60E-03 | Reactome |
| Viral
carcinogenesis | 207 | 20 (9.7) | 9.29E-03 | 20 (9.7) | 9.29E-03 | KEGG |
| Nuclear envelope
breakdown | 16 | 4 (25.0) | 9.45E-03 | 4 (25.0) | 9.45E-03 | Reactome |
| Renin-angiotensin
system | 17 | 4 (23.5) | 9.53E-03 | 4 (23.5) | 9.53E-03 | KEGG |
| Apoptotic cleavage
of cellular proteins | 40 | - | - | 10 (26.3) | 1.48E-05 | Reactome |
| Apoptosis | 109 | - | - | 16 (15.1) | 9.17E-05 | Reactome |
| Complement and
coagulation cascades | 69 | - | - | 11 (16.2) | 6.21E-04 | KEGG |
| Complement
cascade | 79 | - | - | 11 (14.5) | 1.60E-03 | Reactome |
Characteristics of potential cancer
markers
The secreted and membrane proteins in cancer tissues
are potential markers that may be detected in blood with altered
expression. The membrane organization analysis showed that 137
(17.7%) of the 773 upregulated proteins were secreted proteins,
with 242 (31.3%) of the 773 upregulated proteins identified as
membrane proteins (including 33 type I, 94 type II and 115 type
III). In the downregulated protein profiles, 126 (13.7%) proteins
were identified to be secreted proteins and 338 (36.9%) proteins
were identified to be membrane proteins (including 50 type I, 122
type II and 166 type III) (Table
II).
 | Table IISummary of secreted and membrane
proteins in upregulated and downregulated human breast cancer
proteins. |
Table II
Summary of secreted and membrane
proteins in upregulated and downregulated human breast cancer
proteins.
| | Membrane |
|---|
| |
|
|---|
| Secreted, n | Type I, n | Type II, n | Type III, n |
|---|
| Upregulated | 137 | 33 | 94 | 115 |
| Downregulated | 126 | 50 | 122 | 166 |
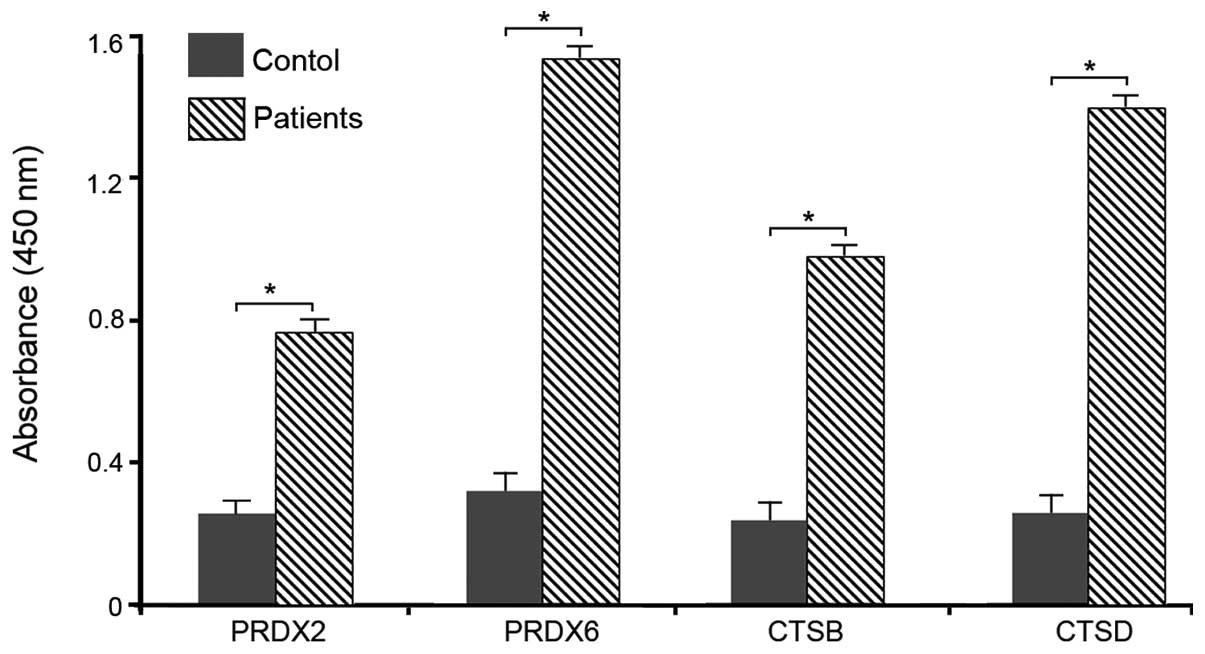
Validation of selected secreted
proteins
ELISA was performed to confirm the altered
expression levels of PRDX2, PRDX6, CTSB and CTSD in the serum among
the breast cancer patients and healthy volunteers as a
representative sample in order to validate the serum levels. The
results indicated that the four proteins were upregulated in the
serum of breast cancer patients (Fig.
3).
Discussion
Breast cancer is one of the most common types of
cancer among females caused by the accumulation of gene mutations
combined with altered gene regulation and protein pathways.
Therefore, the identification of markers to predict, diagnose and
treat breast cancer is of critical importance. By constructing
protein profiles associated with breast cancer, proteins with
altered expression can be identified to further investigate and
decipher the complex signaling networks involved in tumorigenicity
and cancer progression. In the present study, a differentially
expressed protein profile associated with human breast cancer was
constructed by quantitatively comparing the credible
immunohistochemistry results of breast cancer tissues with those of
normal breast tissues. The results provided a novel global analysis
of human breast cancer markers.
As predicted, certain well-known breast cancer
markers, such as the anterior gradient homolog 2 (11), ERBB2 (12) and Rho-associated protein kinase-2
(13) which are the overexpressed
in breast cancer, were included in the present study. Based on the
GO and PANTHER analyses, the differentially expressed proteins in
human breast cancer were grouped according to their major
biological functions. The functional category is useful for
investigating the mechanisms of breast cancer formation and
progression. The leading function was metabolism (19.2% of
proteins), which is an emerging hallmark of cancer (14). The following functions, such as
binding, transportation, signaling transduction and cell cycle
functions, are known to be associated with tumorigenesis (15). In addition, the functions of cell
motility and adhesion are involved in the progression of cancer
invasion and metastasis (16,17).
The enriched functional terms identified in the
upregulated breast cancer proteins may account for cancer
development and progression. In addition, several functional terms
are considered to be involved in breast cancer, for example, PRDX
is reportedly overexpressed in breast cancer (18) and may be used as a marker for breast
cancer (19). In the present study,
four PRDX members (PRDX1, 2, 4 and 6) were identified to exhibit
upregulated levels of expression in the breast cancer tissues.
Peroxidase enzymes are considered to be important in eliminating
the peroxides generated during cancer metabolism, and PRDX1 and 2
in MCF-7 breast cancer cells exhibit important functions as
inhibitors of cell death during the cellular response to oxidative
stress (20). In addition, the
overexpression of PRDX6 leads to a more invasive phenotype and
metastatic potential of human breast cancer (21). An additional protein family of
interest is the CTS proteins; in the present study, five CTS
proteins (CTSB, CTSC, CTSD, CTSH and CTSZ) were identified to be
upregulated in breast cancer. CTSs are overexpressed in breast
cancer and, as a result, have been suggested to be biological
markers for prognosis (22).
Furthermore, CTSB and CTSH have been reported to be overexpressed
in inflammatory breast cancer, as well as involved in cancer
progression and invasion (23). The
downregulated proteins in human breast cancer predominantly belong
to the structural constituents of the muscle and cytoskeleton, as
well as small molecule or antigen binding. These proteins are
significantly involved in transport, protein activation and cell
adhesion. In addition, the low expression levels of specific
proteins have also been associated with cancer progression, such as
the signal transducer and activator of transcription-5a, which
showed reduced expression in primary breast cancer and is
subsequently an independent marker of poor prognosis (24).
As predicted, in the present study, several
well-known cancer pathways, such as glycolysis, the cell cycle and
the phosphoinositide 3-kinase-Akt signaling pathway (25), were identified and confirmed the
reliability of the pathway analyses used. The most enriched pathway
with upregulated proteins was glycolysis, which indicated that
breast cancer relies on the production of ATP by glycolysis for
proliferation and progression. The overall activities of the
pathways determine the invasive and metastatic phenotype of cancer
cells. Thus, pathological analysis of the constituents of the
pathway and development of the inhibitors directed at the pathway
are likely to have clinical benefits in the diagnosis, prognosis
and treatment of breast cancer. These complex pathways and networks
are highly regulated and the alteration of specific molecules may
lead to the development of cancer.
The membrane proteins are considered to be
potentially effective therapeutic targets and the secreted proteins
may serve as biomarkers for cancer (26). Thus, these proteins were screened
using the Membrane Organization tool (LOCATE) and were found to
exhibit different localization characteristics, including
intracellular proteins (that executed the previously described
functions) and extracellular proteins, which may be useful
biomarkers. A total of 137 secreted proteins and 242 membrane
proteins were found to be upregulated in human breast cancer.
Generally, cancer cells decrease the number of cell-cell
interactions and increase the number of cell-extracellular matrix
interactions, which subsequently results in cancer metastasis.
Therefore, proteins secreted from cancer cells, such as CTS and
PRDX, may serve as promising biomarkers of cancer cell migration,
invasion and angiogenesis. Analyzing the expression of these
proteins in blood specimens may aid in the determination of a
diagnosis of breast cancer, as a molecular diagnostic tool. Certain
secreted and membrane proteins serve as signals for cell
communication and control cell proliferation, differentiation and
other physiological functions. For example, the secreted proteins
conjugative transfer region 1 and stanniocalcin 2 serve as
potential prognostic markers in breast cancer (27).
In conclusion, the present study constructed and
characterized a novel protein profile associated with human breast
cancer. However, further studies are warranted to confirm the
enriched functions and pathways. These results may be used as a
reliable resource to identify the altered pathways in human breast
cancer, as well as potential cancer targets for the early
diagnosis, therapeutic targets and disease response markers of
breast cancer.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (no. 81300533) and Shandong
Provincial Natural Science Foundation, China (no. ZR2013HQ002).
References
|
1
|
Misek DE and Kim EH: Protein biomarkers
for the early detection of breast cancer. Int J Proteomics.
2011:3435822011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu FJ, Hua XF and Wang WJ: A new
bioinformatics insight into human cancer-associated proteins. Oncol
Rep. 27:1932–1936. 2012.PubMed/NCBI
|
|
3
|
Kamel D, Brady B, Tabchy A, Mills GB and
Hennessy B: Proteomic classification of breast cancer. Curr Drug
Targets. 13:1495–1509. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Goncalves A and Bertucci F: Clinical
application of proteomics in breast cancer: state of the art and
perspectives. Med Princ Pract. 20:4–18. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hondermarck H, Tastet C, El
Yazidi-Belkoura I, Toillon RA and Le Bourhis X: Proteomics of
breast cancer: the quest for markers and therapeutic targets. J
Proteome Res. 7:1403–1411. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kadowaki M, Sangai T, Nagashima T,
Sakakibara M, Yoshitomi H, Takano S, Sogawa K, Umemura H, Fushimi
K, Nakatani Y, Nomura F and Miyazaki M: Identification of
vitronectin as a novel serum marker for early breast cancer
detection using a new proteomic approach. J Cancer Res Clin Oncol.
137:1105–1115. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Issaq HJ, Waybright TJ and Veenstra TD:
Cancer biomarker discovery: Opportunities and pitfalls in
analytical methods. Electrophoresis. 32:967–975. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sawyers CL: The cancer biomarker problem.
Nature. 452:548–552. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hua XF, Wang XB and Liu FJ: Functional
analysis of human cancer-associated genes and their association
with the testes and epididymis. Oncol Lett. 6:811–816.
2013.PubMed/NCBI
|
|
10
|
Ordóñez NG: Application of
immunohistochemistry in the diagnosis of epithelioid mesothelioma:
a review and update. Hum Pathol. 44:1–19. 2013.PubMed/NCBI
|
|
11
|
Fritzsche FR, Dahl E, Pahl S, Burkhardt M,
Luo J, Mayordomo E, Gansukh T, Dankof A, Knuechel R, Denkert C,
Winzer KJ, Diete M and Kristiansen G: Prognostic relevance of AGR2
expression in breast cancer. Clin Cancer Res. 12:1728–1734. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tan M and Yu D: Molecular mechanisms of
erbB2-mediated breast cancer chemoresistance. Adv Exp Med Biol.
608:119–129. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu S, Goldstein RH, Scepansky EM and
Rosenblatt M: Inhibition of rho-associated kinase signaling
prevents breast cancer metastasis to human bone. Cancer Res.
69:8742–8751. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Soga T: Cancer metabolism: key players in
metabolic reprogramming. Cancer Sci. 104:275–281. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Basten SG and Giles RH: Functional aspects
of primary cilia in signaling, cell cycle and tumorigenesis. Cilia.
2:62013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Palmer TD, Ashby WJ, Lewis JD and Zijlstra
A: Targeting tumor cell motility to prevent metastasis. Adv Drug
Deliv Rev. 63:568–581. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Behrens J: The role of cell adhesion
molecules in cancer invasion and metastasis. Breast Cancer Res
Treat. 24:175–184. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Noh DY, Ahn SJ, Lee RA, Kim SW, Park IA
and Chae HZ: Overexpression of peroxiredoxin in human breast
cancer. Anticancer Res. 21:2085–2090. 2001.PubMed/NCBI
|
|
19
|
Karihtala P, Mäntyniemi A, Kang SW,
Kinnula VL and Soini Y: Peroxiredoxins in breast carcinoma. Clin
Cancer Res. 9:3418–3424. 2003.PubMed/NCBI
|
|
20
|
Bae JY, Ahn SJ, Han W and Noh DY:
Peroxiredoxin I and II inhibit H2O2-induced
cell death in MCF-7 cell lines. J Cell Biochem. 101:1038–1045.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chang XZ, Li DQ, Hou YF, Wu J, Lu JS, Di
GH, Jin W, Ou ZL, Shen ZZ and Shao ZM: Identification of the
functional role of peroxiredoxin 6 in the progression of breast
cancer. Breast Cancer Res. 9:R762007. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nomura T and Katunuma N: Involvement of
cathepsins in the invasion, metastasis and proliferation of cancer
cells. J Med Invest. 52:1–9. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Decock J, Obermajer N, Vozelj S, Hendrickx
W, Paridaens R and Kos J: Cathepsin B, cathepsin H, cathepsin X and
cystatin C in sera of patients with early-stage and inflammatory
breast cancer. Int J Biol Markers. 23:161–168. 2008.PubMed/NCBI
|
|
24
|
Peck AR, Witkiewicz AK, Liu C, Klimowicz
AC, Stringer GA, Pequignot E, Freydin B, Yang N, Ertel A, Tran TH,
Girondo MA, Rosenberg AL, Hooke JA, Kovatich AJ, Shriver CD, Rimm
DL, Magliocco AM, Hyslop T and Rui H: Low levels of Stat5a protein
in breast cancer are associated with tumor progression and
unfavorable clinical outcomes. Breast Cancer Res. 14:R1302012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fu-Jun L, Shao-Hua J and Xiao-Fang S:
Differential proteomic analysis of pathway biomarkers in human
breast cancer by integrated bioinformatics. Oncol Lett.
4:1097–1103. 2012.PubMed/NCBI
|
|
26
|
Arcinas A, Yen TY, Kebebew E and Macher
BA: Cell surface and secreted protein profiles of human thyroid
cancer cell lines reveal distinct glycoprotein patterns. J Proteome
Res. 8:3958–3968. 2009. View Article : Google Scholar
|
|
27
|
Esseghir S, Kennedy A, Seedhar P, Nerurkar
A, Poulsom R, Reis-Filho JS and Isacke CM: Identification of NTN4,
TRA1, and STC2 as prognostic markers in breast cancer in a screen
for signal sequence encoding proteins. Clin Cancer Res.
13:3164–3173. 2007. View Article : Google Scholar : PubMed/NCBI
|