Introduction
Breast cancer is one of the common types of cancer
in females. The incidence of breast cancer increases each year, and
the proportion of affected young females also increases, posing a
serious threat to the health of the population (1). Overexpression of the human epidermal
growth factor receptor 2 (HER2) gene has been confirmed to closely
correlate with the prognosis of breast cancer patients and with the
effects of chemotherapy and hormonal therapy. The HER2 gene encodes
a 185-kDa transmembrane receptor with tyrosine kinase activity,
without a known ligand (2). HER2 is
involved in the regulation of cell growth, survival and
differentiation. Based on the definition by the American Society of
Clinical Oncology and College of American Pathologists (ASCO/CAP)
(3), it is estimated that ~14.7% of
breast carcinomas exhibit HER2 genetic heterogeneity (4). For the assessment of the HER2 gene
copy number, it has been suggested that systems assessing the
HER2/chromosome 17 centromere (CEP17) ratio, such as dual- or
single-color fluorescence in situ hybridization (FISH) or
chromogenic in situ hybridization (CISH), provide a more
accurate evaluation of HER2 amplification than single-probe
systems. These methods have successfully identified patients who
will benefit from trastuzumab therapy in clinical trials (5). Approximately 8% of breast cancers
exhibit increased copy numbers of CEP17 using FISH (i.e., average
CEP17 >3.0 per nucleus), and these cancers possess chromosome 17
polysomy (3,6,7).
Abnormalities of chromosome 17 are important
molecular genetic events in tumorigenesis, particularly in breast
cancer (8). Several important
genes, including the oncogenic genes HER2 and TOP2A and the
tumor suppressive gene p53, are essential in the development and
progression of breast cancer (9).
The polysomy of chromosome 17, one of the major types of
abnormality of chromosome 17, is frequent and identified in 20–40%
of invasive breast carcinomas (10). An increased HER2 gene copy number
has been reported to be associated with polysomy 17, a contributing
factor for HER2 protein overexpression (11,12).
Studies have also shown that chromosome 17 polysomy may be
associated with HER2 gene expression, the prognosis of breast
cancer patients and sensitivity to chemotherapy (13). However, several other studies
identified no effects of polysomy 17 on HER2 protein expression
(10), or found that only a small
number of cases were affected (14,15).
If polysomy 17 is an effective factor influencing
immunohistochemistry (IHC) scores, a number of positive IHC cases
caused by the polysomy may be missed when following the algorithm
for FISH analysis (3). As a result,
an accurate assessment of HER2 status and chromosome 17 polysomy is
of great importance when identifying patients who are eligible for
trastuzumab therapy (16,17).
In the majority of studies, thin paraffin tissue
sections (4 μm) have been used for FISH assay, which is defined as
a routine procedure in the International Standard Guide for
analyzing HER2 amplification and chromosome 17 polysomy (3). However, the average diameter of a
nucleus is >6 μm and the diameter of a tumor cell nucleus is
often much greater. It may be assumed that, due to tissue
sectioning, nuclei are not intact in 4-μm-thick sections, causing a
possible loss of some genetic material. This may lead to inaccurate
results, particularly in the evaluation of polysomy 17 by gene copy
numbers using FISH, as a number of copies may be in an adjacent
section. As a result, using whole, intact nuclei for FISH may
improve the accuracy of the results.
In the present study, FISH analysis of whole nuclei
(WNFISH) and IHC were used to analyze HER2 gene amplification and
HER2 protein expression in 109 breast cancer specimens.
Materials and methods
Case selection
One hundred and nine patients (aged 33 to 83 years;
median, 49.5 years old) with invasive breast ductal carcinoma who
were treated at the General Hospital of Shenyang Military Area
Command (Shenyang, China) between 2010 and 2011 were selected for
this retrospective study. HER2 gene status was evaluated in the 109
formalin-fixed paraffin-embedded (FFPE) tissues by WNFISH with
nuclei extracted from the FFPE tissue blocks. The study was
approved by the Ethics Committee of the General Hospital of
Shenyang Military Area Command. Written informed consent was
obtained from all patients according to the instructions of the
Ethics Committee of the General Hospital of Shenyang Military Area
Command.
Preparation of whole nuclei
A tissue microarray (TMA) was constructed using the
FFPE breast cancer tissue blocks. For each specimen, four tissue
cores were collected using a blunt core needle, 0.6 mm in diameter
(18). The needle was a component
of a Manual Tissue Arrayer Beecher Instruments Inc., Silver Spring,
MD, USA) and was pushed through the entire paraffin tissue block.
The cut tissue core remaining in the needle was then forced into a
1.5-ml polypropylene microcentrifuge tube.
Xylene (1 ml) was added into the tube twice, each
time for 20 min with light agitation. Subsequently, 1.0 ml
dehydrated ethanol was added twice, each time for 3 min with light
agitation. Then, 80%, 70% and 50% ethanol (1 ml of each) was
successively added into the tube, each for 3 min with light
agitation. The ethanol was discarded, and the tube was dried at
45°C for 10 min to evaporate the remaining ethanol. Enzymatic
digestion was then performed by adding 300 μl freshly prepared
proteinase K solution [0.01% proteinase K, 30 mAnson-U/mg, in 0.05
mol/l Tris(hydroxymethyl)aminomethane hydrochloride (pH 7.0), 0.01
mol/l ethylenediaminetetraacetic acid disodium salt and 0.01 mol/l
sodium chloride] purchased from Merck KGaA (Darmstadt, Germany) to
the microcentrifuge tube. The sample was then incubated at 37°C for
2 h. To aid the enzymatic digestion, the sample was vortexed for 3
sec at 20-min intervals during this incubation period.
The solution was mixed and centrifuged at 300 × g
for 5 sec to deposit the tissue mass. The suspension containing the
nuclei was sampled using a pipette. The nuclei were washed by
resuspension and vortexing in 100 μl phosphate-buffered saline
(PBS). The PBS solution was removed and the nuclei were fixed by
resuspension and vortexing twice in freshly prepared fixative
(three parts methanol and one part glacial acetic acid). The
fixative was removed and the nuclei were resuspended in 100 μl
distilled water. The nuclei density was calculated on a cell
counting plate and adjusted to 1×104 cells/μl using
distilled water.
The pretreated nuclei suspension (1×104
cells/μl) was pipetted onto poly-L-lysine-coated slides (Shanghai
Baoman Biotechnology Co., Ltd., Shanghai, China). The slides were
heated at 65°C for 1 h, and the nuclei were then ready for the
consecutive experiments after air drying.
WNFISH
WNFISH analysis was performed using a commercially
available, Food and Drug Administration-approved test kit
(PathVysion HER-2 DNA Probe kit; Abbott Molecular, Downers Grove,
IL, USA). The hybridization mixture included a centromere
17-specific green-labeled DNA probe and HER2/neu-specific
orange-labeled DNA probe.
The slides coated with whole nuclei were dried in an
oven at 65°C for 1 h, and fixed with methanol-glacial acetic acid
(3:1) for 1 h. After air drying, the slides were placed in citrate
buffer (pH 6.0) and incubated for 10 min in a microwave oven, then
transferred to freshly prepared 0.4% pepsin solution (0.16 g
pepsin, 2,850 U/mg solid, in 40 ml of 0.9% sodium chloride, pH 1.5)
and dehydrated through a series of graded ethanol solutions. After
dehydration, 10 μl HER2/CEN-17 probe mix was applied to the tissue
and covered with a coverslip. The sections were placed in a HYBrite
instrument (Vysis, Inc., Downers Grove, IL, USA), denatured at 82°C
for 10 min, and hybridized at 45°C for 16 h. Following
hybridization, the slides were washed in stringent wash buffer
briefly at room temperature, and then in a 65°C citrate buffer
solution for 10 min. The slides were dehydrated, air-dried and
counterstained with 4′,6-diamidino-2-phenylindole.
The samples were analyzed under a ×60 oil immersion
objective using an Olympus BX61 fluorescence microscope (Olympus,
Tokyo, Japan) with the appropriate filters.
FISH signals were assessed by two independent
assessors examining 30 non-overlapping nuclei for each sample. The
number of chromosome 17 signals and HER2 signals was recorded for
each case. Calculation of the mean number of HER2 signals, CEP17
signals and the HER2/CEP17 ratio was performed. HER2 gene
amplification status was classified according to the ASCO/CAP
criteria (13). The results were
calculated as a HER2/CEP17 ratio. Negative HER2 gene amplification
was defined as a HER2/CEP17 ratio of <1.8. Equivocal HER2 gene
amplification was defined as a HER2/CEP17 ratio between 1.8 and
2.2. Positive HER2 gene amplification was defined as a HER2/CEP17
ratio of ≥2.2.
An average count of chromosome 17 ≥2.6 per nucleus
was considered as polysomy (19).
IHC
A tissue microarray was constructed from the 109
FFPE breast cancer tissue blocks according to a previously
described procedure (20). Four
tissue cores were selected from the defined regions to construct a
TMA in a 30×30 matrix with 16 cores in the last row as a location
indicator. Sections (3 μm thick) were cut from the TMA blocks for
morphological observations and IHC staining.
The TMA slides were stained with polyclonal rabbit
anti-human c-erbB-2 oncoprotein antibody (A0485; DakoCytomation,
Carpinteria, CA, USA), using the standard streptavidin-biotin
complex method (21), with
appropriate positive and negative controls, according to the
manufacturer’s instructions. HER2 immunoreactivity was interpreted
based on the new ASCO/CAP recommendations (3) and scored as 0 (no staining), 1+ (weak
and incomplete membrane staining), 2+ (strong, complete membrane
staining in ≤30% of tumor cells or weak/moderate heterogeneous
complete membrane staining in ≥10% of tumor cells) or 3+ (strong,
complete, homogeneous membrane staining in >30% of tumor
cells).
Statistical analysis
The χ2 test was used for nonparametric
data (chromosomal ploidy status, HER2 gene amplification, HER2
protein expression, nuclear atypia and lymph node metastasis). SPSS
software, version 13.0 (SPSS, Inc., Chicago, IL, USA) was used for
statistical analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
FISH and IHC analyses
The CEP17 and HER2 statuses evaluated by WNFISH were
available for all 109 samples. Among the 109 cases, the WNFISH
method detected HER2 amplification (HER2 ratio ≥2.2) in 30 cases,
equivocal amplification (ratios between 1.8 and 2.2) in 19 cases,
and no HER2 amplification (ratio <1.8) in 60 cases. The WNFISH
method detected CEP17 polysomy (CEP17 number ≥2.6) in 37 cases and
no polysomy (CEP17 number <2.6) in 72 cases. Polyploidy was
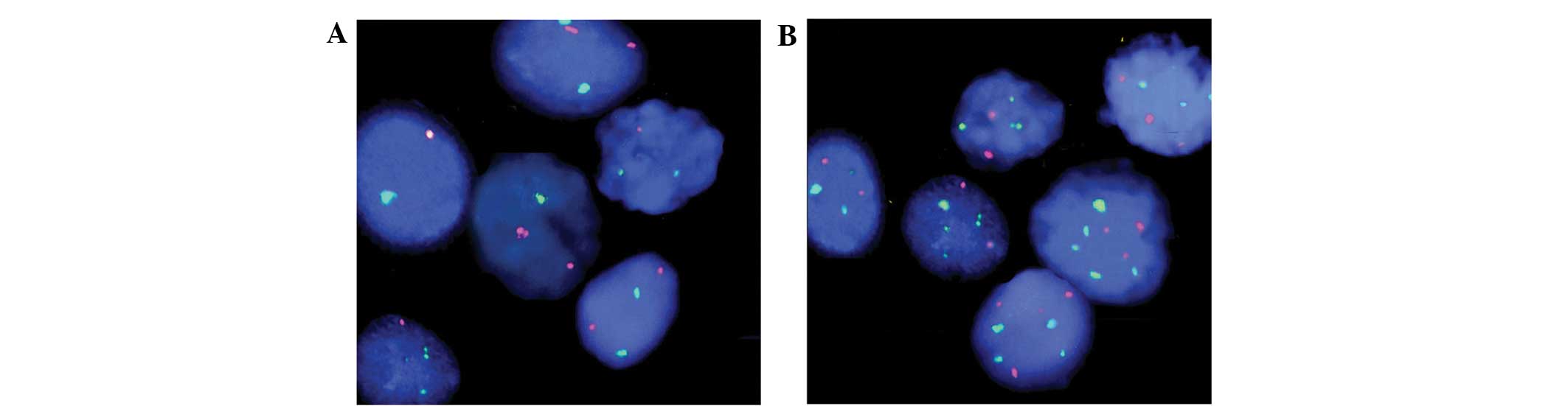
associated with tumor cell pleomorphism and HER2 gene variability,
as detected in the FISH analysis (Fig.
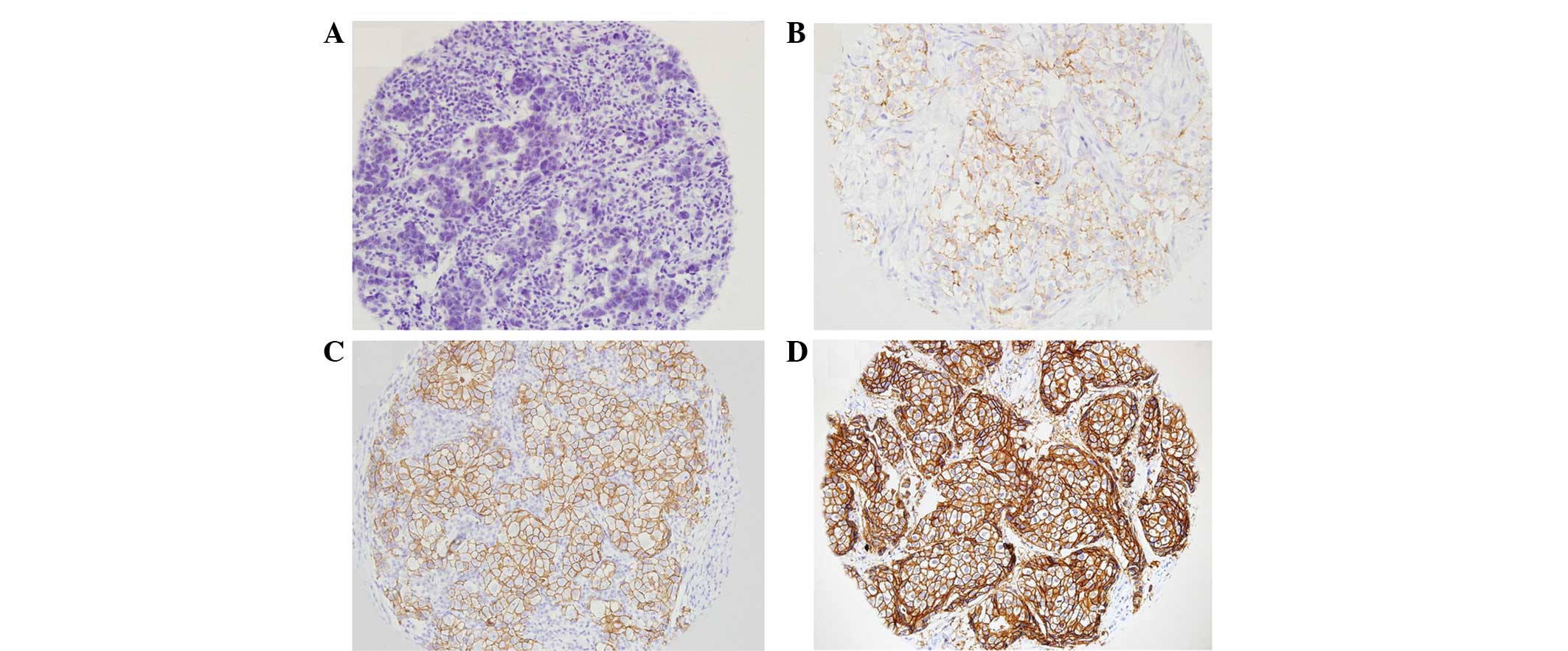
1). Among the 109 cases analyzed by TMA and IHC, 31 cases were
found to be HER2-negative, 14 cases were scored 1+, 23 cases were
scored 2+ and 41 cases were scored 3+. Representative images of IHC
staining for each score are shown in Fig. 2.
Chromosome 17 polysomy and HER2 gene
amplification
Among the 37 cases with chromosome 17 polysomy, HER2
gene amplification was detected in 18 cases (48.6%), while HER2
gene amplification was detected in only 12 cases (16.7%) among the
72 cases without chromosome 17 polysomy (P=0.002; Table I).
 | Table ICorrelation between chromosome 17
polysomy and HER2 gene amplification and HER2 protein
expression. |
Table I
Correlation between chromosome 17
polysomy and HER2 gene amplification and HER2 protein
expression.
| Polysomy 17 | N | HER2 amplification
status | P-value | HER2 protein
expression | P-value |
|---|
|
|
|---|
| Non-amplifieda | Equivocalb | Amplifiedc | − | + | 2+ | 3+ |
|---|
| + | 37 | 15 | 4 | 18 | 0.002 | 3 | 4 | 9 | 21 | 0.003 |
| − | 72 | 45 | 15 | 12 | | 28 | 10 | 14 | 20 | |
Chromosome 17 polysomy and HER2 protein
expression
The correlation between chromosome 17 polysomy and
HER2 protein expression is shown in Table I. Among the 37 cases with chromosome
17 polysomy, 3+ HER2 expression was detected in 21 cases (56.8%),
while 3+ HER2 expression was detected in only 20 cases (27.8%)
among the 72 cases without chromosome 17 polysomy (P=0.003).
Chromosome 17 polysomy, nuclear atypia
and lymph node metastasis
The correlation between chromosome 17 polysomy and
nuclear atypia and lymph node metastasis is shown in Table II. In 70.3% (26/37) of the breast
cancer cases with polyploidy, the cancer cells had large, unusual
or megakaryocytic nuclei with a high degree of nuclear atypia,
while no nuclear atypia was observed in 41.7% (30/72) of the breast
cancer cases without polyploidy (P=0.008). In cases with chromosome
17 polysomy, the incidence of lymph node metastasis was 73.0%
(27/37), while the incidence of lymph node metastasis was only
36.1% (26/72) in cases without chromosome 17 polysomy
(P<0.001).
 | Table IICorrelation between chromosome 17
polysomy and nuclear atypia and lymph node metastasis |
Table II
Correlation between chromosome 17
polysomy and nuclear atypia and lymph node metastasis
| Polysomy 17 | N | Nuclear atypia | P-value | Metastasis | P-value |
|---|
|
|
|---|
| Low-grade | High-grade | + | − |
|---|
| + | 37 | 11 | 26 | 0.008 | 27 | 10 | 0.0005 |
| − | 72 | 42 | 30 | | 26 | 46 | |
Discussion
The incidence of chromosome 17 polysomy in invasive
breast carcinoma is ~20–40% (10).
Watters et al (22) reported
that cancer cells in ~50% of invasive breast carcinoma cases are
aneuploid. This genetic change may be associated with poor
prognosis in certain breast cancer patients. In that study, 75% of
chromosome 17 polysomy cases were accompanied by HER2 gene
amplification. Risio et al (23) observed that patients with chromosome
monomers and HER2 gene amplification may not respond to trastuzumab
treatment. Hammock et al (24) demonstrated that a 3+ IHC result
without HER2 gene amplification, which is observed in ~50% of
patients, was caused by chromosome 17 polysomy. However, the
authors identified there was no marked correlation between IHC 2+
and polysomy. A number of multicenter studies (8,25,26)
have demonstrated that there was an evident correlation between IHC
3+ and HER2 gene amplification and the sensitivity of patients to
targeted therapy. Cases with negative FISH results and IHC 3+ were
defined as ‘central HER2-negative’ and mRNA in situ
hybridization was performed on these cases, identifying that the
positive rate of HER2 mRNA was low and confirming that the ‘central
HER2-negative’ cases were truly HER2-negative ones.
Thin tissue section FISH is the routinely used
method for the detection of gene status in paraffin-embedded breast
cancer samples. The average thickness of the sections is 4 μm,
which is less than the average diameter of a cell nucleus (6 μm).
Thus, in the majority of instances, only a portion of a given
nucleus is observed in the tissue sections. Thus, sectioning the
tissues into 4-μm-thick sections may cause the loss of nuclear
integrity, which may ultimately lead to detection bias in a FISH
assay. In the present study, in order to preserve the structure and
genetic material of the cells, whole, intact nuclei were extracted
from the paraffin-embedded tissues for FISH analysis, and the HER2
gene and CEP17 statuses were detected. It may be assumed that the
results obtained through this approach accurately identify the gene
status for better predicting the prognosis and response to targeted
therapy of patients.
The results show that there was an increase in HER2
gene amplification and HER2 protein expression in the cases with
chromosome 17 polysomy, demonstrating that HER2 overexpression in
certain breast carcinoma cases may be due to increases in the HER2
gene copy number caused by chromosome 17 amplification. Among the
109 breast cancer specimens assessed using FISH, 37 cases (33.9%)
were revealed to have chromosome 17 polysomy. This number was
smaller than that in the study by Watters et al (22). In the present study, the proportion
of cases with chromosome 17 polysomy was not greater than that
previously observed, despite the fact that intact nuclei were used
for the FISH analysis. The cause of this discrepancy may be due to
the small sample size or the different source of samples selected
for analysis in the present study.
In the cases with chromosome 17 amplifications, the
incidence of nuclear atypia and lymph node metastasis was higher
than that in the cases without chromosome 17 amplification. This
result indicates that chromosome 17 amplification may correlate
with poor prognosis in certain breast cancer patients, which is in
agreement with several other studies (13,22).
However, further studies are required to investigate the
correlation between chromosome 17 amplification and the efficacy of
targeted therapy in breast cancer.
Additional studies should aim to explore the
associations between chromosome 17 polysomy and the follow-up data
of patients after trastuzumab treatment. These future studies may
provide information on the efficacy and drug resistance to
molecular targeted therapy observed in certain breast cancers.
In conclusion, performing HER2 FISH analysis on
whole, intact nuclei from paraffin-embedded tissues is feasible,
and provides results correlating with those of IHC. Chromosome 17
polysomy correlates with nuclear atypia and with a higher rate of
lymph node metastases.
Acknowledgements
This study was supported by the Science Fund Project
of the General Hospital of Shenyang Military Area Command (grant
no. 09Y-Z12) and the Natural Science Foundation of Liaoning
Province (grant no. 2013020217).
References
|
1
|
McPherson K, Steel CM and Dixon JM: Breast
cancer-epidemiology, risk factors, and genetics. BMJ. 321:624–628.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gschwind A, Fischer OM and Ullrich A: The
discovery of receptor tyrosine kinases: targets for cancer therapy.
Nat Rev Cancer. 4:361–370. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wolff AC, Hammond ME, Schwartz JN, et al:
American Society of Clinical Oncology/College of American
Pathologists guideline recommendations for human epidermal growth
factor receptor 2 testing in breast cancer. J Clin Oncol.
25:118–145. 2007. View Article : Google Scholar
|
|
4
|
Akiyama T, Sudo C, Ogawara H, et al: The
product of the c-erbB-2 gene: a 185-kilodalton glycoprotein with
tyrosine kinase activity. Science. 232:1644–1646. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dal Lago L, Durbecq V, Desmedt C, et al:
Correction for chromosome-17 is critical for the determination of
true Her-2/neu gene amplification status in breast cancer. Mol
Cancer Ther. 5:2572–2579. 2006.PubMed/NCBI
|
|
6
|
Ma Y, Lespagnard L, Durbecq V, et al:
Polysomy 17 in HER-2/neu status elaboration in breast cancer:
effect on daily practice. Clin Cancer Res. 11:4393–4399. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Reddy JC, Reimann JD, Anderson SM and
Klein PM: Concordance between central and local laboratory HER2
testing from a community-based clinical study. Clin Breast Cancer.
7:153–157. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reinholz MM, Bruzek AK, Visscher DW, et
al: Breast cancer and aneusomy 17: implications for carcinogenesis
and therapeutic response. Lancet Oncol. 10:267–277. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang W and Yu Y: The important molecular
markers on chromosome 17 and their clinical impact in breast
cancer. Int J Mol Sci. 12:5672–5683. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Downs-Kelly E, Yoder BJ, Stoler M, et al:
The influence of polysomy 17 on HER2 gene and protein expression in
adenocarcinoma of the breast: a fluorescent in situ hybridization,
immunohistochemical, and isotopic mRNA in situ hybridization study.
Am J Surg Pathol. 29:1221–1227. 2005. View Article : Google Scholar
|
|
11
|
Bose S, Mohammed M, Shintaku P and Rao PN:
Her-2/neu gene amplification in low to moderately expressing breast
cancers: possible role of chromosome 17/Her-2/neu polysomy. Breast
J. 7:337–344. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Farabegoli F, Ceccarelli C, Santini D, et
al: c-erbB-2 over-expression in amplified and non-amplified breast
carcinoma samples. Int J Cancer. 84:273–277. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pritchard KI, Munro A, O’Malley FP, et al:
Chromosome 17 centromere (CEP17) duplication as a predictor of
anthracycline response: evidence from the NCIC Clinical Trials
Group (NCIC CTG) MA.5 Trial. Breast Cancer Res Treat. 131:541–551.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lal P, Salazar PA, Ladanyi M and Chen B:
Impact of polysomy 17 on HER-2/neu immunohistochemistry in breast
carcinomas without HER-2/neu gene amplification. J Mol Diagn.
5:155–159. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Varshney D, Zhou YY, Geller SA and Alsabeh
R: Determination of HER-2 status and chromosome 17 polysomy in
breast carcinomas comparing HercepTest and PathVysion FISH assay.
Am J Clin Pathol. 121:70–77. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rosenberg CL: Polysomy 17 and HER-2
amplification: true, true, and unrelated. J Clin Oncol.
26:4856–4858. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vanden Bempt I, Van Loo P, Drijkoningen M,
et al: Polysomy 17 in breast cancer: clinicopathologic significance
and impact on HER-2 testing. J Clin Oncol. 26:4869–4874.
2008.PubMed/NCBI
|
|
18
|
Jiang HY, Zhang SQ and Zhao T: A new
method to make nuclei or cell microarrays. Diagn Mol Pathol.
15:109–114. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tse CH, Hwang HC, Goldstein LC, et al:
Determining true HER2 gene status in breast cancers with polysomy
by using alternative chromosome 17 reference genes: implications
for anti-HER2 targeted therapy. J Clin Oncol. 29:4168–4174. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li X, Wan L, Shen H, et al: Thyroid
transcription factor-1 amplification and expressions in lung
adenocarcinoma tissues and pleural effusions predict patient
survival and prognosis. J Thorac Oncol. 7:76–84. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Birner P, Oberhuber G, Stani J, et al:
Evaluation of the United States Food and Drug
Administration-approved scoring and test system of HER-2 protein
expression in breast cancer. Clin Cancer Res. 7:1669–1675.
2001.PubMed/NCBI
|
|
22
|
Watters AD, Going JJ, Cooke TG and
Bartlett JM: Chromosome 17 aneusomy is associated with poor
prognostic factors in invasive breast carcinoma. Breast Cancer Res
Treat. 77:109–114. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Risio M, Casorzo L, Redana S and
Montemurro F: HER2 gene-amplified breast cancers with monosomy of
chromosome 17 are poorly responsive to trastuzumab-based treatment.
Oncol Rep. 13:305–309. 2005.PubMed/NCBI
|
|
24
|
Hammock L, Lewis M, Phillips C and Cohen
C: Strong HER-2/neu protein overexpression by immunohistochemistry
often does not predict oncogene amplification by fluorescence in
situ hybridization. Hum Pathol. 34:1043–1047. 2003.
|
|
25
|
Pritchard KI, Shepherd LE, O’Malley FP, et
al: HER2 and responsiveness of breast cancer to adjuvant
chemotherapy. N Engl J Med. 354:2103–2111. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ejlertsen B, Jensen MB, Nielsen KV, et al:
HER2, TOP2A, and TIMP-1 and responsiveness to adjuvant
anthracycline-containing chemotherapy in high-risk breast cancer
patients. J Clin Oncol. 28:984–990. 2010. View Article : Google Scholar : PubMed/NCBI
|