Introduction
Meningiomas are one of the most frequent
intracranial tumors. The majority of meningiomas are benign lesions
of World Health Organization (WHO) grade I (GI), however, certain
histopathological variants are associated with more aggressive
clinical behavior and correspond to WHO GII and III (1). The GII and III meningiomas have an
increased risk of local recurrence. The standard treatment for
high-grade meningiomas involves surgical resection with optional
radiotherapy, and radiosurgery in the case of an incomplete
resection. No pharmacological agents are routinely used for the
treatment of meningiomas. The epithelial growth factor receptor
(EGFR) is one of the most extensively studied molecular targets for
anti-cancer therapy and is also considered as a target for
meningioma treatment (2). This
receptor is expressed in most meningioma patients and was initially
identified to be involved in tumor development (3). The results of studies on EGFR
expression in meningiomas are not entirely consistent. The
percentage of EGFR-immunopositive tumors range between 50% (n=85)
(4) and 89% (n=132) (5). In addition to the expression status,
receptor activation has also been shown in patients (6,7). In
the IOMM-Lee meningioma cell line, the EGFR pathway was shown to be
involved in radiation-induced progression (8). EGFR activates several downstream
pathways, primarily those of mitogen-activated protein kinase
(MAPK), phosphatidylinositol-4,5-bisphosphate 3-kinase/protein
kinase B (PI3K/AKT) and phospholipase C, which have been found to
play a role in meningioma pathogenesis (7,9).
Based on this background, two of the low molecular
weight anti-EGFR inhibitors, gefitinib and erlotinib, were
introduced in phase II clinical trials in patients with recurrent
meningioma (North American Brain Tumor Consortium Trials 00–01 and
01–03). In these studies, small groups of patients were included
(n=9 and n=16, respectively) and no clear effect of the treatment
was observed (10).
The data from previous clinical trials on the
treatment of other tumor types showed that clinical response may
depend on the presence of the mutations in EGFR (11) and in genes encoding downstream
proteins involved in the signal transduction, including Kirsten rat
sarcoma viral oncogene homolog (KRAS), v-Raf murine sarcoma
viral oncogene homolog B1 (BRAF) or
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α
(PIK3CA) (12). Mutations of
the EGFR tyrosine kinase (TK) domains lead to constitutive
kinase activation and were shown to be a good predictive marker in
non-small cell lung cancer, while KRAS and BRAF were
negative prognostic markers in colorectal cancer (12,13).
PIK3CA encodes the catalytic subunit of the PI3K protein and
is affected by gain of function mutations in a significant ratio of
solid tumors (14). The role of
PIK3CA mutations in anti-EGFR therapy has not been
definitively clarified. In colorectal cancer they coexist with
KRAS mutations and are considered to be co-responsible for
the resistance to targeted therapy (12). The mutations in the aforementioned
downstream proteins generally cause activation of the MAPK or AKT
pathways independent of the ligand binding or receptor status, and
in lung cancer they have been shown to be mutually exclusive with
EGFR mutations (15).
Previous meningioma studies have shown that
EGFR does not undergo gene amplification (16,17),
but to the best of our knowledge the point mutations in the TK
domain of the receptor, which is encoded by exons 18–21, have not
been analyzed. Each of the BRAF, KRAS and PI3K
genes has been analyzed once previously (18–20).
The present study aimed to assess the occurrence of the mutations
in the EGFR TK domain and in the selected downstream genes,
KRAS, BRAF and PIK3CA, in a relatively large
group of meningioma patients.
Materials and methods
Patients and tissue samples
Tissue samples used for DNA isolation and genomic
sequencing consisted of archival formalin-fixed, paraffin-embedded
(FFPE) meningioma specimens from patients who underwent surgical
tumor resection at the Department of Neurosurgery, M.
Sklodowska-Curie Memorial Oncology Center (Warsaw, Poland) between
2007 and 2012. In total, 55 meningioma samples, including 20 GI, 22
GII and 13 GIII, were used in the study. All tissue samples
underwent histopathological examination that involved a review of
the initial diagnosis and grading according to the 2007 WHO
guidelines (1). The representative
tissue samples were selected for molecular analysis. Patient
characteristics are listed in Table
I. Written informed consent was obtained from the patients and
the study was approved by the local ethics committee of M.
Sklodowska-Curie Memorial Cancer Center and Institute of Oncology
(Warsaw, Poland).
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Characteristics | Value |
|---|
| Patients, n | 55 |
| Male | 21 |
| Female | 34 |
| Age, years |
| Range | 21–80 |
| Median | 55 |
| Grade, n |
| GI | 30 |
| Meningioma
menigotheliale | 7 |
| Meningioma
fibroblasticum | 4 |
| Meningioma
transitionale | 13 |
| Other | 6 |
| GII meningioma
atypicum | 16 |
| GIII meningioma
anaplasticum | 9 |
Identification of genomic mutations
FFPE tissue samples were manually macrodissected and
DNA was isolated with the use of a QIAamp DNA mini kit (Qiagen,
Hilden, Germany). The purity and concentration of the DNA samples
were assessed with the use of NanoDrop 1000 spectrophotometer
(Thermo Scientific, Rockford, IL, USA) and with PicoGreen dsDNA
quantitation assay (Invitrogen Life Technologies, Carlsbad, CA,
USA).
DNA was amplified by polymerase chain reaction (PCR)
containing 1× PCR buffer, 2 mM MgCl2, 250 μM of each
oligonucleotide, 0.15 μM of each PCR primer and 0.5 units of
FastStart Taq DNA Polymerase (Roche Diagnostics, Mannheim,
Germany). The GeneAmp 9700 PCR system (Applied Biosystems, Foster
City, CA, USA) was used with the following cycling conditions:
Initial denaturation at 94°C for 3 min, followed by 40 cycles of 30
sec at 94°C, 30 sec at 57ºC and 30 sec at 72°C, and a final
elongation for 7 min at 72°C. The PCR products were analyzed by
electrophoresis in a 2% agarose gel, visualized with the use of
ethidium bromide and subjected to DNA sequencing. Briefly, 2.5 μl
of the PCR product was purified with the use of ExoStar (GE
Healthcare Life Sciences, Pittsburgh, PA, USA) and labeled with
BigDye Terminator v.3.1 (Applied Biosytems) according to the
manufacturer’s instructions. The ABI PRISM 3300 Genetic Analyzer
(Applied Biosystems) was used for capillary electrophoresis and the
sequence readout.
Results
All the DNA regions involved in the sequence
analysis (EGFR exons 18–21, KRAS exon 1, BRAF
exon 15 and PI3K exons 9 and 20) were successfully amplified
and sequenced for all the patients enrolled. No mutations were
identified in either EGFR, KRAS or BRAF. There
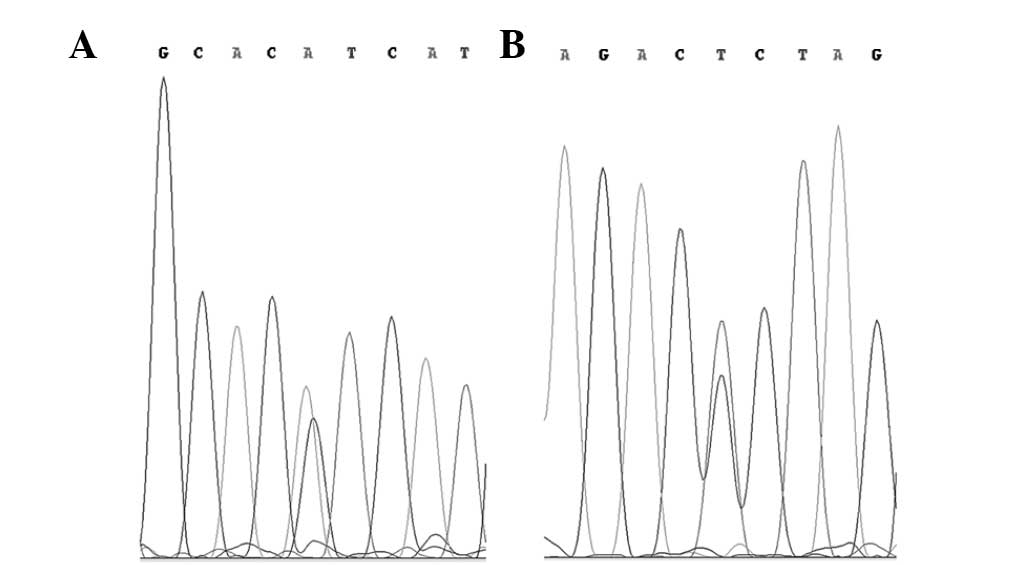
were two PIK3CA exon 20 mutations identified. These were the
missense mutation A>AG, 1047H>H/R in codon 3140, and the
silent mutation C>CT, 1025T>T in codon 3075 (Fig. 1). These samples were resected from a
54-year-old male patient with a GII atypical meningioma in the
cerebellopontine angle and from a 53-year-old female patient with a
GIII anaplastic meningioma located in the cerebellopontine angle,
respectively.
Discussion
The results of the present study are consistent with
previously reported studies that also did not identify an incidence
of mutations in KRAS (17)
or BRAF (18). The lack of
mutations in the EGFR TK domain and the downstream proteins
that have been observed may indicate a low importance of this
pathway in the growth of meningiomas, despite a previously
suggested hypothesis (6). This is
in line with previous studies that report no incidence of
EGFR gene amplification in meningiomas (16), as these molecular aberrations have a
similar effect on signal transduction. This is also consistent with
a previous study reporting that EGFR expression occurs less
frequently in malignant meningiomas than in GI patients (5), and EGFR-positive patients have an
improved survival prognosis compared with EGFR-negative patients
(4).
The purpose of the mutation screening in the present
study was to assess whether it is possible to use the molecular
background to identify meningioma patients who may have a
potentially improved response to anti-EGFR treatment, as is
possible in the case of other tumors. The results show that known
predictive markers have no value in meningiomas, and the lack of
EGFR and downstream protein mutations may partially explain
the results of the clinical trials with the anti-EGFR inhibitors,
erlotinib and gefitinib (10).
All the aforementioned data indicate that despite
the fact that EGFR is expressed in meningeal tumors, it does not
contribute to tumor development or progression, as was hypothesized
initially (9). This in turn
indicates that the EGFR pathway is not a promising target for the
treatment of meningiomas.
In the present study, the only identified genetic
aberrations were the mutations of the kinase domain of
PIK3CA that were observed in two patients with GII and GIII
tumors. The genetic changes, 1047H>H/R and 1025T>T, have
already been reported in the COSMIC (Catalog of Somatic Mutations
in Cancer) database (database ID 775 and 21451, respectively). The
1047H>H/R missense mutation is the most common mutation reported
in tumor samples and accounts for 24.4% of all variations of the
PIK3CA coding sequence submitted to the COSMIC database.
This mutation has revealed strong oncogenic potential in
vivo (20) and recently was
also shown to be associated with the response to PI3K/AKT pathway
inhibitors in clinical trials (21). The second identified genomic
variation does not affect the amino acid sequence and is less
frequent in cancer patients, with a frequency of 0.3% PIK3CA
mutations reported in the COSMIC database.
The occurrence of the PIK3CA mutations in
meningiomas was investigated in a previous study, and one of the
patients with anaplastic meningioma was identified as a mutation
carrier (19). The combination of
data from the previous and current studies indicates a 3.4% (3/87)
frequency of PIK3CA variations in atypical and anaplastic
meningiomas. This can be described as rare, but the PI3K/AKT
pathway may also be deregulated by the mutations in phosphatase and
tensin homolog (PTEN); a PI3K inhibitor. In meningiomas,
these gene mutations have previously been shown to occur with a
frequency similar to the PIK3CA mutations (22). In total, two of the PTEN
mutations were identified in GIII tumors and one in a
radiation-induced meningioma. No PIK3CA or PTEN
mutations were found in any GI patients (19,22).
This indicates the involvement of this pathway in tumor
progression. The phosphorylation in the PI3K/AKT pathway was
previously observed as being more frequent in GII and III
meningiomas, and pharmacological inhibition of this pathway in
primary malignant meningioma cells resulted in reduced
proliferation and survival (7).
In the light of all the presented facts it appears
that PI3K and its co-regulators may be more reasonable molecular
targets in meningiomas than EGFR. Recently, a number of small
molecular inhibitors that target PI3K have been reported in
clinical trials (21), and
mutations of PIK3CA/PTEN have been show to have a predictive
value in the administration of these compounds (23,24).
We believe that these results should encourage clinicians to
consider the possible use of this pharmacological treatment in
malignant meningiomas.
Acknowledgements
This study was supported by the National Science
Centre (grant no. UMO-2011/01/D/NZ5/02798).
References
|
1
|
Louis DN, Ohgaki H, Wiestler OD, et al:
The 2007 WHO classification of tumours of the central nervous
system. Acta Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Normanno N, De Luca A, Bianco C, et al:
Epidermal growth factor receptor (EGFR) signaling in cancer. Gene.
366:2–16. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torp SH, Helseth E, Dalen A and Unsgaard
G: Expression of epidermal growth factor receptor in human
meningiomas and meningeal tissue. APMIS. 100:797–802. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Smith JS, Lal A, Harmon-Smith M, Bollen AW
and McDermott MW: Association between absence of epidermal growth
factor receptor immunoreactivity and poor prognosis in patients
with atypical meningioma. J Neurosurg. 106:1034–1040. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wernicke AG, Dicker AP, Whiton M, et al:
Assessment of epidermal growth factor receptor (EGFR) expression in
human meningioma. Radiat Oncol. 5:462010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Carroll RS, Black PM, Zhang J, Kirsch M,
Percec I, Lau N and Guha A: Expression and activation of epidermal
growth factor receptors in meningiomas. J Neurosurg. 87:315–323.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mawrin C, Sasse T, Kirches E, et al:
Different activation of mitogen-activated protein kinase and Akt
signaling is associated with aggressive phenotype of human
meningiomas. Clin Cancer Res. 11:4074–4082. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kargiotis O, Chetty C, Gogineni V, et al:
uPA/uPAR downregulation inhibits radiation-induced migration,
invasion and angiogenesis in IOMM-Lee meningioma cells and
decreases tumor growth in vivo. Int J Oncol. 33:937–947.
2008.PubMed/NCBI
|
|
9
|
Johnson M and Toms S: Mitogenic signal
transduction pathways in meningiomas: novel targets for meningioma
chemotherapy? J Neuropathol Exp Neurol. 64:1029–1036. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Norden AD, Raizer JJ, Abrey LE, et al:
Phase II trials of erlotinib or gefitinib in patients with
recurrent meningioma. J Neurooncol. 96:211–217. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lynch TJ, Bell DW, Sordella R, et al:
Activating mutations in the epidermal growth factor receptor
underlying responsiveness of non-small-cell lung cancer to
gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
De Roock W, Claes B, Bernasconi D, et al:
Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy
of cetuximab plus chemotherapy in chemotherapy-refractory
metastatic colorectal cancer: a retrospective consortium analysis.
Lancet Oncol. 11:753–762. 2010.PubMed/NCBI
|
|
13
|
Brand TM, Iida M and Wheeler DL: Molecular
mechanisms of resistance to the EGFR monoclonal antibody cetuximab.
Cancer Biol Ther. 11:777–792. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Samuels Y, Wang Z, Bardelli A, et al: High
frequency of mutations of the PIK3CA gene in human cancers.
Science. 304:5542004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kosaka T, Yatabe Y, Endoh H, Kuwano H,
Takahashi T and Mitsudomi T: Mutations of the epidermal growth
factor receptor gene in lung cancer: biological and clinical
implications. Cancer Res. 64:8919–8923. 2004. View Article : Google Scholar
|
|
16
|
Diedrich U, Lucius J, Baron E, Behnke J,
Pabst B and Zoll B: Distribution of epidermal growth factor
receptor gene amplification in brain tumours and correlation to
prognosis. J Neurol. 242:683–688. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chaffanet M, Chauvin C, Lainé M, et al:
Comparative analysis of the NF2, TP53, PTEN, KRAS, NRAS and HRAS
genes in sporadic and radiation-induced human meningiomas. Int J
Cancer. 94:218–221. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schindler G, Capper D, Meyer J, et al:
Analysis of BRAF V600E mutation in 1,320 nervous system tumors
reveals high mutation frequencies in pleomorphic xanthoastrocytoma,
ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta
Neuropathol. 121:397–405. 2011. View Article : Google Scholar
|
|
19
|
Pang JC, Chung NY, Chan NH, Poon WS,
Thomas T and Ng HK: Rare mutation of PIK3CA in meningiomas. Acta
Neuropathol. 111:284–285. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bader AG, Kang S and Vogt PK:
Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc
Natl Acad Sci USA. 103:1475–1479. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Janku F, Wheler JJ, Naing A, et al: PIK3CA
mutation H1047R is associated with response to PI3K/AKT/mTOR
signaling pathway inhibitors in early-phase clinical trials. Cancer
Res. 73:276–284. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peters N, Wellenreuther R, Rollbrocker B,
Hayashi Y, Meyer-Puttlitz B, Duerr EM, Lenartz D, Marsh DJ, Schramm
J, Wiestler OD, Parsons R, Eng C and von Deimling A: Analysis of
the PTEN gene in human meningiomas. Neuropathol Appl Neurobiol.
24:3–8. 1998. View Article : Google Scholar
|
|
23
|
Willems L, Tamburini J, Chapuis N, Lacombe
C, Mayeux P and Bouscary D: PI3K and mTOR signaling pathways in
cancer: new data on targeted therapies. Curr Oncol Rep. 14:129–138.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Martini M, Ciraolo E, Gulluni F and Hirsch
E: Targeting PI3K in cancer: Any good news? Front Oncology.
3:1082013. View Article : Google Scholar : PubMed/NCBI
|