Introduction
In 2013, colorectal cancer had the third highest
incidence of new cases and the third highest rate of cancer
mortality in the USA, with 142,280 and 50,830 individuals,
respectively (1). The occurrence
and development of this lethal disease is a multi-step process
involving multiple gene mutations. The diversity and complexity of
somatic mutational processes that underlie carcinogenesis in humans
is being revealed through mutational patterns hidden within cancer
genomes (2). A variety of genomic
consortia, including The Cancer Genome Atlas (TCGA) and the
International Cancer Genome Consortium, are attempting to catalog
all somatic mutations occurring in major cancer types. In addition,
the Catalogue Of Somatic Mutations In Cancer (COSMIC) has served as
a central repository designed to store and exhibit somatic mutation
information derived from the cancer genome consortia and from the
literature (3–5). Driving the massive data collection is
the use of next-generation sequencing (NGS), which has the ability
to probe millions of DNA fragments for mutations and is
subsequently enabling clinicians to more accurately gauge the risk
of developing cancer and tailor therapies to treat cancers with
specific genetic mutations (6). The
Ion Torrent Personal Genome Machine (PGM; Invitrogen Life
Technologies, Carlsbad, CA, USA) presents an emerging NGS approach
that relies on non-optical semiconductor sequencing technology with
a rapid turnaround time (7). The
deep coverage achieved by the PGM makes it possible to detect
somatic mutations in tumor cells with low allele frequency, which
may not be detected by conventional Sanger sequencing.
Notably, much of the data reported thus far by the
Cancer Genomic Consortium using NGS has focused on sequencing from
the primary tumor, with limited data on ‘druggable’ mutations
present in the metastases. The current study used this emerging
technology to detect somatic mutations in formalin-fixed,
paraffin-embedded (FFPE) tissues obtained from the metastatic
nodules of metastatic colorectal cancer (mCRC) patients, in order
to identify somatic alterations suitable for anticancer drug
treatment.
Patients and methods
Patients
FFPE tissues obtained from patients with mCRC were
collected from The First Affiliated Hospital, School of Medicine,
Zhejiang University (Hangzhou, China). All patients provided
written informed consent and the study protocol was approved by the
Institutional Ethics Committee of The First Affiliated Hospital.
Patient information, including age, gender, diagnosis, positive
lymph node number, response rate, disease-free survival following
primary surgery, overall survival following salvage chemotherapy,
number of metastasic organs and chemotherapy regimen were recorded.
Chemotherapy efficacy evaluation was performed according to the
Response Evaluation Criteria in Solid Tumors guidelines, version
1.1 (8).
NGS sequencing
DNA preparation was performed, as described
previously (9). The DNA was then
sequenced using the PGM (Invitrogen Life Technologies) according to
the manufacturer’s instructions. For the targeted amplification of
known cancer genes, the Ion AmpliSeq™ Cancer Panel (Invitrogen Life
Technologies), which is designed to detect 739 COSMIC mutations in
604 loci from 46 oncogenes and tumor suppressor genes using as
little as 10 ng of input DNA, was used. Next, a template was
prepared using the Ion PGM 200 Xpress template kit (Invitrogen Life
Technologies) and sequencing was performed using the Ion Sequencing
kit version 2.0 on an Ion 316 chip. Data were analyzed using the
Ampliseq™ Variant Caller plug-in within the Ion Torrent Suite
software (Invitrogen Life Technologies). The sequences of all
primers and probes are available on request.
Validation Sanger sequencing of KRAS and
FGFR3
The sequencing template used for KRAS was a 170-bp
polymerase chain reaction (PCR) fragment of the KRAS gene,
generated using the following primers: Forward,
5′-AAGGCCTGCTGAAAATGACTG-3′ and reverse,
5′-AGAATGGTCCTGCACCAGTAA-3′ [Generay Biotech (Shanghai) Co., Ltd.,
Shanghai, China]. The PCR conditions used were as follows: 40
cycles of predenaturation for five min at 95°C, denaturation at
95°C for 20 sec, annealing at 60°C for 20 sec and elongation at
72°C for 20 sec, followed by a final extension at 72°C for five
min.
The sequencing template used for FGFR3 was a 296-bp
PCR fragment of the FGFR3 gene, generated using the following
primers: Forward, 5′-GTGTGTATGCAGGCATCCTCAGC-3′ and reverse,
5′-ATGGTGAGCAGAGACGAGGAGAGG-3′ [Generay Biotech (Shanghai) Co.,
Ltd.]. The PCR conditions used were as follows: 40 cycles of
predenaturation for five min at 95°C, denaturation at 95°C for 20
sec, annealing at 62°C for 20 sec and elongation at 72°C for 20
sec, followed by a final extension at 72°C for five min.
The PCR products were then purified using the shrimp
alkaline phosphatase/exonuclease PCR clean method [New England
Biolabs (UK) Ltd., Hitchin, UK]. Next, the purified samples (2 μl)
were used directly for a sequencing reaction using the Big Dye
Terminator cycle sequencing mix, version 3.1 (Applied Biosystems,
Carlsbad, CA, USA). Sequencing reactions were then performed for
the two DNA strands using the PCR oligonucleotides (3.2 pmol) as
respective primers. Dye purification was performed using
alcohol/sodium acetate precipitation, and subsequent sequence
analysis was conducted using an ABI 3130 genetic analyzer (Applied
Biosystems).
Gene pathway analysis
Ingenuity pathway analysis (IPA; Qiagen, Valencia,
CA, USA) was used for core analysis to identify the existing
metastasis network.
Statistical analysis
Statistical analyses were conducted using SPSS
version 20.0 (IBM, Armonk, NY, USA). All tests of significance were
two-sided and P≤0.05 was considered to indicate a statistically
significant difference. Cox regression analysis was used to
investigate a potential correlation between the alteration numbers
and clinical factors, including age, gender, diagnosis, positive
lymph node number, response rate, disease-free survival following
primary surgery, overall survival following salvage chemotherapy,
number of metastasic organs and chemotherapy regimen.
Results
Patient characteristics
A total of 10 mCRC patients were enrolled in the
current study between April 2007 and August 2010 (Table I). The median age was 60 years
(range, 37–73 years) and the patients consisted of five males and
five females.
 | Table IPatient characteristics and clinical
outcome. |
Table I
Patient characteristics and clinical
outcome.
| Patient number | Gender | Age, years | Primary location | Positive lymph
node | Response rate
(first-line chemotherapy) | Disease-free
survival, days | Overall survival,
days |
|---|
| 23 | Male | 60 | Rectum | 4 | PR | 1125 | 449 |
| 24 | Female | 64 | Colon | 2 | SD | 594 | Alive |
| 25 | Male | 60 | Rectum | 4 | PD | 403 | 126 |
| 26 | Female | 59 | Rectum | / | SD | 518 | Alive |
| 27 | Male | 73 | Colon | 0 | PD | 1212 | 74 |
| 28 | Female | 51 | Rectum | 0 | PD | 814 | 64 |
| 29 | Female | 69 | Rectum | 2 | PD | 1096 | Alive |
| 30 | Male | 68 | Colon | / | PR | / | 852 |
| 31 | Male | 37 | Colon | / | PR | / | 420 |
| 32 | Female | 57 | Rectum | / | PR | / | Alive |
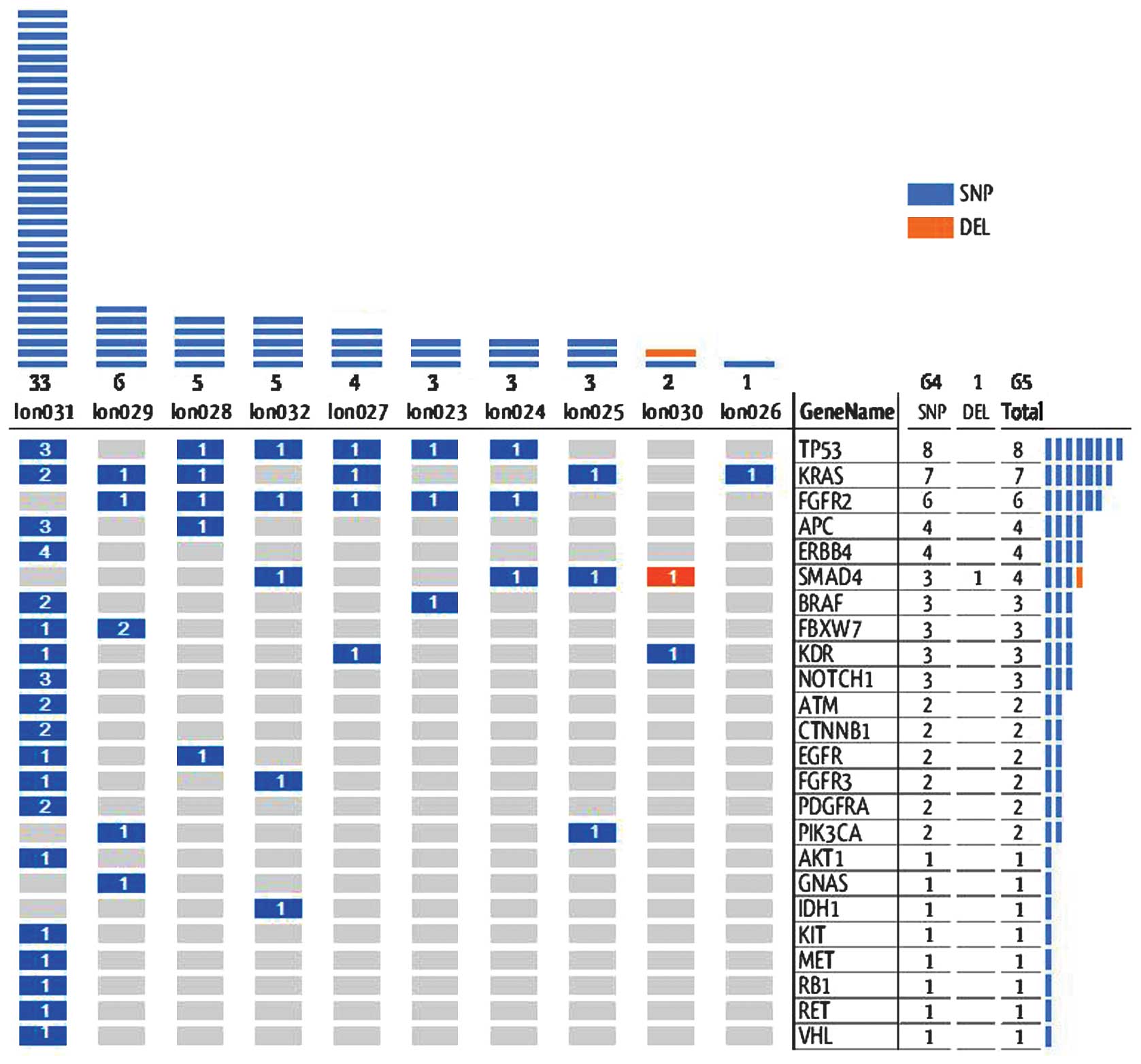
Overall gene alterations
Among the 10 specimens, 65 genetic alterations were
identified in 24 genes, following the exclusion of germline
mutations according the single nucleotide polymorphism (SNP)
database, as shown in Fig. 1.
Among these alterations, 41% were present in the
COSMIC database. These alterations confirmed by COSMIC were all
SNPs, divided into missense (83%) and nonsense (17%) changes.
Notably, four genes exhibited >1 alteration; APC (n=2), FBXW7
(n=2), TP53 (n=3) and KRAS (n=5). No clinical factors were found to
significantly correlate with the alteration numbers in patients by
statistical analysis.
Sanger sequencing validation
Sanger sequencing of KRAS revealed that all results
were consistent with PGM, with the exception of one. Sample 31 was
wild-type, however, PGM identified the G13D mutation (c.38G>A;
COSMIC 532). PGM also revealed that the percentage of mutations at
the cell level of this sample was 26.6%. Sanger sequencing of FGFR3
revealed that all samples were wild-type with seven consecutive ‘C’
repeats (GCCTGCGCAGCCCCCCCAAGAAA). However, PGM revealed a ‘C’ to
‘G’ change in eight samples, with the sequence
‘GCCTGCGCAGGCCCCCCAAGAAA.
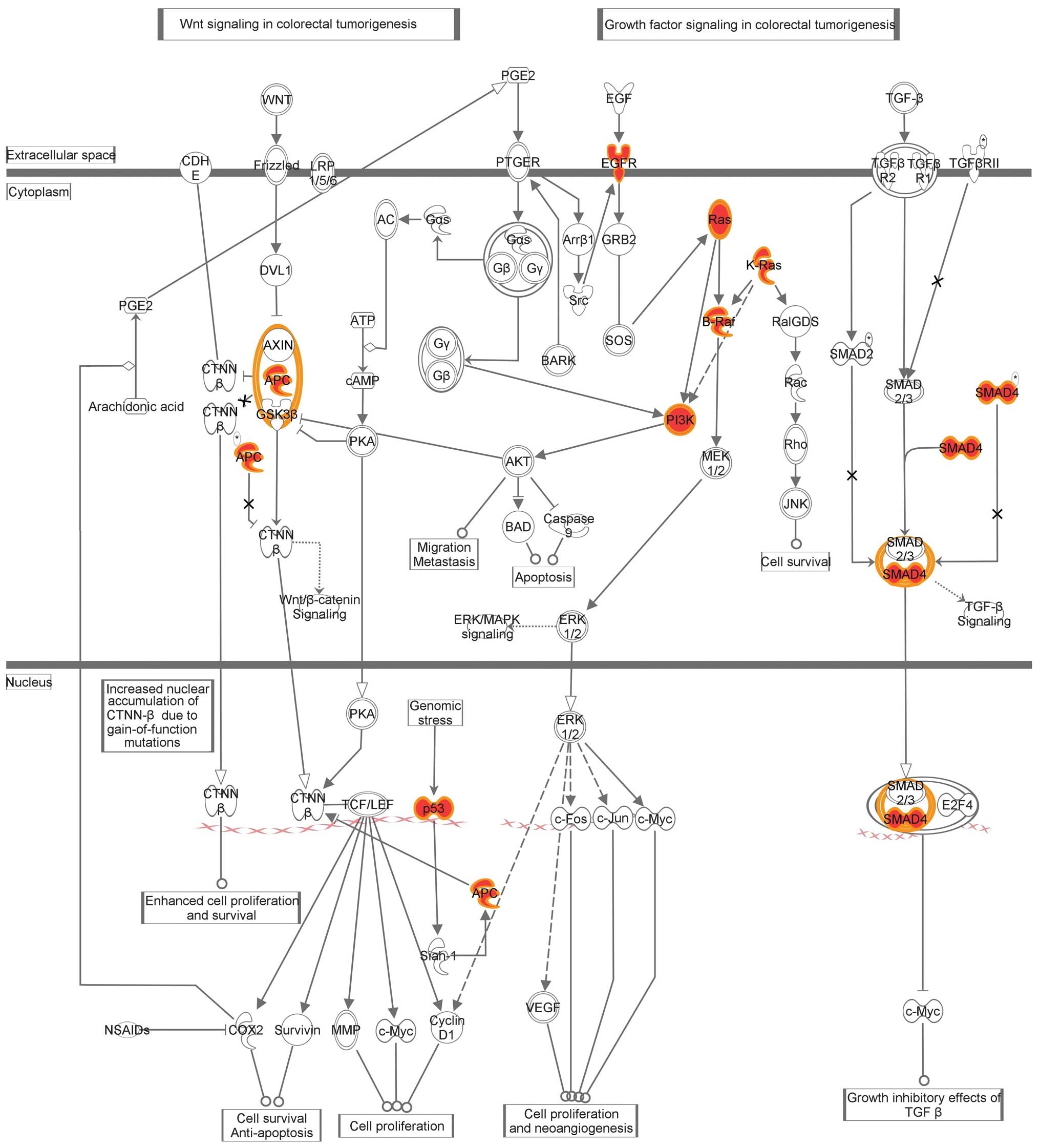
Pathway analysis of detected somatic
mutations
The IPA identified ‘colorectal cancer metastasis
signaling’ as the most commonly mutated canonical pathway, which
includes Wnt, phosphoinositide 3-kinase (PI3K)/AKT and transforming
growth factor (TGF)-β/SMAD signaling (Fig. 2).
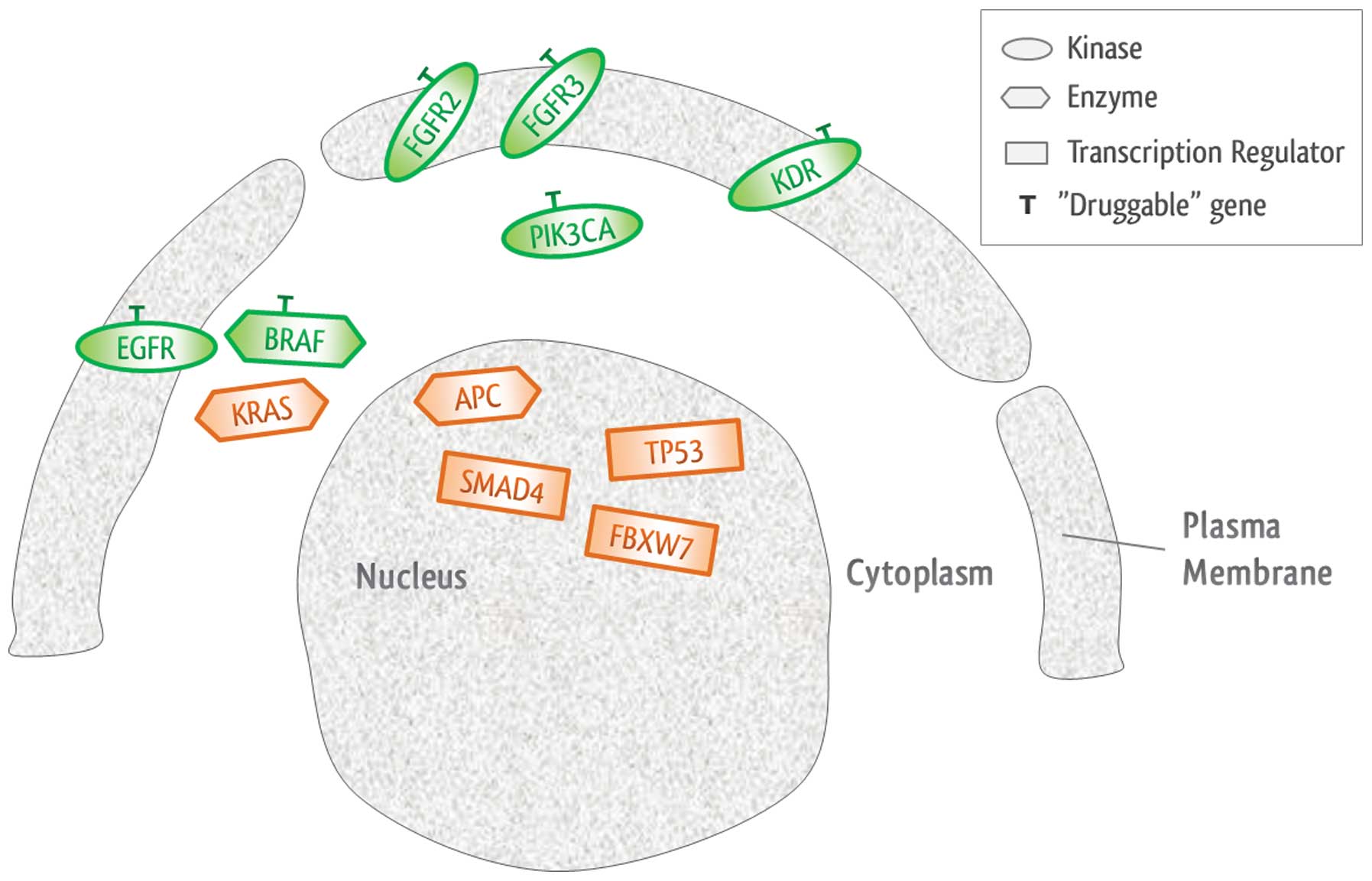
Notably, 90% (9/10) patients harbored at least one
‘druggable’ alteration (range, 1–6 alterations) that has been
associated with a clinical treatment option or is currently being
investigated in clinical trials of novel targeted therapies, as
shown in Table II. In addition,
IPA clarified that there were six ‘druggable’ genes with specific
target drugs, and 90% of samples exhibited at least one of them, as
shown in Fig. 3.
 | Table IIIngenuity pathway analysis identified
genes and corresponding drugs. |
Table II
Ingenuity pathway analysis identified
genes and corresponding drugs.
| Gene | Location | Type | Drug(s) |
|---|
| APC | Nucleus | Enzyme | |
| BRAF | Cytoplasm | Enzyme | Vemurafenib,
sorafenib |
| EGFR | Plasma membrane | Kinase | Cetuximab,
panitumumab and BMS-599626, among others |
| FBXW7 | Nucleus | Transcription
regulator | |
| FGFR2 | Plasma membrane | Kinase | Palifermin |
| FGFR3 | Plasma membrane | Kinase | Pazopanib |
| KDR | Plasma membrane | Kinase | Sunitinib, axitinib
and AEE 788, among others |
| KRAS | Cytoplasm | Enzyme | |
| PIK3CA | Cytoplasm | Kinase | SF-1126, PX-866 and
NVP-BEZ235, among others |
| SMAD4 | Nucleus | Transcription
regulator | |
| TP53 | Nucleus | Transcription
regulator | |
Discussion
Oncology is progressing away from
organ-of-origin-based management strategies towards the more
refined strategy of using targeted therapies driven by a tumor’s
molecular characteristics. At present, NGS aids in ‘building
bridges’ between molecular screening and clinical reality, with an
accuracy of 96.1% in comparison to Sanger sequencing (10). Furthermore, PGM is aiming to become
a ‘point-of-care’ NGS platform (11). In the present study, PGM identified
65 genetic alterations in 24 genes using as little as 10 ng of
input DNA of each sample. In addition, PCM has the ability to
reveal alterations at very low allele frequencies, a major
limitation of conventional Sanger sequencing. In patient 31, as
Sanger sequencing revealed a ‘wide-type’ result prior to
chemotherapy, cetuximab was administered to the patients, which
achieved a good response. However, the PGM also revealed a KRAS
G13D mutation, presenting an noteworthy situation, as a randomized
clinical trial previously demonstrated that tumors with a KRAS G13D
mutation may also be sensitive to cetuximab in colorectal cancer
patients (12).
A limitation of the PGM is the large number of
false-positive indels, which result from homopolymer errors. To
overcome this, the current study used the Ampliseq™ Variant Caller
plug-in within the Ion Torrent Suite software, which found only one
deletion alteration among the 65 somatic variants detected. In
addition, due to the lack of matched germline samples, the SNP
database was used to exclude germline variants prior to the
generation of the final results, However, difficulties remain in
differentiating somatic mutations from rare germline inherited
SNPs. An unexpected false-positive indel in FGFR3 was also
observed, which was possibly due to its seven consecutive identical
base ‘C’ repeats, which are prone to errors in PGM analysis.
To obtain a pathway-centric analysis of the data,
the current study used IPA, which combines advanced pathway
enrichment analysis with the pathway topological analysis to aid in
the identification of the most relevant metabolic pathways involved
in diseases and cellular processes (13). IPA identified significant cancer
pathways, including those of Wnt signaling, PI3K/AKT and
TGF-β/SMAD, which are known to be frequently activated in cancer.
In addition, mutations in the APC gene have been identified as the
initiating event in the inherited and spontaneous forms of mCRC.
Furthermore, when this tumor suppressor gene is mutated, its
ability to regulate the Wnt pathway is lost (14). The past decade has revealed that the
PI3K/AKT signaling pathway is among the most highly mutated
pathways in human carcinogenesis (15). Mutations in this pathway occur in
the majority of cancers contributing to the resistance to
apoptosis, the deregulation of proliferation and changes in the
metabolism characteristics of transformed cells (16). Finally, the function of members of
the TGF-β family is exerted via specific kinase receptors and
intracellular SMAD transcription factors, including the common
mediator Smad4. The initiation of adenocarcinomas of the
gastrointestinal tract and squamous cell carcinomas of the skin can
involve the loss of SMAD4 (17).
Notably, 11 genes, including the expected APC, BRAF,
KRAS, PIK3CA and TP53 genes, were mutated in at least two samples
of the current study, which is similar to the gene list revealed by
the TCGA Network in 2012 (18).
This phenomenon indicates that almost all colorectal
cancer patients may have the chance to be treated with at least one
target drug according to their ‘druggable’ genes. For example, in
the present study, vemurafenib was considered to target patients
with the BRAF V600E mutation (19).
In addition, the anti-epidermal growth factor receptor, cetuximab,
has been proven to be of great success in mCRC treatment (20). Other examples of drug and targeted
gene pairs are as follows: Palifermin and FGFR2, pazopanib and
FGFR3, AEE 788 and KDR, and BEZ235 and PIK3CA (21–23).
The majority of somatic mutations in this tumor
class have great potential to provide a variety of target drugs for
cancer patients. However, challenges remain in translating
sequencing information into clinical practice. Therefore,
identification of genetic factors that affect the response to
treatment are essential to identify and develop next generation
medicines that target the ‘druggable’ alterations of patients.
Acknowledgements
The authors would like to thank Dr Anwu Zhou, Mrs.
Wenying Huang and Dr Yankai Jia from Genewiz, Inc (South
Plainfield, NJ, USA) for their technical support. This study was
supported by the Major Scientific Project of Zhejiang Province
(grant no. 2012C13014-2), the National Science and Technology Major
Project (grant no. 2012ZX10002017), the National Natural Science
Foundation of China (grant nos. 81201557 and 81272679), the
Zhejiang Natural Science Foundation (grant no. LY13H160007) and the
Zhejiang Medicines and Health Science and Technology Project (grant
no. 201348801).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar
|
|
2
|
Alexandrov LB, Nik-Zainal S, Wedge DC,
Campbell PJ and Stratton MR: Deciphering signatures of mutational
processes operative in human cancer. Cell Rep. 3:246–259. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bamford S, Dawson E, Forbes S, et al: The
COSMIC (Catalogue of Somatic Mutations in Cancer) database and
website. Br J Cancer. 91:355–358. 2004.PubMed/NCBI
|
|
4
|
Forbes SA, Bindal N, Bamford S, et al:
COSMIC: mining complete cancer genomes in the Catalogue of Somatic
Mutations in Cancer. Nucleic Acids Res. 39:D945–D950. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alexandrov LB, Nik-Zainal S, Wedge DC, et
al; Australian Pancreatic Cancer Genome Initiative; ICGC Breast
Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain.
Signatures of mutational processes in human cancer. Nature.
500:415–421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
O’Hanlon LH: How next-generation
sequencing could change cancer care. J Natl Cancer Inst.
105:836–838. 2013.PubMed/NCBI
|
|
7
|
Rothberg JM, Hinz W, Rearick TM, et al: An
integrated semiconductor device enabling non-optical genome
sequencing. Nature. 475:348–352. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nishino M, Jackman DM, Hatabu H, et al:
New Response Evaluation Criteria in Solid Tumors (RECIST)
guidelines for advanced non-small cell lung cancer: comparison with
original RECIST and impact on assessment of tumor response to
targeted therapy. AJR Am J Roentgenol. 195:W221–W228. 2010.
View Article : Google Scholar
|
|
9
|
Schewe C, Goldmann T, Grosser M, et al:
Inter-laboratory validation of PCR-based detection of
Mycobacterium tuberculosis in formalin-fixed,
paraffin-embedded tissues. Virchows Arch. 447:573–585. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hadd AG, Houghton J, Choudhary A, et al:
Targeted, high-depth, next-generation sequencing of cancer genes in
formalin-fixed, paraffin-embedded and fine-needle aspiration tumor
specimens. J Mol Diagn. 15:234–247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Quail MA, Smith M, Coupland P, et al: A
tale of three next generation sequencing platforms: comparison of
Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC
Genomics. 13:3412012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tejpar S, Celik I, Schlichting M,
Sartorius U, Bokemeyer C and Van Cutsem E: Association of KRAS G13D
tumor mutations with outcome in patients with metastatic colorectal
cancer treated with first-line chemotherapy with or without
cetuximab. J Clin Oncol. 30:3570–3577. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu S, Gao Y, Dong X, et al: Serum
metabonomics coupled with Ingenuity Pathway Analysis characterizes
metabolic perturbations in response to hypothyroidism induced by
propylthiouracil in rats. J Pharm Biomed Anal. 72:109–114. 2013.
View Article : Google Scholar
|
|
14
|
Cho KH, Baek S and Sung MH: Wnt pathway
mutations selected by optimal beta-catenin signaling for
tumorigenesis. FEBS Lett. 580:3665–3670. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Engelman JA, Luo J and Cantley LC: The
evolution of phosphatidylinositol 3-kinases as regulators of growth
and metabolism. Nat Rev Genet. 7:606–619. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vivanco I and Sawyers CL: The
phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang G and Yang X: Smad4-mediated TGF-beta
signaling in tumorigenesis. Int J Biol Sci. 6:1–8. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cancer Genome Atlas Network. Comprehensive
molecular characterization of human colon and rectal cancer.
Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chapman PB, Hauschild A, Robert C, et al;
BRIM-3 Study Group. Improved survival with vemurafenib in melanoma
with BRAF V600E mutation. N Engl J Med. 364:2507–2516. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lièvre A, Bachet JB, Le Corre D, et al:
KRAS mutation status is predictive of response to cetuximab therapy
in colorectal cancer. Cancer Res. 66:3992–3995. 2006.PubMed/NCBI
|
|
21
|
Okamoto K, Neureiter D, Alinger B, et al:
The dual EGF/VEGF receptor tyrosine kinase inhibitor AEE788
inhibits growth of human hepatocellular carcinoma xenografts in
nude mice. Int J Oncol. 33:733–742. 2008.PubMed/NCBI
|
|
22
|
Traxler P, Allegrini PR, Brandt R, et al:
AEE788: a dual family epidermal growth factor receptor/ErbB2 and
vascular endothelial growth factor receptor tyrosine kinase
inhibitor with antitumor and antiangiogenic activity. Cancer Res.
64:4931–4941. 2004. View Article : Google Scholar
|
|
23
|
Serra V, Markman B, Scaltriti M, et al:
NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and
inhibits the growth of cancer cells with activating PI3K mutations.
Cancer Res. 68:8022–8030. 2008. View Article : Google Scholar : PubMed/NCBI
|