Introduction
Gastric cancer is one of the most common malignant
tumors of the gastrointestinal tract, exhibiting high morbidity and
mortality rates, particularly in Northeast Asia, including China,
Japan and South Korea (1,2). The pathogenesis of gastric cancer
remains unclear, however, social-economic environment, lifestyle,
nutrition, education, smoking and Helicobacter pylori
infection are all associated with its occurrence (3–6).
Human cytomegalovirus (HCMV) is a ubiquitous
β-herpes virus that may cause the infection of multiple cell types
in human hosts. The virus persists in 30–100% of the population
worldwide, particularly in certain areas of Africa and Asia, via
three different infection modes: Acute, persistent and latent
infections (7,8). Asymptomatic infection, caused by
latent state HCMV in healthy individuals, may increase the risk of
atherosclerosis and age-related immune senescence (9,10).
Furthermore, severe or acute disease may be induced in
immunocompromised hosts, including acquired immunodeficiency
syndrome patients and transplant recipients, due to the
reactivation of latent HCMV (11,12).
An increased number of cases of gastrointestinal diseases caused by
HCMV infection have been reported, including ulcerative colitis and
esophageal ulcers (13,14). In addition, emerging evidence has
indicated that HCMV infection may be associated with human
malignancies, including colon and prostate cancer (15,16).
However, the association between gastric cancer and HCMV remains
unclear.
The HCMV genome encodes >200 predicted open
reading frames (ORFs) and is comprised of unique long (UL), unique
short and other repeated sequences (8,17). The
locus spanning UL133–UL138, but particularly UL138, within the ULb’
region is considered to be important for viral latency (18–21).
The reactivation of latent virus (viral replication) may be
estimated by detecting the expression of the immediate-early (IE)
gene (22,23). In the present study, the expression
of UL133–UL138 and IE1 (UL123) were investigated in gastric cancer
and corresponding normal tissues using nested polymerase chain
reaction (PCR), and the clinical association between gastric cancer
and HCMV infection was evaluated.
Materials and methods
Patients and specimens
The paired tissue samples used in the present study
consisted of gastric adenocarcinoma and corresponding normal
tissues, which were obtained by negative resection margin. The
samples were snap-frozen in liquid nitrogen within 30 min of
resection and stored for RNA/DNA extraction. All specimens were
obtained from patients diagnosed with gastric cancer (n=60) by
endoscopic biopsy who underwent surgery at The First Affiliated
Hospital of Wenzhou Medical University (Wenzhou, China) between
February 2011 and December 2012. No patients received radiation or
chemotherapy prior to surgery. The histopathological diagnosis of
gastric adenocarcinoma was confirmed following surgery by the
Department of Pathology according to the criteria of the World
Health Organization (24). Table I shows the clinicopathological
features of the cancer patients according to the National
Comprehensive Cancer Network (2012) guidelines (25). Informed written consent was obtained
from all patients and the study was approved by the Human Research
Ethics Committee of The First Affiliated Hospital of Wenzhou
Medical University.
 | Table IPrimers and amplification conditions
for PCR. |
Table I
Primers and amplification conditions
for PCR.
| Primers (5′ to
3′) | PCR conditions | |
|---|
|
|
| |
|---|
| Name | Forward | Reverse | Annealing
temperature, °C | Annealing time,
sec | Cycles, n | Size, bp |
|---|
| UL133 |
TACCTGCCGATGGGTTCGCTACT |
GGTTTGTCTTTCGCCCTACCTTTCTT | 65 | 30 | 38 | 324 |
| UL135 |
ATGGTGTGGCTGTGGCTCGGCGTCGGGCTCCTCGa |
TCAGGTCATCTGCATTGACTCGGCGTCCTTCATGa | 65 | 30 | 35 | 927 |
|
GGATGGTCTGCCGATAGATAAACCCGb |
CGCTGGCCGAGGACGACAAAGAb | 57 | 30 | 35 | 143 |
| UL136 |
ATGTCAGTCAAGGGCGTGGAGATGCa |
TTACGTAGCGGGAGATACGGCGTTCa | 60 | 30 | 35 | 723 |
|
GCGGTGTTTCACGTTATCTGTGCb |
ATGGCTCGCCGTCTGCTTCTb | 65 | 30 | 35 | 191 |
| UL138 |
ATGGACGATCTGCCGCTGAAa |
TCACGTGTATTCTTGATGATa | 57 | 30 | 35 | 510 |
|
GCTTACCACTGGCACGACACCTb |
TACTCCCCGTACAGCTCGCAACb | 57 | 30 | 35 | 89 |
| IE1 |
AGCCTTCCCTAAGACCACCAAT |
CATAGCAGCACAGCACCCGACA | 60 | 30 | 32 | 290 |
Homology and similarity analysis
A total of 18 HCMV genomes were found and downloaded
from the National Center for Biotechnology Information GenBank
(http://www.ncbi.nlm.nih.gov/) and also
from the University College London virus database (http://www.biochem.ucl.ac.uk/bsm/virus_database/VIDA_table_herpesviridae_cg.html).
The similarity of each UL133–UL138 and IE1 coding sequence was
examined using Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi)
and aligned by clustalx2 (ftp://ftp.ebi.ac.uk/pub/software/clustalw2/). The
corresponding coding sequences in the varying strains were then
used for homology analysis and specific primer design.
RNA isolation and nested PCR
Total RNA was extracted from frozen tissue specimens
using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA,
USA) according to the manufacturer’s instructions. Next, the
first-strand complementary DNA was reverse transcribed using 1 μg
total RNA and the reverse transcription kit (Toyobo Co., Ltd.,
Osaka, Japan) according to the manufacturer’s instructions. The
corresponding primers and the conditions of PCR amplification, as
performed by the pre-programmed Thermal Cycler (Bio-Rad, Hercules,
CA, USA), are listed in Table I.
Following amplification, the PCR products were subjected to
electrophoresis in 2% agarose gel, stained with ethidium bromide,
and images were captured using an ultraviolet light
transilluminator (Bio-Rad). The UL133–UL138 and IE1 PCR
amplification products were then purified, cloned into a pMD19-T
Vector (Takara Bio, Inc., Shiga, Japan) and transformed in
Escherichia coli DH 5α (Novagen, Merck KGaA, Darmstadt,
Germany). Finally, the positive colonies (three monoclones per
sample) were sequenced using M13 sequencing primers [Sangon Biotech
(Shanghai) Co., Ltd., Shanghai, China] by the 3730xl DNA Analyzer
(Applied Biosystems, Carlsbad, CA, USA) to confirm the PCR
specificity.
Statistical analysis
Statistical analyses were performed to investigate
the differences in UL133–138 and IE1 expression between specimens
using the χ2 test, and Fisher’s exact test was used for
samples with small sample numbers. Logistic regression analysis was
used to assess the effect of those loci in the cancer tissues.
P<0.05 was considered to indicate a statistically significant
difference and all analyses were performed using SPSS version 16.0
(SPSS, Inc., Chicago, IL, USA).
Results
Homological analysis of UL133–138 and IE1
coding sequences in different HCMV strains
To establish nested PCR, firstly the similarities
between the UL133–138 and IE1 coding sequences were analyzed in
different HCMV isolates. The data indicated that the UL133–138 and
IE1 coding sequences exhibited a relatively high similarity among
the 18 HCMV strains (data not shown). The homologies of the
nucleotide sequences were 94.39±2.02 (range, 91.12–100), 98.57±0.69
(range, 97.52–100), 98.65±0.60 (range, 97.79–100), 97.96±0.95
(range, 96.47–100) and 97.53±1.25 (range, 95.26–100), respectively.
Based on these observations, specific primers were designed to
detect the expression of these genes in neoplastic and normal
gastric tissues.
Accuracy and specificity of the nested
PCR assay
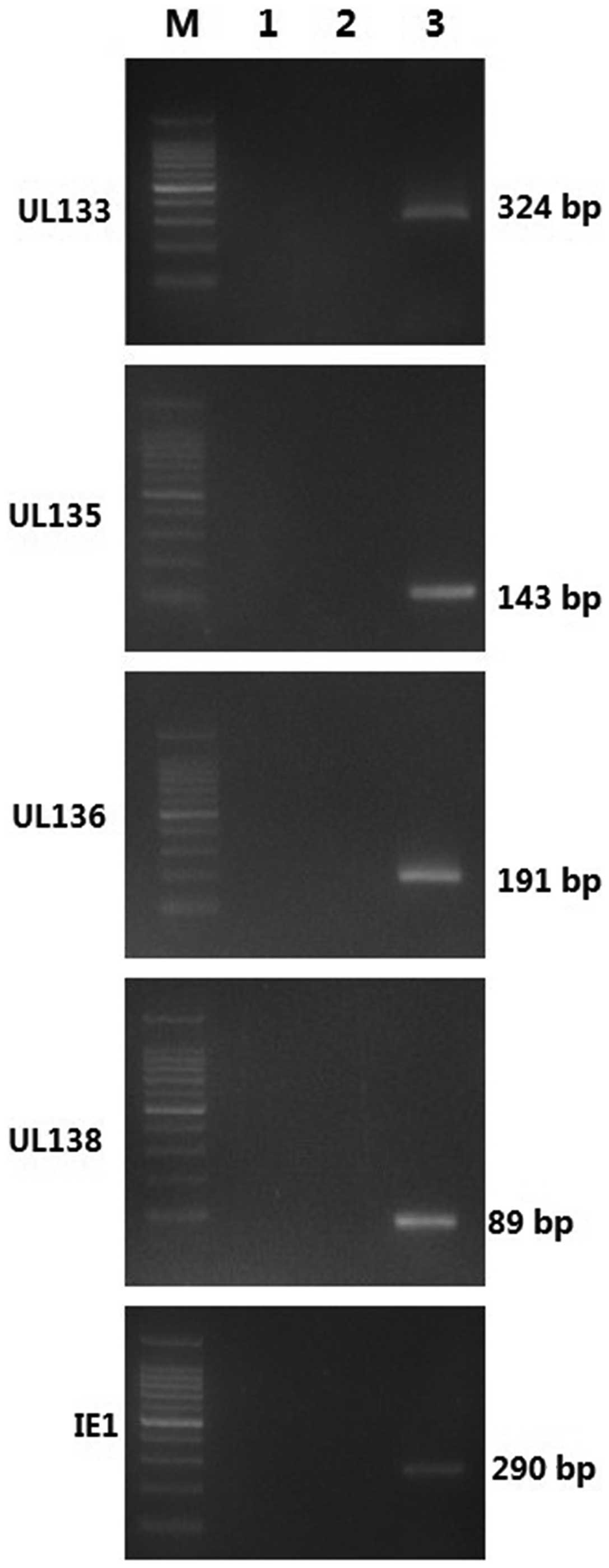
To assess the accuracy and specificity of nested
PCR, a minus-reverse transcription (RT) control was set up, in
which RT templates were replaced by water or RNA. As shown in
Fig. 1, electrophoresis revealed a
single band of UL133–UL138 and IE1 at the appropriate positions
(324 bp for UL133, 143 bp for second stage UL135, 191 bp for second
stage UL136, 89 bp for second stage UL138 and 290 bp for IE1,
respectively). No PCR product was observed in the minus-RT control.
Additional sequencing further confirmed the specificity and
accuracy of the nested PCR assay.
UL133–138 and IE1 expression in gastric
cancer and adjacent normal gastric tissues
The nested PCR method described previously was used
to further investigate the expression of IE1 and individual genes
in the UL133–UL138 locus in paired neoplastic and normal gastric
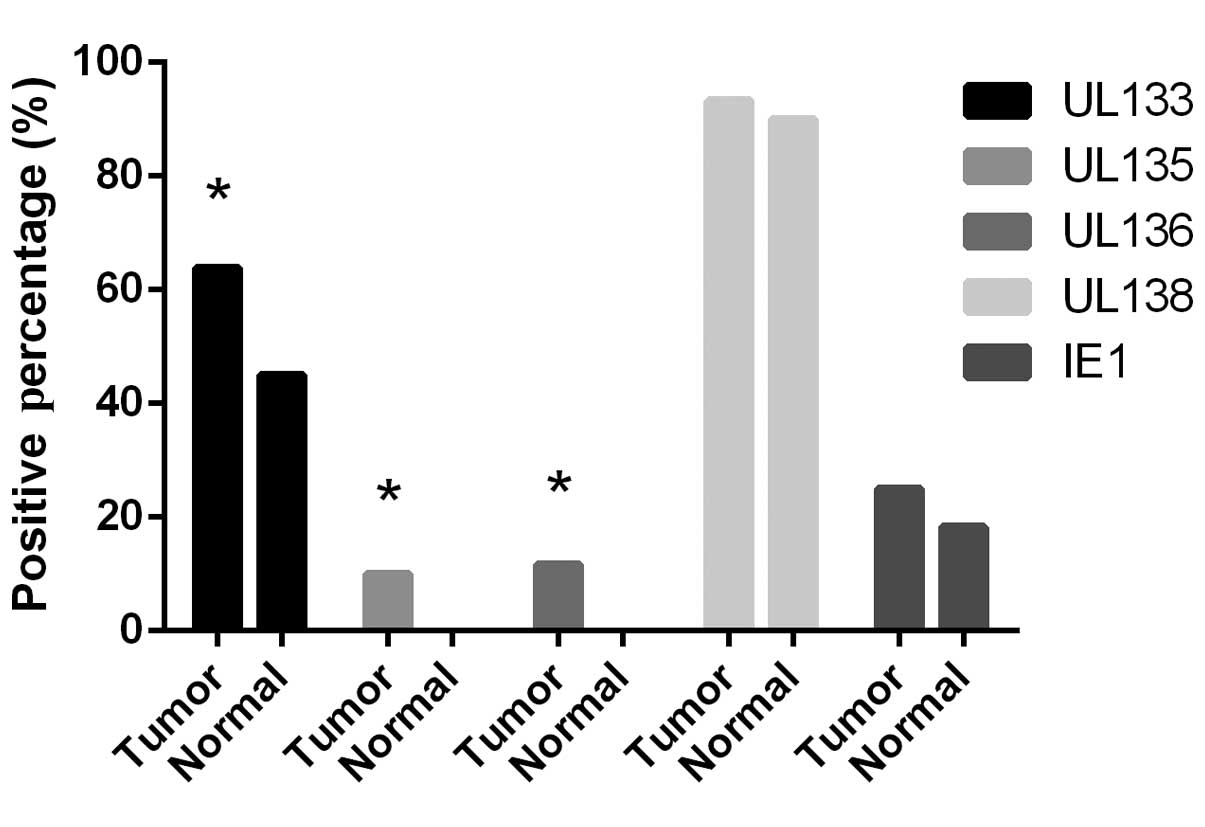
tissues. As shown in Fig. 2, the
detection rate of IE1 was 25.00% (15/60) in the cancer tissues and
18.33% (11/60) in the normal tissues. No significant differences
were identified between the malignant and normal tissues (P=0.375).
The UL133 expression rate was found to be 68.33% (41/60) in the
gastric cancer tissues, which was significantly higher than that in
the corresponding normal tissues (45.00%; 27/60; P=0.01). Notably,
the expression of UL135 and UL136 was only positive in the gastric
cancer samples, in six cases (10%) for UL135 and seven cases
(11.67%) for UL136 (P=0.027 and 0.013, respectively). The highest
level of expression was detected from the UL138 gene in the tumor
and normal tissues. The expression of UL138 in the tumor and
corresponding normal tissues was 93.33% (56/60) and 90.00% (54/60),
respectively, and no significant difference was identified between
the two groups (P=0.509).
Correlation between the UL133–138 locus
and clinical analysis
No significant differences were identified between
the expression of UL133, UL135, UL136, UL138 and IE1 and the
clinicopathological features of gastric cancer, including
pathological differentiation type, tumor-node-metastasis staging,
diameter of tumor, and patient age and gender (Table II). Further analysis of the pattern
of infection was performed, and as a result, seven infection
patterns in the gastric cancer tissues and three patterns in the
corresponding normal tissues were summarized (Table III). The pattern of UL138 alone
accounted for 18.97% (11/58) of all patterns observed in the cancer
tissues, while the same pattern was detected in 52.63% (30/57) of
the normal tissues. Furthermore, a statistically significant
difference was identified in this pattern transcript in different
tissues (P<0.01). The UL133 + UL138 infection pattern occupied
the largest proportion, detected in 60.34% (35/58) of the cancer
tissues, which was similar to the phenomenon observed in the normal
tissues (24/57). However, 69.64% (39/56) of UL138 positive cases
were detected in the UL133 transcripts in the gastric cancer
tissues, while the ratio in normal tissues was 44.44% (24/54)
(P=0.008). The patterns, including UL135 or/and UL136, were found
in 17.24% (10/58) of transcripts and were only detected in cancer
tissues.
 | Table IIClinicopathological features and HCMV
mRNA expression of 60 patients. |
Table II
Clinicopathological features and HCMV
mRNA expression of 60 patients.
| UL133 | UL135 | UL136 | UL138 | IE1 |
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
features | Tumor, n | Normal, n | P-value | Tumor, n | Normal, n | P-value | Tumor, n | Normal, n | P-value | Tumor, n | Normal, n | P-value | Tumor, n | Normal, n | P-value |
|---|
| Gender | | | 0.784 | | | - | | | - | | | 0.947 | | | 0.462 |
| Male | 26 | 18 | | 3 | 0 | | 5 | 0 | | 37 | 36 | | 9 | 5 | |
| Female | 15 | 9 | | 3 | 0 | | 2 | 0 | | 19 | 18 | | 6 | 6 | |
| Age, years | | | 0.869 | | | - | | | - | | | 0.881 | | | 0.428a |
| <60 | 16 | 10 | | 1 | 0 | | 3 | 0 | | 21 | 21 | | 7 | 3 | |
| ≥60 | 25 | 17 | | 5 | 0 | | 4 | 0 | | 35 | 33 | | 8 | 8 | |
| Tumor size, cm | | | 0.884 | | | - | | | - | | | 0.978 | | | 0.951 |
| <5 | 22 | 14 | | 3 | 0 | | 2 | 0 | | 32 | 31 | | 7 | 5 | |
| ≥5 | 19 | 13 | | 3 | 0 | | 5 | 0 | | 24 | 23 | | 8 | 6 | |
|
Differentiation | | | 0.500 | | | - | | | - | | | 0.731 | | | 0.426a |
| Well/moderate | 17 | 9 | | 0 | 0 | | 0 | 0 | | 20 | 21 | | 5 | 6 | |
| Poor/none | 24 | 18 | | 6 | 0 | | 7 | 0 | | 36 | 33 | | 10 | 5 | |
| TNM stage | | | 0.698a | | | - | | | - | | | 0.791 | | | 1.000a |
| I | 6 | 6 | | 0 | 0 | | 0 | 0 | | 11 | 8 | | 0 | 0 | |
| II | 12 | 6 | | 1 | 0 | | 0 | 0 | | 15 | 16 | | 6 | 4 | |
| III | 23 | 15 | | 5 | 0 | | 7 | 0 | | 30 | 30 | | 9 | 7 | |
| IV | 0 | 0 | | 0 | 0 | | 0 | 0 | | 0 | 0 | | 0 | 0 | |
 | Table IIIAnalysis of the different
transcription patterns of the UL133–138 locus and the
clinicopathological features of the normal and tumor tissues. |
Table III
Analysis of the different
transcription patterns of the UL133–138 locus and the
clinicopathological features of the normal and tumor tissues.
| | | Pathological
differentiation, n | TNM stage, n | Diameter of tumor
(cm), n |
|---|
| | |
|
|
|
|---|
| Tissue | Patterns of
infection | Count | P | M-P | M | W | IV | III | II | I | Large (≥5) | Small (<5) |
|---|
| Tumor |
| UL133 | 2 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 2 | 0 |
| UL138 | 11 | 5 | 2 | 4 | 0 | 0 | 3 | 3 | 5 | 5 | 6 |
| UL133 + UL138 | 35 | 15 | 4 | 9 | 7 | 0 | 18 | 11 | 6 | 14 | 21 |
| UL135 + UL138 | 2 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 2 |
| UL136 + UL138 | 4 | 3 | 1 | 0 | 0 | 0 | 3 | 1 | 0 | 2 | 2 |
| UL133 + UL135 +
UL138 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| UL133 + UL135 +
UL136 + UL138 | 3 | 2 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 3 | 0 |
| Normal |
| UL133 | 3 | 2 | 0 | 1 | 0 | 0 | 3 | 0 | 0 | 3 | 0 |
| UL138 | 30 | 8 | 9 | 8 | 5 | 0 | 18 | 10 | 2 | 13 | 17 |
| UL133 + UL138 | 24 | 13 | 3 | 5 | 3 | 0 | 12 | 6 | 6 | 10 | 14 |
Discussion
Increasing evidence indicates that HCMV may be
associated with certain human malignancies, including cancers of
the brain, colon, breast and prostate (15,16,26,27),
and rhabdomyosarcoma (28).
However, no studies have investigated the association between HCMV
and gastric cancer. In addition, similar to other herpes viruses, a
key biological property of HCMV is latent infection, during which,
the viral genome persists in the absence of the production of an
infectious virus and only a subset of viral genes are expressed
(29,30). Few studies have explored the status
of HCMV infection in cancerous tissues.
Previous studies have indicated that the UL138 ORF
is highly conserved in clinical strains and is thus important in
latent HCMV infection (31). The
UL133–UL138 locus has also been found to suppress viral replication
thereby promoting latent infection (20). However, the IE gene is critical for
the detection of viral DNA replication, viral reactivation and the
differentiation of monocytes to macrophages (22,23).
In the present study, it was confirmed that gastric cancer is
associated with latent HCMV infection, and that the genes of IE1
and the UL133–UL138 locus are expressed in the epithelium of
neoplastic and normal tissues. Furthermore, the detection of the
expression of UL133, UL135, UL136 genes, which are associated with
HCMV latency, were found to be significantly different in the
normal and cancer tissues, whereas no significant difference was
identified in the expression of IE1, which correlates with viral
replication, between malignant and normal tissues. The transcripts
of UL135 and UL136 were only detected in the gastric cancer
tissues. Notably, the majority of the positive UL135 and UL136
transcripts were found concentrated in poorly-differentiated
tissues, and in the tissue of positive patients at stage III of
gastric cancer. These results indicate that UL136 and UL135 are
risk factors associated with the development of gastric cancer. In
addition, the results confirmed that latent HCMV infection, but not
replication infection, may have a close association with the
occurrence and development of neoplastic differentiation and the
staging progress. However, the low detection rate of transcription
that was observed limits further explanations (20).
At present, there is no conclusive evidence that the
virus exhibits an oncogenic role, as normal cells do not appear to
be transformed following infection (32,33).
Numerous studies support the theory that HCMV may be oncomodulatory
in the neoplastic process (33–35).
This theory suggests that HCMV infection may induce cellular
responses that provide favorable conditions for the growth of
neoplastic cells. In addition, it has been reported that HCMV
infection may affect cell signaling pathways (36), release inflammatory cytokines
(37), promote immune evasion
(38,39), inhibit cancer cell apoptosis
(33), cause DNA mutations and
deregulate the cell cycle of infected cells (40,41).
Therefore, the combined effects of HCMV genes orchestrated by viral
and cellular mechanisms during HCMV infection may present a
significant mechanism in the neoplastic process.
Previous studies have reported that in the UL133–138
locus, three overlapping transcripts (3.6, 2.7 and 1.4 kb) encode
four putative ORFs, UL133, UL135 and UL136 upstream of UL138, by
canonical and stress-inducible alternative mechanisms of
translation initiation (21). We
hypothesized that UL135 and UL136 would be more detectable than
UL133 transcripts, however, they exhibited a lower detection rate
than UL133. In addition, the expression of UL135 and UL136 was not
detected in the adjacent normal gastric tissues. The pattern of
UL133 + UL138 exhibited the largest proportion in cancer tissues. A
total of 69.64% (35/58) of UL138-positive cases were also detected
in the UL133 transcripts of the gastric cancer tissues, while the
expression in the normal tissues was 44.44% (24/54; P=0.008). In
the corresponding normal tissues, the pattern of UL138 alone
accounted for the majority of the expression patterns detected,
which may indicate low levels or no HCMV latent infection in the
majority of the normal tissues. Previous studies have revealed an
association between the UL133 and UL138 proteins in fibroblasts,
and the results of the present study further confirmed the
interaction between UL133 and UL138 in gastric cancer tissues
(19,20). Furthermore, the analysis of the HCMV
gene transcription patterns may improve our understanding of the
association between HCMV infection and gastric cancer.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81001343), the Zhejiang
Provincial Natural Science Foundation of China (grant nos. Y2100909
and Y2100660) and the Wenzhou Science and Technology Bureau (grant
no. H20100028).
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108.
2005.
|
|
2
|
Eurosurveillance editorial team. WHO
launches the World Health Statistics 2012. Euro Surveill.
17:201752012.
|
|
3
|
Yang L: Incidence and mortality of gastric
cancer in China. World J Gastroenterol. 12:17–20. 2006.
|
|
4
|
Ladeiras-Lopes R, Pereira AK, Nogueira A,
et al: Smoking and gastric cancer: systematic review and
meta-analysis of cohort studies. Cancer Causes Control. 19:689–701.
2008.
|
|
5
|
Møller H, Heseltine E and Vainio H:
Working group report on schistosomes, liver flukes and Helicobacter
pylori. Int J Cancer. 60:587–589. 1995.
|
|
6
|
Bertuccio P, Rosato V, Andreano A, et al:
Dietary patterns and gastric cancer risk: a systematic review and
meta-analysis. Ann Oncol. 24:1450–1458. 2013.
|
|
7
|
Krech U: Complement-fixing antibodies
against cytomegalovirus in different parts of the world. Bull World
Health Organ. 49:103–106. 1973.
|
|
8
|
Dunn W, Chou C, Li H, et al: Functional
profiling of a human cytomegalovirus genome. Proc Natl Acad Sci
USA. 100:14223–14228. 2003.
|
|
9
|
Streblow DN, Orloff SL and Nelson JA: Do
pathogens accelerate atherosclerosis? J Nutr. 131:S2798–S2804.
2001.
|
|
10
|
Vasto S, Colonna-Romano G, Larbi A, Wikby
A, Caruso C and Pawelec G: Role of persistent CMV infection in
configuring T cell immunity in the elderly. Immun Ageing.
4:22007.
|
|
11
|
Rowshani AT, Bemelman FJ, van Leeuwen EM,
van Lier RA and ten Berge IJ: Clinical and immunologic aspects of
cytomegalovirus infection in solid organ transplant recipients.
Transplantation. 79:381–386. 2005.
|
|
12
|
Cheung TW and Teich SA: Cytomegalovirus
infection in patients with HIV infection. Mt Sinai J Med.
66:113–124. 1999.
|
|
13
|
Yi F, Zhao J, Luckheeram RV, et al: The
prevalence and risk factors of cytomegalovirus infection in
inflammatory bowel disease in Wuhan, Central China. Virol J.
10:432013.
|
|
14
|
Jang HJ, Kim AS and Hwang JB:
Cytomegalovirus-associated esophageal ulcer in an immunocompetent
infant: When should ganciclovir be administered? Korean J Pediatr.
55:491–493. 2012.
|
|
15
|
Chen HP, Jiang JK, Chen CY, et al: Human
cytomegalovirus preferentially infects the neoplastic epithelium of
colorectal cancer: a quantitative and histological analysis. J Clin
Virol. 54:240–244. 2012.
|
|
16
|
Samanta M, Harkins L, Klemm K, Britt WJ
and Cobbs CS: High prevalence of human cytomegalovirus in prostatic
intraepithelial neoplasia and prostatic carcinoma. J Urol.
170:998–1002. 2003.
|
|
17
|
Murphy E, Rigoutsos I, Shibuya T and Shenk
TE: Reevaluation of human cytomegalovirus coding potential. Proc
Natl Acad Sci USA. 100:13585–13590. 2003.
|
|
18
|
Grainger L, Cicchini L, Rak M, et al:
Stress-inducible alternative translation initiation of human
cytomegalovirus latency protein pUL138. J Virol. 84:9472–9486.
2010.
|
|
19
|
Petrucelli A, Umashankar M, Zagallo P, Rak
M and Goodrum F: Interactions between proteins encoded within the
human cytomegalovirus UL133–UL138 locus. J Virol. 86:8653–8662.
2012.
|
|
20
|
Umashankar M, Petrucelli A, Cicchini L, et
al: A novel human cytomegalovirus locus modulates cell
type-specific outcomes of infection. PLoS Pathog.
7:e10024442011.
|
|
21
|
Petrucelli A, Rak M, Grainger L and
Goodrum F: Characterization of a novel Golgi apparatus-localized
latency determinant encoded by human cytomegalovirus. J Virol.
83:5615–5629. 2009.
|
|
22
|
Taylor-Wiedeman J, Sissons P and Sinclair
J: Induction of endogenous human cytomegalovirus gene expression
after differentiation of monocytes from healthy carriers. J Virol.
68:1597–1604. 1994.
|
|
23
|
Reeves MB, MacAry PA, Lehner PJ, Sissons
JG and Sinclair JH: Latency, chromatin remodeling, and reactivation
of human cytomegalovirus in the dendritic cells of healthy
carriers. Proc Natl Acad Sci USA. 102:4140–4145. 2005.
|
|
24
|
Flucke U, Monig SP, Baldus SE, et al:
Differences between biopsy- or specimen-related Laurén and World
Health Organization classification in gastric cancer. World J Surg.
26:137–140. 2002.
|
|
25
|
Ajani JA, Bentrem DJ, Besh S, et al;
National Comprehensive Cancer Network. Gastric cancer, version
2.2013: featured updates to the NCCN Guidelines. J Natl Compr Canc
Netw. 11:531–546. 2013.
|
|
26
|
Lau SK, Chen YY, Chen WG, et al: Lack of
association of cytomegalovirus with human brain tumors. Mod Pathol.
18:838–843. 2005.
|
|
27
|
Harkins LE, Matlaf LA, Soroceanu L, et al:
Detection of human cytomegalovirus in normal and neoplastic breast
epithelium. Herpesviridae. 1:82010.
|
|
28
|
Cinatl J Jr, Cinatl J, Radsak K, et al:
Replication of human cytomegalovirus in a rhabdomyosarcoma cell
line depends on the state of differentiation of the cells. Arch
Virol. 138:391–401. 1994.
|
|
29
|
Nachmani D, Stern-Ginossar N, Sarid R and
Mandelboim O: Diverse herpesvirus microRNAs target the
stress-induced immune ligand MICB to escape recognition by natural
killer cells. Cell Host Microbe. 5:376–385. 2009.
|
|
30
|
Bego MG and St Jeor S: Human
cytomegalovirus infection of cells of hematopoietic origin:
HCMV-induced immunosuppression, immune evasion, and latency. Exp
Hematol. 34:555–570. 2006.
|
|
31
|
Qi Y, He R, Ma YP, Sun ZR, Ji YH and Ruan
Q: Human cytomegalovirus UL138 open reading frame is highly
conserved in clinical strains. Chin Med Sci J. 24:107–111.
2009.
|
|
32
|
Cinatl J, Scholz M, Kotchetkov R, Vogel JU
and Doerr HW: Molecular mechanisms of the modulatory effects of
HCMV infection in tumor cell biology. Trends Mol Med. 10:19–23.
2004.
|
|
33
|
Michaelis M, Doerr HW and Cinatl J: The
story of human cytomegalovirus and cancer: increasing evidence and
open questions. Neoplasia. 11:1–9. 2009.
|
|
34
|
Soroceanu L and Cobbs CS: Is HCMV a tumor
promoter? Virus Res. 157:193–203. 2011.
|
|
35
|
Cinatl J Jr, Vogel JU, Kotchetkov R and
Wilhelm Doerr H: Oncomodulatory signals by regulatory proteins
encoded by human cytomegalovirus: a novel role for viral infection
in tumor progression. FEMS Microbiol Rev. 28:59–77. 2004.
|
|
36
|
Chan G, Bivins-Smith ER, Smith MS and
Yurochko AD: NF-kappaB and phosphatidylinositol 3-kinase activity
mediates the HCMV-induced atypical M1/M2 polarization of monocytes.
Virus Res. 144:329–333. 2009.
|
|
37
|
Nachtwey J and Spencer JV: HCMV IL-10
suppresses cytokine expression in monocytes through inhibition of
nuclear factor-kappaB. Viral Immunol. 21:477–482. 2008.
|
|
38
|
Loenen WA, Bruggeman CA and Wiertz EJ:
Immune evasion by human cytomegalovirus: lessons in immunology and
cell biology. Semin Immunol. 13:41–49. 2001.
|
|
39
|
Michelson S: Human cytomegalovirus escape
from immune detection. Intervirology. 42:301–307. 1999.
|
|
40
|
Castillo JP and Kowalik TF: HCMV
infection: modulating the cell cycle and cell death. Int Rev
Immunol. 23:113–139. 2004.
|
|
41
|
Shen Y, Zhu H and Shenk T: Human
cytomagalovirus IE1 and IE2 proteins are mutagenic and mediate
‘hit-and-run’ oncogenic transformation in cooperation with the
adenovirus E1A proteins. Proc Natl Acad Sci USA. 94:3341–3345.
1997.
|