Introduction
Lung cancer is the most prevalent cause of
cancer-related mortality worldwide, and non-small cell lung cancer
(NSCLC) is responsible for ~80% of all lung cancer cases (1). microRNAs (miRNAs or miRs) are a class
of endogenous, non-coding, single-stranded RNAs with a length of
20–25 nucleotides. miRNA genes are transcribed in the nucleus to
primary transcripts (2). Following
digestion by Drosha, precursor miRNA is formed and transported to
the cytoplasm, where it is digested by Dicer to produce mature
miRNA (3,4). miRNAs are believed to regulate other
genes by hybridizing to complementary sequences in the 3′
untranslated region (3′UTR) of target mRNA, resulting in mRNA
degradation or translational inhibition (5).
Previously, numerous studies have demonstrated that
miRNAs play a significant role in the development and progression
of NSCLC. A study by Yanaihara et al (6) reported that patients with lung cancer
with a high expression of miR-155 or low expression of miR-let-7a-2
exhibited a worse prognosis. The miRNA-200 family members have been
shown to affect E-cadherin expression and epithelial-to-mesenchymal
transition, which is an essential early step in tumor metastasis
(7). A study by Ceppi et al
(8) reported that the loss of
miR-200c expression induced an aggressive, invasive and
chemoresistant phenotype in NSCLC. Understanding the specific roles
of miRNAs in NSCLC progression could aid in identifying predictive
markers and devising novel therapeutic strategies for patients.
In the present study, the potential roles of miRNA
in invasion and metastasis of NSCLC were investigated. First, miRNA
microarray analysis was performed to identify the various
expressions of miR-339-5p in different NSCLC cells. Subsequently,
NSCLC cell migration and invasion assays were performed in
vitro. Finally, the tissue samples were used to validate these
results. Quantitative polymerase chain reaction (qPCR) analysis was
performed in three independent experiments, each using two
independent samples. miRNA expression data are presented as fold
difference relative to U6 based on the following equation: RQ =
2−ΔΔCt.
Materials and methods
Cell culture
Paired high-metastatic human pulmonary giant cell
carcinoma cells, 95D, and low-metastatic human pulmonary giant cell
carcinoma cells, 95C, were provided by the laboratory of the
Department of Respiratory Diseases [Chinese People’s Liberation
Army (PLA) General Hospital, Beijing, China] and grown in RPMI-1640
medium (Gibco, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS;
Gibco) at 37°C, in a humidified atmosphere of 95% air and 5%
CO2. This study was approved by the ethics committee of
The 309th Hospital of Chinese People’s Liberation Army (Beijing,
China)
NSCLC tissue specimens
A total of 60 surgical NSCLC tissue specimens and
paired adjacent normal lung tissues (NAT) were obtained from the
Chinese PLA General Hospital and Chinese PLA 309th Hospital
(Beijing, China). Patients who had received any chemotherapy or
radiation therapy prior to surgery or had rheumatic disease, acute
infection, human immunodeficiency virus or other types of cancer
were excluded from the present study. Clinical stage was determined
according to the American Joint Commission on Cancer and Union for
International Cancer Control 2007 tumor-node-metastasis (TNM)
staging criteria (9). RNA was
extracted and qPCR was performed.
Isolation of total RNA
RNA of the NSCLC cells and fresh tissue samples were
extracted using the mirVana RNA isolation kit (Am1560; Ambion,
Austin, TX, USA) according to the manufacturer’s instructions. RNA
quality and quantity was determined by spectrophotometry (ND-1000;
NanoDrop Technologies, Wilmington, DE, USA).
miRNA microarray
The total RNA was phosphorylated and dimethyl
sulfoxide was added to dephosphorylate the RNA. Subsequently, the
mixture was assembled and the reaction was labeled and incubated
for 2 h at 16°C. The sample was then dried with a vacuum
concentrator for 1–2 h at 45–55°C. The hybridization mixture was
then assembled according to the manufacturer’s instructions. It was
subsequently delivered to the chip, which was covered with Agilent
human miRNA array V12.0 (Agilent Technologies, Inc., Santa Clara,
CA, USA), prior to drying for 20 h at 55°C and 20 × g. Finally, the
miRNA microarray was washed and scanned by an Agilent microarray
scanner (Agilent Technologies, Inc.).
qPCR
RNA samples of lung cancer cells or tissue samples
were subjected to reverse transcription reactions using the TaqMan
microRNA reverse transcription kit (4366596; Ambion). Subsequently
the cDNA was amplified by qPCR using the TaqMan Assay (miRNA339-5p,
4427975; and U6, 439547; Ambion) with the TaqMan Universal Master
Mix (4369016; Ambion) in three independent experiments, each using
three independent samples. U6 small nuclear RNA was used as an
internal control. miRNA expression data are presented as the fold
difference relative to U6 based on the following equation: RQ =
2−ΔΔCt, where RQ is the relative quantity.
Transient miRNA transfection
95C and 95D cells (1×106) were seeded and
grown overnight in six-well plates. The following day, the cells
were transfected with either the miR-339-5p mimic, 2′-O-methylated
single-stranded miR-339-5p antisense oligonucleotides (ASO) or the
control oligonucleotides (GenePharma Co., Ltd., Shanghai, China)
using Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad,
CA, USA), according to the manufacturer’s instructions. The miRNA
mimics are small double-stranded RNA oligonucleotides, with the
sequence 5′-UCCCUGUCCUCCAGGAGCUCACGUGAGCUCCUGGAGGACAGG GAUU-3′. The
ASO sequence was 5′-CGUGAGCUCCUGGAGGACAGGGA-3′. The negative
control RNA was used to eliminate the potential
non-sequence-specific effects, and the sequences were
non-homologous to any human genome sequences. Those sequences were
5′-UUCUCCGAACGUGUCACGUTT-3′ (sense), 5′-ACGUGACACGUUCGUAGAATT-3′
(antisense; a negative control for the miRNA mimic) and
5′-CAGUACUUUUGUGUAGUACAA-3′ (a negative control for the mRNA
antisense transfection).
Cell migration and invasion assay
A Transwell insert (24-well insert, pore size 8 μm;
Corning, Inc., Corning, NY, USA) was used to investigate the effect
of miR-339-5p on the migration and invasion of the 95C and 95D
cells in vitro.
For the cell migration assay, 4×104 cells
were resuspended in serum-free RPMI-1640 and placed in the top
portion of the chamber. The lower chamber was filled with 10% FBS
as the chemoattractant and incubated at 37°C in 5% CO2
for 24 h. Subsequently, the cells on the upper surface of the
membrane were removed using cotton buds with phosphate-buffered
saline, and the cells on the lower surface of the insert were fixed
in 75% methanol and stained with 0.1% crystal violet. The images of
five random fields of each insert were captured under a light
microscope at a magnification of ×200 (Nikon Corporation, Tokyo,
Japan). The cells in the images were counted, and the data were
summarized as the means ± SD and presented as a percentage of the
controls. Assays were conducted in duplicate in three independent
experiments.
For the invasion assay, the inserts were previously
covered with 100 μl of the mixture, which contained pre-cooled
serum-free RPMI-1640 and Matrigel (1:10; BD Biosciences, San Diego,
CA, USA), and were allowed to solidify at 37°C in 5% CO2
for 3 h. Following this, 5×104 cells were resuspended in
serum-free RPMI-1640 and placed in the top portion of the chamber
and then the remainder of the invasion assay followed the protocol
for the cell migration assay.
Statistical analysis
All statistical analyses were performed using SPSS
13.0. The paired-samples t-test was used to analyze significant
differences in has-miR-339-5p expression between NSCLC and NAT
tissues. The χ2 test was used to determine the
correlation between has-miR-33p-5p expression and
clinicopathological variables. The two-sided Fisher’s exact test
was used to determine the association between has-miR-339-5p
expression and clinicopathological variables when the number of
tumors analyzed was less than five. The Mann-Whitney U test was
used for clinical-stage ranked data analysis. Spearman’s
correlation analysis was used to determine the correlation between
miR-339-5p expression and clinical stage and lymph node metastasis
status. Other results were analyzed using the independent samples
t-test. Results were considered to indicate a statistically
significant difference at values of P<0.05.
Prediction of miR339-5p target genes
Three miRNA databases (http://www.microrna.org/microrna/home.do; http://pictar.mdc-berlin.de; and http://www.targetscan.org) were searched for
prediction of miR-339-5p target genes.
Results
Differential miRNAs between 95C and 95D
cells by microarray analysis
Agilent human miRNA array V12.0, which contained 855
probes based on Sanger miRBase release 13.0, was used to scan
miRNAs that were differentially expressed between 95C and 95D
cells. The cells were paired pulmonary giant cells with low or high
metastatic capacities, respectively. In total, 44 miRNAs exhibited
significantly differential expression, among which miR-339-5p was
focused on as it is one of the most evidently altered miRNAS and
has previously been reported to be associated with the metastasis
of breast cancer (10) (Table I).
 | Table IFold changes of the miRNA expression
between 95C and 95D cells. |
Table I
Fold changes of the miRNA expression
between 95C and 95D cells.
| Systematic name | Fold-change (95C vs.
95D) | Regulation (95C vs.
95D) |
|---|
| hsa-miR-146b-3p | 5.7694 | Up |
| hsa-miR-513a-3p | 6.2644 | Up |
| hsa-miR-155 | 2.1363 | Up |
| hsa-miR-338-5p | 2.0583 | Up |
| hsa-miR-588 | 8.1228 | Up |
| hsa-miR-924 | 4.3476 | Down |
| hsa-miR-494 | 2.1707 | Down |
| hsa-miR-339-5p | 8.0135 | Up |
|
hsa-miR-150* | 5.3655 | Down |
|
hsa-miR-1226* | 2.3421 | Down |
|
hsa-miR-493* | 3.7905 | Up |
| hsa-let-7g | 2.2086 | Up |
|
hsa-miR-30e* | 2.3531 | Up |
|
hsa-miR-106b* | 2.3968 | Down |
| hsa-miR-1299 | 5.1606 | Up |
| hsa-miR-1915 | 2.1175 | Down |
|
hsa-miR-30a* | 2.2098 | Up |
| hsa-let-7b | 2.0122 | Up |
| hsa-miR-28-5p | 2.5076 | Up |
| hsa-miR-1287 | 8.3396 | Down |
| hsa-miR-26b | 2.0724 | Up |
| hsa-miR-1290 | 3.2220 | Up |
| hsa-miR-892a | 6.3257 | Up |
| hsa-miR-7 | 2.2993 | Up |
| hsa-miR-1826 | 10.5247 | Down |
| hsa-let-7f | 2.0995 | Up |
| hsa-miR-198 | 16.6237 | Down |
| hsa-miR-658 | 23.5899 | Down |
| hsa-miR-1246 | 2.9612 | Up |
| hsa-miR-299-5p | 2.0088 | Up |
|
hsa-miR-29b-1* | 2.1965 | Up |
| hsa-miR-760 | 7.0691 | Up |
| hsa-miR-923 | 2.2522 | Down |
| hsa-miR-630 | 4.5866 | Down |
| hsa-miR-501-5p | 2.0365 | Up |
| hsa-miR-324-5p | 2.0318 | Down |
| hsa-miR-196a | 2.2921 | Up |
| hsa-miR-196b | 2.4324 | Up |
| hsa-miR-513b | 3.3342 | Down |
|
hsa-miR-129* | 2.3897 | Up |
|
hsa-miR-92a-2* | 20.1876 | Down |
| hsa-miR-625 | 2.2465 | Up |
| hsa-miR-624 | 4.8870 | Up |
| hsa-miR-1268 | 6.8535 | Up |
qPCR verification of miR-339-5p
expression between 95C and 95D cells
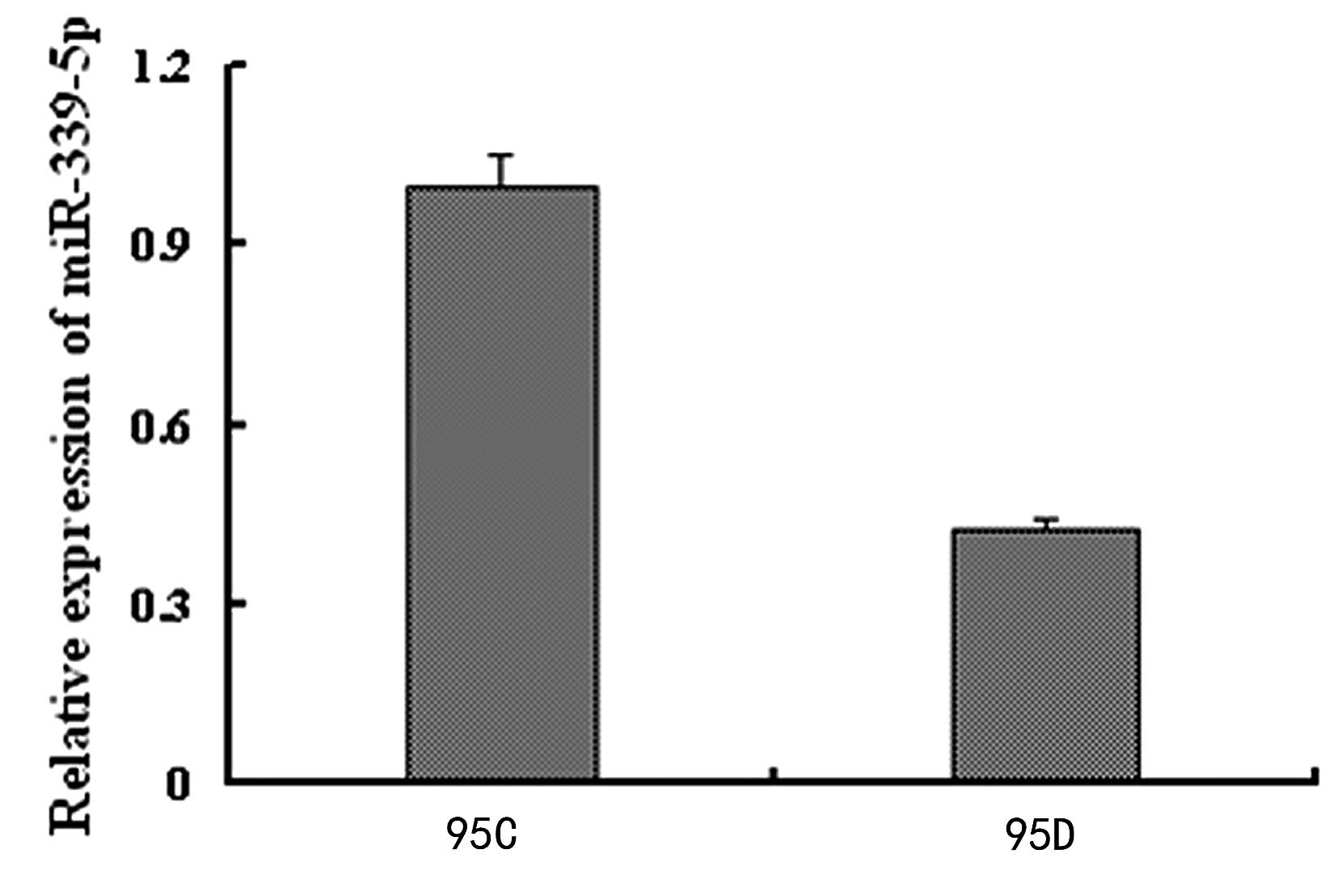
qPCR was further used to validate that the
miR-339-5p expression of the 95C cells was significantly higher, by
3.4662 fold, compared with that of 95D cells (Fig 1).
Effects of miR-339-5p on cell migration
and invasion
qPCR was used to confirm that transfection was
successful. Expression in transfected cells was normalized to that
of untreated cells, and U6 expression was used as an internal
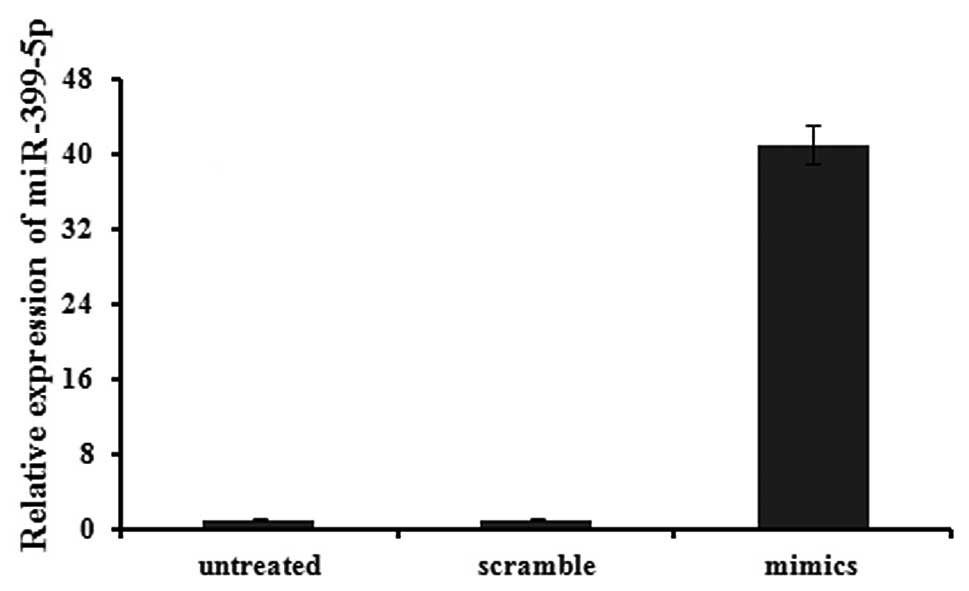
standard. The expression of has-miR-339-5p was clearly increased,
by 41.07 fold, by transfection of miR-339-5p mimics into 95D cells
after 24 h (Fig. 2).
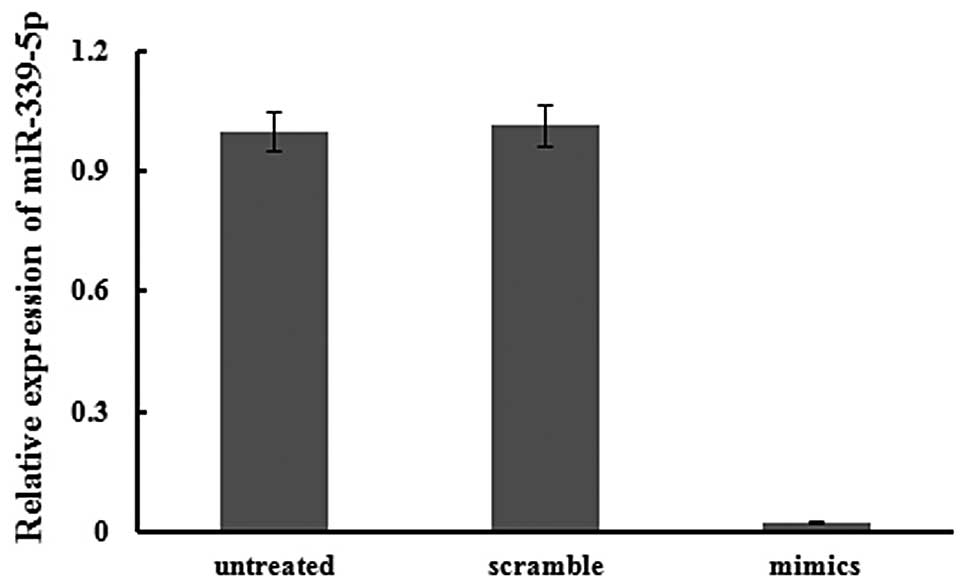
Simultaneously, the expression of has-miR-339-5p was markedly
decreased, by 38.73 fold, by transfection of the ASO of miR-339-5p
into 95C cells after 24 h (Fig.
3).
Effects of miR-339-5p on NSCLC cell
migration and invasion
A loss-of-function approach was adopted for the
analysis of the effects of miR-339-5p on NSCLC cell migration and
invasion. The expression of has-miR-339-5p was decreased by
transfection of 2′-O-methylated single-stranded miR-339-5p ASO into
95C cells, which was validated by qPCR. There were no variations
between untreated cells and cells transfected with the scramble
oligonucleotide (P=0.814). However, the number of migrating cells
that were transfected with the ASO was significantly increased
(P<0.01) (Fig 4A). For the
invasion analysis, there was no difference between untreated cells
and cells transfected with the scramble oligonucleotide. However,
the number of invading cells that were transfected with the ASO was
significantly increased (P<0.01) (Fig 4B).
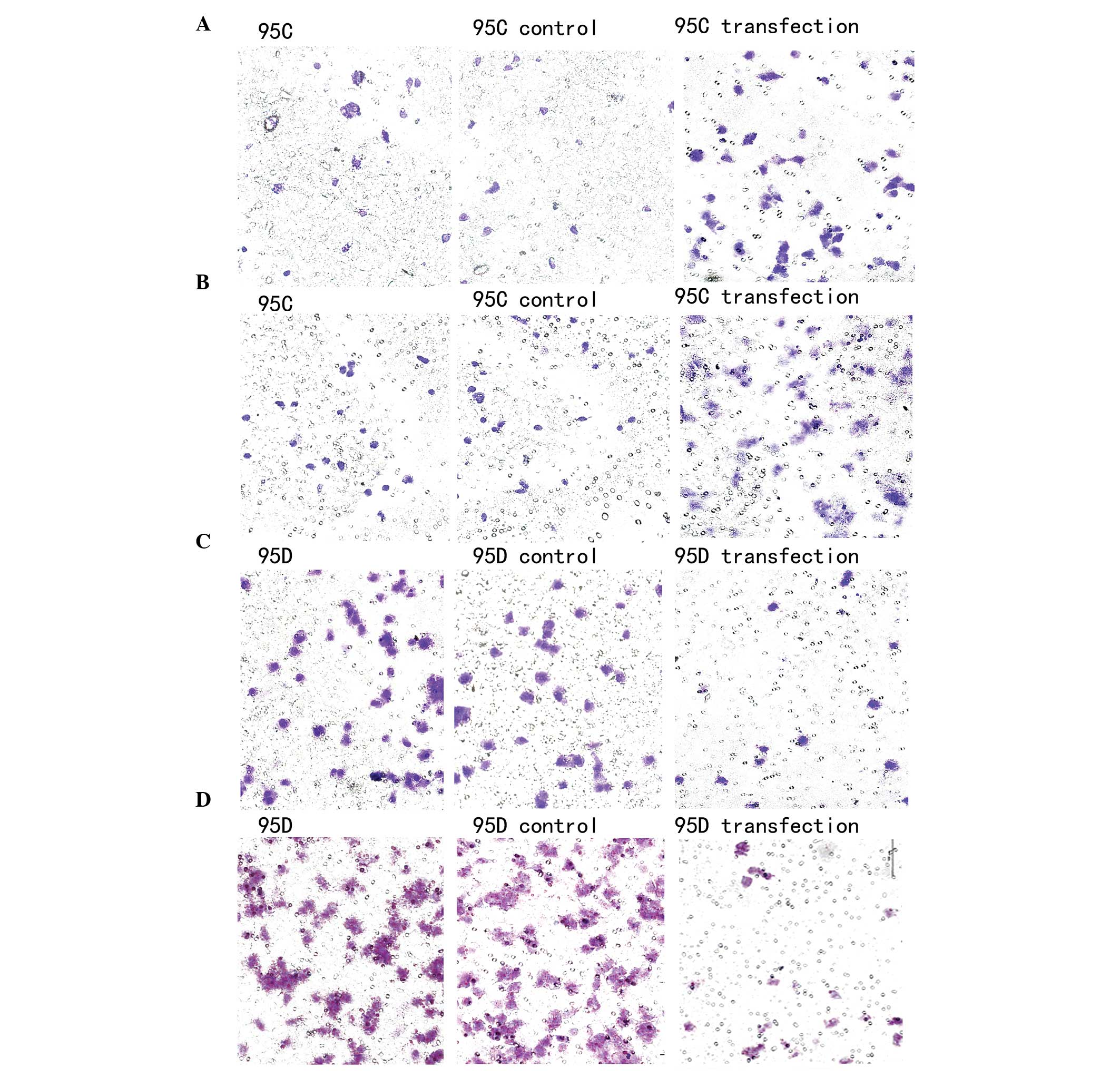 | Figure 4Migration and invasion assays of the
95C and 95D cells. (A) In the migration assay, there were no
differences between untreated 95C cells and the control cells which
were transfected with the scramble oligonucleotide (35±3 and 37±4
cells, P=0.814). However, the number of migrating cells that were
transfected with the antisense oligonucleotides (ASO) was
significantly increased (56±5 cells, P<0.01). (B) In the
invasion assay, there were no differences between untreated 95C
cells and the control cells which were transfected with the
scramble oligonucleotide (25±2 and 22±4 cells, P=0.768). However,
the number of invading cells that were transfected with the ASO was
significantly increased (50±3 cells, P<0.01). (C) In the
migration assay, there were no differences between untreated 95D
cells and the control cells which were transfected with the
scramble oligonucleotide (55±3 and 51±4 cells, P=0.814). However,
the number of migrating cells that were transfected with the mimics
was significantly decreased (10±5 cells, P<0.01). (D) In the
invasion assay, there were no differences between untreated 95D
cells and the control cells which were transfected with the
scramble oligonucleotide (70±2 and 67±4 cells, P=0.768). However,
the number of invading cells that were transfected with the mimics
was significantly decreased (12±3 cells, P<0.01). |
A gain-of-function approach was then adopted. The
expression of has-miR-339-5p was increased by transfection of the
miR-339-5p mimics into 95D cells and validated by qPCR. There were
no variations between untreated cells and cells transfected with
the scramble oligonucleotide (P=0.814). However, the number of
migrating cells that were transfected with the mimics was
significantly increased (P<0.01) (Fig 4C). For the invasion analysis, there
was no variation between untreated cells and cells transfected with
the scramble oligonucleotide (P=0.768). However, the number of
invading cells that were transfected with the mimics was
significantly increased (P<0.01) (Fig 4D).
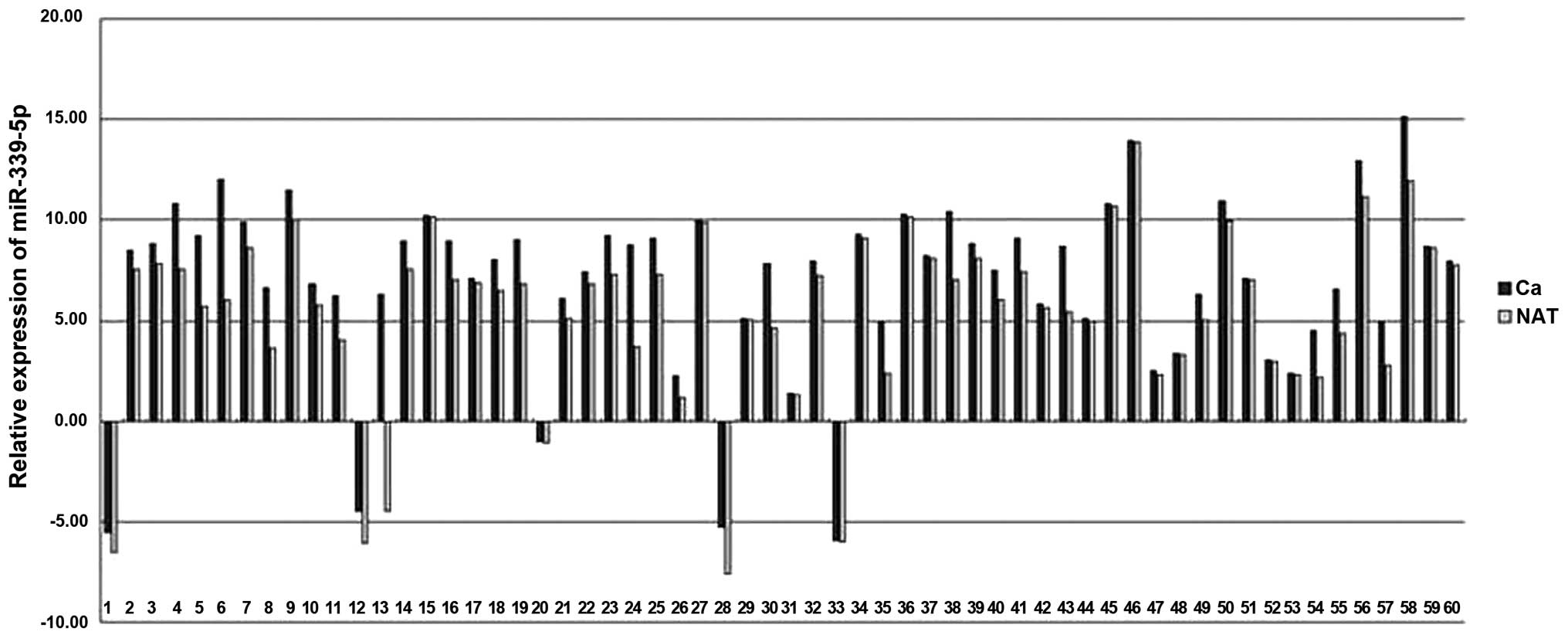
Decreased expression of miR-339-5p in
NSCLC cancer tissues
In the patients with NSCLC, miR-339-5p expression
was decreased in cancer tissues in comparison with matched LACs
(Fig 5). The mean expression levels
of miR-339-5p in NSCLCs was decreased by ~1.9 fold compared with
NATs (minimum, 33.78; and maximum, 1.03).
Association of has-miR-339-5p relative
quantitative expression (cancer tissue expression/normal tissue
expression) in NSCLCs and NATs with clinicopathological features of
NSCLCs
To determine the effects of has-miR-339-5p
expression on tumor progression and metastasis, the patients with
lung cancer were divided into two groups based on the mean level of
the ratio of miR-339-5p relative expression (carcinoma/NAT) in 60
NSCLCs (mean =0.5249). The two groups were defined as high-relative
and low-relative expression (≥0.5249 or <0.5249). The
associations between miR-339-5p relative expression and
clinicopathological variables for lung cancer are shown in Table II. Associations between miR-339-5p
expression and lymph node metastasis were observed to be
statistically significant (P<0.001, two-sided Fisher’s exact
test). Changes in expression of miR-339-5p were also statistically
significantly associated with clinical stages (P<0.001,
Mann-Whitney test). No correlation was observed between miR-339-5p
expression and gender, age and pathological type (data not
shown).
 | Table IIAssociation between miR-339-5p
relative expression and clinicopathological variables in lung
cancer tissues. |
Table II
Association between miR-339-5p
relative expression and clinicopathological variables in lung
cancer tissues.
| | miR-339-5p
expression (Ca/N) |
|---|
| |
|
|---|
| n | Low expression | High
expression | P-value |
|---|
| Lymph node
metastasis |
| Yes | 23 | 21 | 2 | |
| No | 37 | 16 | 21 | <0.001a |
| Gender |
| Male | 41 | 26 | 15 | |
| Female | 19 | 11 | 8 | 0.682b |
| Age |
| ≤60 | 41 | 25 | 16 | |
| >60 | 19 | 12 | 7 | 0.872b |
| Pathological
type |
|
Adenocarcinoma | 39 | 24 | 15 | 0.978b |
| Squamous cell
carcinoma | 21 | 13 | 8 | |
| Clinical stage |
| I | 25 | 6 | 19 | <0.001c |
| II | 11 | 9 | 2 | |
| III | 20 | 18 | 2 | |
| IV | 4 | 4 | 0 | |
In order to improve the characterization of the
association between miR-339-5p expression, TNM stage and lymph node
metastasis, the data was further analyzed using Spearman’s
correlation test. The results showed a negative correlation between
has-miR-339-5p relative quantitative expression and TNM stage
(r=−0.927, P<0.001) and lymph node metastasis (r=−0.828,
P<0.001).
Bioinformatic analyses
Bioinformatic analyses found that B-cell lymphoma 6
protein (BCL6) and valosin-containing protein (VCP) gene may be
potential miR-339-5p targets.
Discussion
Metastasis is a common event in cancer pathology and
represents the primary cause of cancer-related mortality. The steps
required for metastasis involve significant changes in gene
expression. A number of studies investigating miRNA regulation of
the aforementioned steps have identified several miRNAs that may
promote or inhibit the metastatic potential. It has been reported
that miRNA-10b, miR-21, miR-373, miR-378 and miR-17-92 can promote
breast cancer metastasis (11–15),
while miR-335, miR-206 and let-7 family can inhibit the metastasis
of breast cancer (16,17). Although there have been numerous
studies on the miRNAs associated with lung cancer metastasis
(6,8), but the molecular mechanism is not
clear.
miRNA microarray analysis is a high-throughput rapid
analysis of the miRNA expression profiling method. 95C and 95D
cells were the sublines of human lung giant cell carcinoma maternal
cells (PLA-801) that were isolated by the Department of Pathology,
of the General Hospital of Chinese PLA Hospital (Beijing, China).
The paired cells have the same genetic background and varied
metastatic capacity. In order to study the miRNAs associated with
the NSCLC metastasis, the miRNA microarray was first applied to
find 44 miRNAs whose expression were different in the 95C and 95D
cells. Compared with 95C cells, 29 miRNA expression levels of 95D
increased and 15 miRNAs were downregulated, which included
miRNA-339-5p. The expression of miRNA-339-5p in 95C cells was
eight-fold higher compared with that in the 95D cells.
Subsequently, qPCR detection of the miRNA-339-5p expression levels
of 95C and 95D cells was performed, and the results of which were
consistent with the microarray results. Transfection, migration and
invasion assays were then used to confirm that miRNA-339-5p could
inhibit the NSCLC cell migration and invasion ability.
Finally, qPCR confirmed that the expression of
has-miR-339-5p was decreased significantly in the majority of
NSCLCs. A negative correlation was evaluated between has-miR-339-5p
relative quantitative expression and TNM stage and lymph node
metastasis. These data indicate that has-miR-339-5p could inhibit
NSCLC metastasis.
miRNAs regulate gene expression by binding to
sequences in the 3′UTR of an expressed mRNA, resulting in either
modulation of translation efficiency or degradation of the mRNA. In
order to understand the mechanism of miRNA inhibiting metastasis,
the interaction between miRNA and target genes must be known.
miRecords is one of the most commonly used mRNA target gene
predicting sites (18). The site
integrates information from numerous target gene predictable sites.
The genes that were predicted by the majority of sites from
miRecords were chosen as the target genes in the present study. The
sites must include TargetScan, PicTar and miRanda, which are the
three most commonly used bioinformatic programmes to predict miRNA
target genes (19–21). According to the predicted results
from the webserver databases, BCL6 and VCP were the most likely
target genes predicted. BCL6 is a proto-oncogene located on
chromosome 3q27 that encodes a transcriptional repressor that was
originally characterized as a regulator of B-lymphocyte development
and growth and has been indicated in the pathogenesis of B-cell
lymphoma (22,23). Numerous studies have revealed that
BCL6 is associated with cancer metastasis. A study by Pinto et
al (24) found that BCL6 could
significantly increase the expression of three metastasis-related
genes [chemokine (CXC motif) receptor 4, fms-related tyrosine
kinase 1 and integrin β3] in breast cancer cell lines. A study by
Wu et al (10) revealed that
miR-339-5p could inhibit the expression of BCL-6 mRNA, which is
associated with suppression of the migration and invasion of breast
cancer cells. Further studies are required to find the association
between BCL6 and lung cancer metastasis. VCP (also known as p97) is
a member of the AAA ATPase family. VCP has a pivotal role in the
ubiquitin-degradation of misfolded proteins and also exhibits an
anti-apoptotic function and metastasis via the activation of the
nuclear factor-κB (NF-κB) signaling pathway. Yamamoto et al
(25) found that VCP
(p97)expression is associated with the progression and prognosis of
patients with NSCLC. VCP is also active in the
ubiquitin/proteasome-degradation pathway, which is involved in
proliferation and anti-apoptosis in human cancer cells. Studies
have shown that the expression levels of VCP correlate with the
prognosis and recurrence of specific human cancers, including
hepatocellular, gastric and colorectal carcinomas. In these
studies, VCP expression was found to correlate with the prognosis
of differentiated thyroid carcinoma (26–28).
It is speculated that miR-339-5p inhibits the VCP expression to
inhibit the metastasis of lung cancer, but the exact mechanism is
unclear.
In conclusion, the results of the present study
identified that miR-339-5p was significantly downregulated in the
primary tissues of patients with NSCLC compared with adjacent
normal tissues. The strong correlation between miR-339-5p
expression and the clinical stage indicates that miR-339-5p may be
a novel biomarker involved in lung cancer metastasis, but further
studies are required to reveal the exact mechanism.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012.
|
|
2
|
Lee Y, Ahn C, Han J, et al: The nuclear
RNase III Drosha initiates microRNA processing. Nature.
425:415–419. 2003.
|
|
3
|
Lund E, Güttinger S, Calado A, et al:
Nuclear export of microRNA precursors. Science. 303:95–98.
2004.
|
|
4
|
Lee Y, Jeon K, Lee JT, et al: MicroRNA
maturation: stepwise processing and subcellular localization. EMBO
J. 21:4663–4670. 2002.
|
|
5
|
Rana TM: Illuminating the silence:
understanding the structure and function of small RNAs. Nat Rev Mol
Cell Biol. 8:23–36. 2007.
|
|
6
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006.
|
|
7
|
Gibbons DL, Lin W, Creighton CJ, et al:
Contextual extracellular cues promote tumor cell EMT and metastasis
by regulating miR-200 family expression. Genes Dev. 23:2140–2151.
2009.
|
|
8
|
Ceppi P, Mudduluru G, Kumarswamy R, et al:
Loss of miR-200c expression induces an aggressive, invasive, and
chemoresistant phenotype in non-small cell lung cancer. Mol Cancer
Res. 8:1207–1216. 2010.
|
|
9
|
Sobin LH, Gospodarowicz MK and Wittekind
CH; International Union Against Cancer. TNM Classification of
Malignant Tumours. 7th edition. WileyBlackwell; New York, NY:
2010
|
|
10
|
Wu ZS, Wu Q, Wang CQ, et al: MiR-339-5p
inhibits breast cancer cell migration and invasion in vitro and may
be a potential biomarker for breast cancer prognosis. BMC Cancer.
10:5422010.
|
|
11
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007.
|
|
12
|
Frankel LB, Christoffersen NR, Jacobsen A,
et al: Programmed cell death 4 (PDCD4) is an important functional
target of the microRNA miR-21 in breast cancer cells. J Biol Chem.
283:1026–1033. 2008.
|
|
13
|
Edmonds MD, Hurst DR, Vaidya KS, et al:
Breast cancer metastasis suppressor 1 coordinately regulates
metastasis-associated microRNA expression. Int J Cancer.
125:1778–1785. 2009.
|
|
14
|
Dews M, Homayouni A, Yu D, et al:
Augmentation of tumor angiogenesis by a Myc-activated microRNA
cluster. Nat Genet. 38:1060–1065. 2006.
|
|
15
|
Foekens JA, Sieuwerts AM, Smid M, et al:
Four miRNAs associated with aggressiveness of lymph node-negative,
estrogen receptor-positive human breast cancer. Proc Natl Acad Sci
USA. 105:13021–13026. 2008.
|
|
16
|
Song G, Zhang Y and Wang L: MicroRNA-206
targets notch3, activates apoptosis, and inhibits tumor cell
migration and focus formation. J Biol Chem. 284:31921–31927.
2009.
|
|
17
|
Yu F, Yao H, Zhu P, et al: let-7 regulates
self renewal and tumorigenicity of breast cancer cells. Cell.
131:1109–1123. 2007.
|
|
18
|
Xiao F, Zuo Z, Cai G, et al: miRecords: an
integrated resource for microRNA-target interactions. Nucleic Acids
Res. 37(Database issue): D105–D110. 2009.
|
|
19
|
Krek A, Grün D, Poy MN, et al:
Combinatorial microRNA target predictions. Nat Genet. 37:495–500.
2005.
|
|
20
|
John B, Enright AJ, Aravin A, et al: Human
microRNA targets. PLoS Biol. 2:e3632004.
|
|
21
|
Lewis BP, Shih IH, Jones-Rhoades MW, et
al: Prediction of mammalian microRNA targets. Cell. 115:787–798.
2003.
|
|
22
|
Phan RT, Saito M, Basso K, et al: BCL6
interacts with the transcription factor Miz-1 to suppress the
cyclin-dependent kinase inhibitor p21 and cell cycle arrest in
germinal center B cells. Nat Immunol. 6:1054–1060. 2005.
|
|
23
|
Polo JM, Dell’Oso T, Ranuncolo SM, et al:
Specific peptide interference reveals BCL6 transcriptional and
oncogenic mechanisms in B-cell lymphoma cells. Nat Med.
10:1329–1335. 2004.
|
|
24
|
Pinto AE, André S, Silva G, et al: BCL-6
oncoprotein in breast cancer: loss of expression in disease
progression. Pathobiology. 76:235–242. 2009.
|
|
25
|
Yamamoto S, Tomita Y, Hoshida Y, et al:
Expression level of valosin-containing protein (p97) is correlated
with progression and prognosis of non-small-cell lung carcinoma.
Ann Surg Oncol. 11:697–704. 2004.
|
|
26
|
Yi P, Higa A, Taouji S, et al:
Sorafenib-mediated targeting of the AAA+ ATPase p97/VCP
leads to disruption of the secretory pathway, endoplasmic reticulum
stress, and hepatocellular cancer cell death. Mol Cancer Ther.
11:2610–2620. 2012.
|
|
27
|
Yamamoto S, Tomita Y, Hoshida Y, et al:
Expression level of valosin-containing protein is strongly
associated with progression and prognosis of gastriccarcinoma. J
Clin Oncol. 21:2537–2544
|
|
28
|
Yamamoto S, Tomita Y, Hoshida Y, et al:
Expression of valosin-containing protein in colorectal carcinomas
as a predictor for disease recurrence and prognosis. Clin Cancer
Res. 10:651–657. 2004.
|