Introduction
Hepatocellular carcinoma is a common malignancy and
is the third leading cause of cancer-related mortality worldwide.
Although great breakthroughs have been achieved in the diagnosis
and treatment of hepatocellular carcinoma, recurrence and
metastasis of the disease continue to affect the prognosis of
patients. Therefore, in order to improve the prognosis of
hepatocellular carcinoma patients, there is an urgent requirement
for the development of new therapeutic targets (1).
Human Kin17 is a 45-kDa nuclear protein that is
remarkably conserved during evolution (2–4). The
Kin17 protein is ubiquitously expressed in mammals and is
correlated with vital biological functions (5). There exists a region of 40 residues in
the core of Kin17, which is homologous to RecA protein. Kin17
protein possesses three motifs: A zinc finger, a bipartite nuclear
localization signal and the core domain that is homologous to RecA
protein. To date, the following major roles of Kin17 have been
identified: i) binding to curved DNA and recombination of human
cells (6,7); ii) complementing the functions of H-NS
and controlling gene expression (8); and iii) upregulation by UVC or
ionizing radiation (9–11). Thereby, Kin17 is associated with
mRNA processing, involving gene transcription and cell cycle
regulation. Taking into account the uncontrolled growth and
unlimited replication potential of hepatocellular carcinoma cells,
Kin17 may therefore be involved in the pathogenesis and progression
of hepatocellular carcinoma.
In the present study, the expression of Kin17 was
investigated in hepatocellular carcinoma tissues and hepatocellular
carcinoma cell lines. A series of experimental methods in
vitro and in vivo were used to explore the possible role
of Kin17 in the carcinogenesis of hepatocellular carcinoma. To the
best our knowledge, this is the first study to evaluate the effects
of Kin17 on the growth of hepatocellular carcinoma.
Materials and methods
Cell lines and tissues
Hepatocellular carcinoma cell lines, HepG2,
SMMC-7721 and BEL-7402 (American Type Culture Collection, Manassa,
VA, USA), were maintained in RPMI-1640 (Gibco-BRL, Carlsbad, CA,
USA) medium containing 10% fetal bovine serum (Hangzhou Sijiqing
Biological Engineering Materials Co., Ltd., Hangzhou, China), 100
μg/ml penicillin and 100 μg/ml streptomycin (both Invitrogen Life
Technologies, Carlsbad, CA, USA). Hepatocellular carcinoma cells
were transfected with the full-length human Kin17 cDNA or a control
pcDNA3.0 vector (both Yinru Biological Engineering., Ltd.,
Changsha, China) using Lipofectamine 2000 (Invitrogen Life
Technologies). The colonies were selected with G418 (550 μg/ml) for
3 weeks and expanded. HepG2 and SMMC-7721 cells with overexpression
of Kin17 were classified as the HepG2-Kin17 and SMMC-7721-Kin17
groups, respectively. Similarly, the equivalent
pcDNA3.0-transfected cells were termed as the HepG2-Vector and
SMMC-7721-Vector groups, respectively. Eight fresh, paired
hepatocellular carcinoma and noncancerous tissue samples were used
for the western blot analysis, along with the hepatocellular
carcinoma cell lines. The specimens were obtained from patients at
the Department of Oncology, First Affiliated Hospital of Xinxiang
Medical University (Weihui, China). Informed consent was provided
by each patient, and the study was approved by the ethics committee
of the First Affiliated Hospital of Xinxiang Medical
University.
Western blot analysis
After dicing the paired cancerous and normal
samples, these tissues were extracted in cell lysis buffer
(radioimmunoprecipitation assay; Thermo Fisher Scientific, Waltham,
MA, USA) with protease inhibitors. A total of 1×106
HepG2, SMMC-7721 and BEL-7402 cells were washed twice with ice-cold
phosphate-buffered saline (Fuzhou Maixin Biotechnology Development
Co., Ltd., Fuzhou, China) and lysed with cell lysis buffer at 4°C
for 30 min. The lysates were acquired by centrifugation at 20,000 ×
g for 15 min at 4°C. Equal amounts of proteins from HepG2,
SMMC-7721 and BEL-7402 cells or paired cancerous and normal tissue
samples were boiled for 8 min prior to being loaded onto 10%
polyacrylamide gels (Kangwei Century Co., Ltd., Beijing, China) and
transferred to polyvinylidene fluoride membranes (Millipore,
Billerica, MA, USA). Following incubation with the monoclonal mouse
anti-human Kin17 antibody (1:150 dilution; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA), the horseradish
peroxidase-conjugated secondary polyclonal goat anti-mouse IgG
(1:4500 dilution; Beijing Zhongshan Golden Bridge Biotechnology
Co., Beijing, China) was incubated for 2 h at room temperature.
Finally, Kin17 was visualized by enhanced chemiluminescence
(Electro-Chemi-Luminescence system; Beijing Shengke Ruida
Technology Development Co., Ltd., Beijing, China). The membranes
were then re-blotted with anti-GAPDH antibody for normalization and
confirmation of equal protein loading. Due to the vital roles of
cyclin D1 and p27Kip1 in regulating cell proliferation, the effect
of Kin17 on their expression was detected. CyclinD1 (polyclonal
mouse anti-human cyclinD1 antibody, 1:100 dilution, Santa Cruz
Biotechnology, Inc.) and p27Kip1 (monoclonal mouse anti-human
p27Kip1 antibody, 1:200 dilution, Santa Cruz Biotechnology, Inc.)
expressions were studied according to the method described
previously. Bandscan 5.0 software (Glyko, Novato, CA, USA) was used
to analyze the results of the western blot analysis. Compared with
the BEL-7402 cells, the endogenous expression of Kin17 was lower in
the HepG2 and SMMC-7721 cells. Therefore, the HepG2 and SMMC-7721
cells were selected for studying.
MTT and colony formation assay
For the MTT assay, HepG2-Vector and
SMMC-7721-Vector, as well as HepG2-Kin17 and SMCC-7721-Kin17
hepatocellular carcinoma cells were plated into 96-well plates at a
density of 1×104 cells/well. The following steps were
performed as described previously (12). Briefly, 120 μl dimethylsulfoxide
(Kangwei Century Co., Ltd.) was added to each well and agitated for
15 min (H97-A, Solarbio Science & Technology Co., Ltd, Beijing,
China). The absorbance was measured at a wavelength of 492nm. For
the colony formation assay, the concentration of the cells was
adjusted to 180 cells/dish and the cells were incubated at 37°C.
The culture media was changed every 2 days. After two weeks, cells
were fixed with methanol and stained with Trypan blue (Solarbio
Science & Technology Co., Ltd.). The average number of colonies
in five visual fields was counted under a microscope (YS100, Nikon,
Tokyo, Japan).
In vivo tumor growth assay
For the xenograft experiment, four- to six-week-old
female nude mice (BALB/cA nu/nu) were obtained from the Animal
Center of Xinxiang Medical University (Xinxiang, China).
HepG2-Kin17 (n=4) and SMMC-7721-Kin17 cells were implanted
subcutaneously into the back of the nude mice (BALB/cA nu/nu).
Simultaneously, HepG2-Vector (n=4) and SMMC-7721-Vector cells were
implanted under the same experimental conditions. Each group had
four nude mice.
The maximum (a) and minimum (b) diameters of the
tumors were measured, and tumor volume was calculated according to
the following formula: Tumor volume = a × b2/2. Five
weeks following tumor implantation, the nude mice were euthanized
and the tumors were excised.
Statistical analysis
Statistical analysis was performed using SPSS 13.0
(SPSS, Inc., Chicago, IL, USA). Comparisons between two groups were
conducted by Student’s t-test. Results are expressed as the mean ±
standard deviation. P<0.05 was considered to indicate a
statistically significant difference.
Results
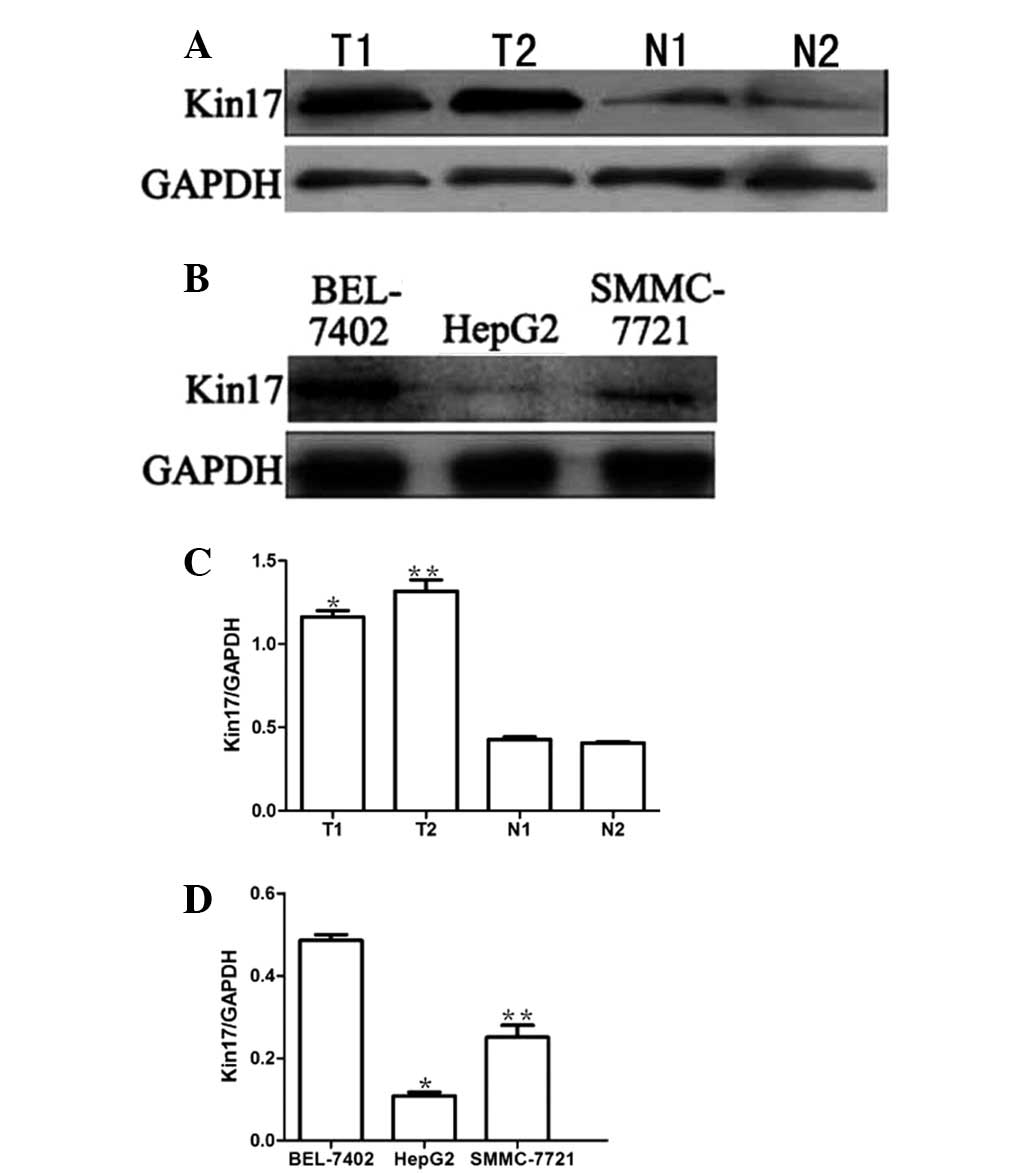
Kin17 expression in hepatocellular
carcinoma tissues and hepatocellular carcinoma cell lines
Kin17 expression was examined by western blotting in
the human hepatocellular carcinoma and normal tissues, and in the
HepG2, SMMC-7721 and BEL-7402 human hepatocellular carcinoma cell
lines. As shown in Fig. 1, Kin17
expression was significantly higher in the hepatocellular carcinoma
tissues (1.347±0.29) compared with that in the corresponding normal
tissues (0.394±0.13) (P<0.05). Furthermore, the expression
levels of Kin17 in the BEL-7402 cells were markedly higher than
those in the HepG2 and SMMC-7721 cells (0.405±0.18 vs. 1.239±0.21
and 0.137±0.11, respectively) (P<0.05).
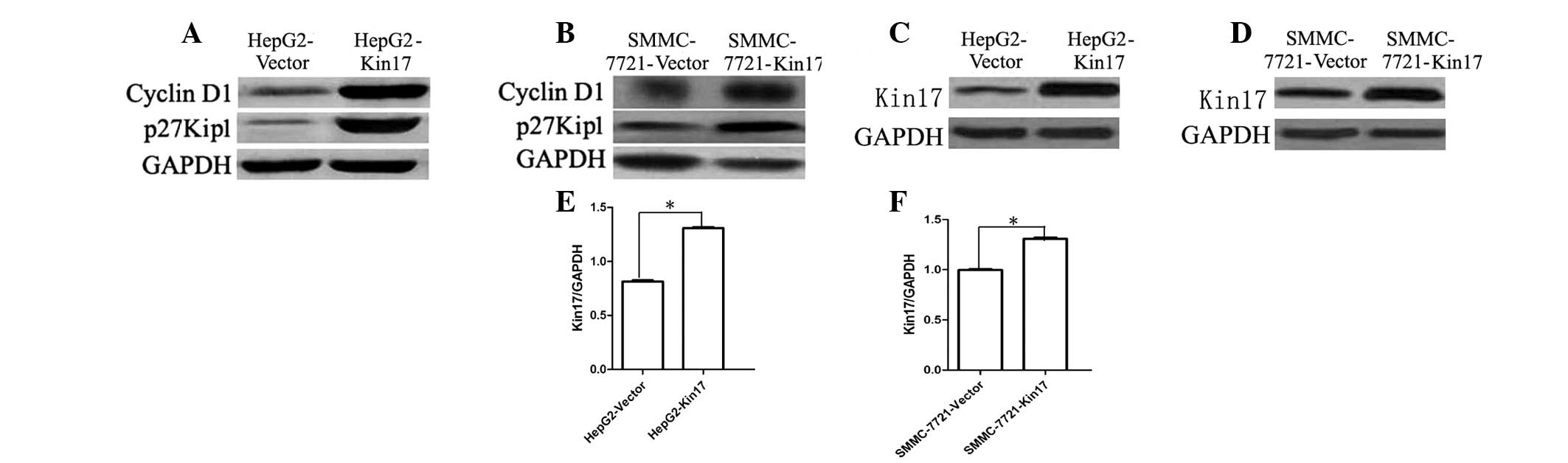
Effects of Kin17 on cyclin D1 and p27Kip1
expression
Western blot analysis revealed that the HepG2-Kin17
and SMMC-7721-Kin17 cells displayed increased expression levels of
cyclin D1 and p27Kip1 protein compared with the HepG2-Vector and
SMMC-7721-Vector cells (P<0.05; Fig.
2). These results demonstrated that the Kin17 might be, at
least in part, associated with cyclin D1 and p27Kip1
expression.
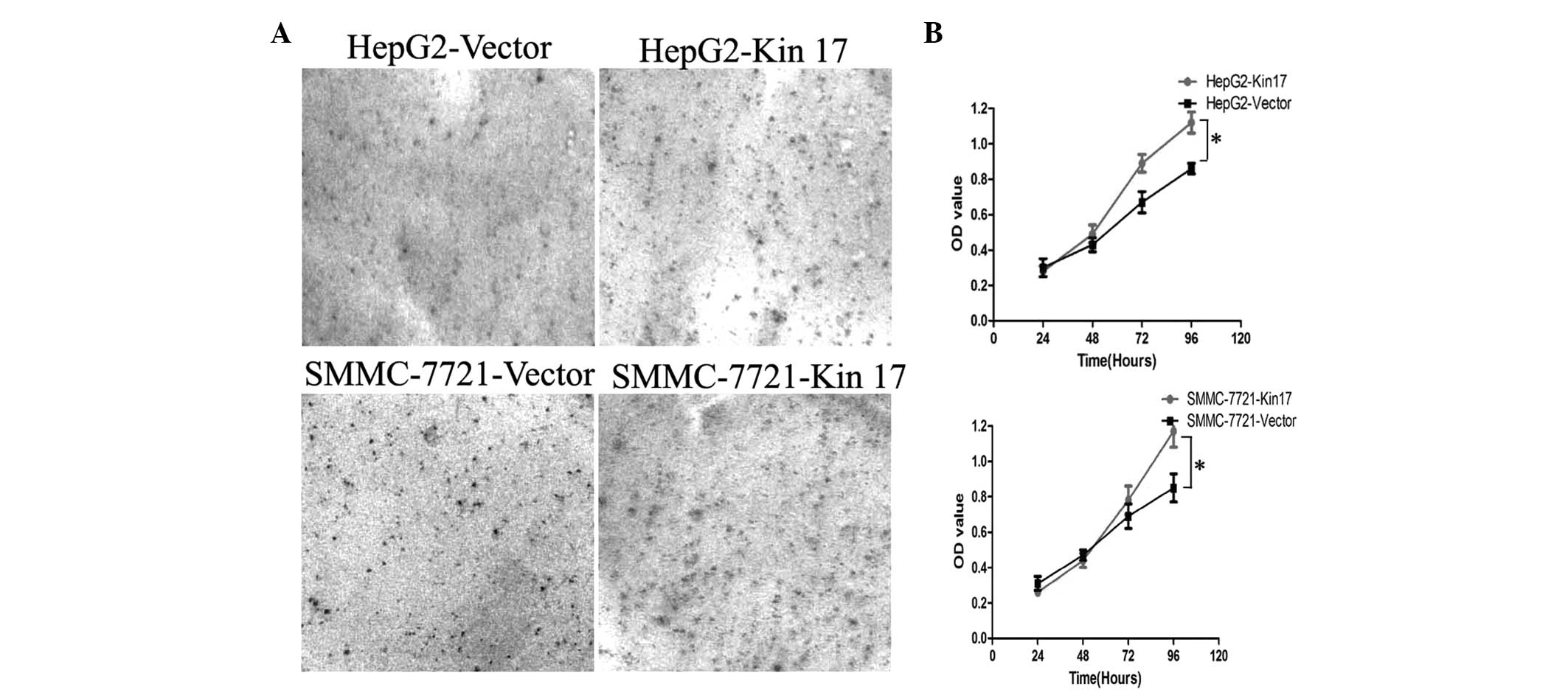
Upregulation of Kin17 expression promotes
the proliferation of hepatocellular carcinoma cells in vitro
Taking into account the low endogenous expression of
Kin17 in HepG2 and SMMC-7721 cells, these two cell lines were
utilized to explore the role of Kin17 on the proliferation of
hepatocellular carcinoma cells. As shown in Fig. 2, the expression of Kin17 was
enhanced in HepG2 and SMMC-7721 cells following transfection with
Kin17 cDNA.
The Colony formation assay demonstrated that the
number of colonies formed by the HepG2-Kin17 group (367±21) was
significantly higher than that of the HepG2-Vector group (189±13)
(P<0.05; Fig. 3A). Similarly,
the number of colonies formed by the SMMC-7721-Kin17 group (394±25)
was greater than that of the SMMC-7721-Vector group (226±16)
(P<0.05), indicating that the results were not specific to HepG2
cells. The MTT assay revealed that overexpression of Kin17 enhanced
the proliferation of HepG2 and SMMC-7721 cells. Three days after
seeding, the cell number of the HepG2-Kin17 group was significantly
higher than that of the HepG2-Vector group (P<0.05) (Fig. 3B). Subsequently, the proliferation
stimulating effect of Kin17 was affirmed in SMMC-7721 cells.
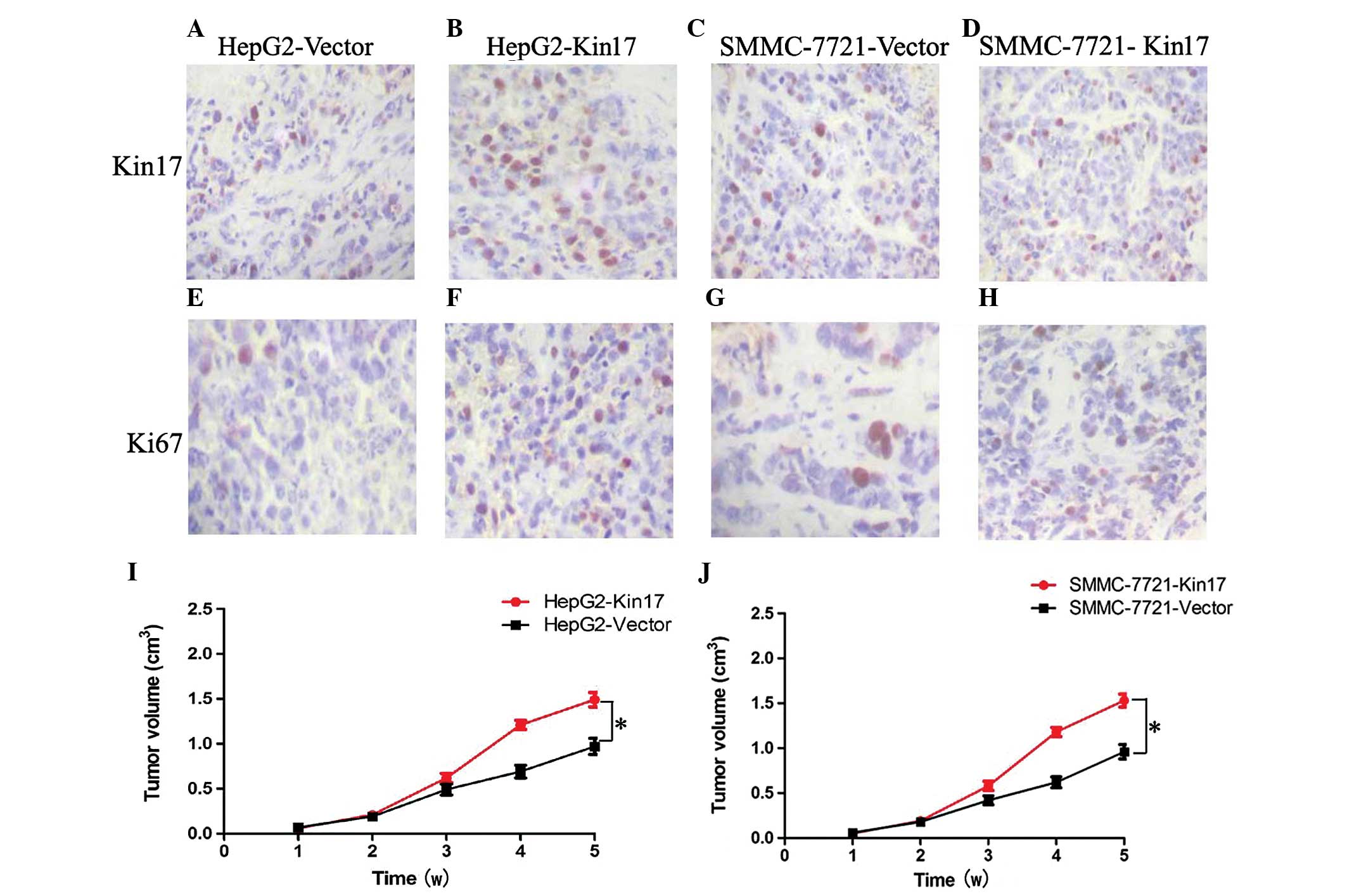
Upregulation of Kin17 expression
accelerated tumor growth in vivo
Compared with the HepG2-Vector (12.6±1.9)and
SMMC-7721-Vector cells (18.8±2.3), Kin17 expression was increased
in the mice transfected with HepG2-Kin17 (29.7±3.1) and
SMMC-7721-Kin17 cells (32.5±4.1) (P<0.05) (Fig. 4A–D). By using Ki-67, it was
identified that the proliferation index (PI) was significantly
increased in the HepG2-Kin17 (20.3±1.8; Fig. 4F) and SMMC-7721-Kin17 (25.9±1.2;
Fig. 4H) groups compared with the
HepG2-Vector (8.2±0.9; Fig. 4E) and
SMMC-7721-Vector (9.7±0.6; Fig. 4G)
groups (P<0.05 for both). Furthermore, mice injected with
HepG2-Kin17 and SMMC-7721-Kin17 cells had a larger tumor volume
than that of animals receiving HepG2-Vector and SMMC-7721-Vector
cells (P<0.05; Fig. 4I and J).
Collectively, these results indicated that upregulation of Kin17
accelerated tumor growth in vivo.
Discussion
The Kin17 protein is expressed in a wide variety of
tissues and is mainly located in the nucleus (6,7,13,14).
It is reported that Kin17 is involved in complex cellular
processes, such as DNA replication and cellular response to DNA
damage (15,16). Moreover, compared with normal human
fibroblasts, elevated levels of Kin17 protein have been found in
immortalized human fibroblasts (15–18).
This finding suggested that Kin17 might be involved in the
tumorigenesis of cancer.
Serum-stimulated mouse fibroblasts showed Kin17 mRNA
expression increased significantly and promoted cell growth
compared with the cells without serum stimulation (3). Similarly, Zeng et al also
revealed overexpression of Kin17 promoted DNA replication and cell
proliferation in breast cancer cells (18). In the present study, Kin17 was
revealed to be overexpressed in hepatocellular carcinoma patient
samples compared with the corresponding normal tissue samples.
Moreover, upregulation of Kin17 expression promoted the growth of
hepatocellular carcinoma cells in vitro and in vivo.
At the cell level, Kin17 promoted cell proliferation and colony
formation. In the hepatocellular carcinoma cell xenograft model,
tumors of mice injected with HepG2-Kin17 and SMMC-7721-Kin17 cells
were larger than those of mice implanted with HepG2-Vector and
SMMC-7721-Vector cells. Moreover, the PI reflected by Ki-67 in the
Kin17-overexpressing hepatocellular carcinoma cells was
significantly higher than that in the control hepatocellular
carcinoma cells. The association between enhanced levels of Kin17
and altered tumorigenic characteristics suggests that Kin17 is
vital for the growth of hepatocellular carcinoma. Paradoxically,
Kannouche et al reported that overproduction of Kin17
inhibited the proliferation of human epithelioid cervical carcinoma
(HeLa) and non-small lung cancer cells (H1299) (19). Further study found that this was
correlated with changes in nuclear morphology and with a decrease
in DNA replication rate. The discrepancy in the results may be due
to the different types of cancer studied.
The molecular mechanism by which Kin17 promotes cell
proliferation is unknown. Cyclin D1 is one of the important
regulators of G1/S transition (20), and p27Kip1 is one of the key
regulators of cell cycle (21). In
the current study, it was identified that Kin17 upregulated cyclin
D1 expression and downregulated p27Kip1 expression. The results
indicated that the effect of Kin17 in promoting the proliferation
of hepatocellular carcinoma cells correlates with the expression of
cyclin D1 and p27Kip1.
In conclusion, the present has study demonstrated
that increased Kin17 expression promotes the growth of
hepatocellular carcinoma in vitro and in vivo.
However, further studies are required to clarify the exact
mechanism of Kin17 in the tumorigenesis of hepatocellular
carcinoma. The important role of Kin17 in the proliferation of
cancer cells may present as a useful target for the treatment of
hepatocellular carcinoma.
References
|
1
|
Tomuleasa C, Soritau O, Rus-Ciuca D, et
al: Isolation and characterization of hepatic cancer cells with
stem-like properties from hepatocellular carcinoma. J
Gastrointestin Liver Dis. 19:61–67. 2010.
|
|
2
|
Aihara H, Ito Y, Kurumizaka H, et al: An
interaction between a specified surface of the C-terminal domain of
RecA protein and double-stranded DNA for homologous pairing. J Mol
Biol. 274:213–221. 1997.
|
|
3
|
Kannouche P, Pinon-Lataillade G, Tissier
A, et al: The nuclear concentration of kin17, a mouse protein that
binds to curved DNA, increases during cell proliferation and after
UV irradiation. Carcinogenesis. 19:781–789. 1998.
|
|
4
|
Kurumizaka H, Aihara H, Ikawa S, et al: A
possible role of the C-terminal domain of the RecA protein. A
gateway model for double-stranded DNA binding. J Biol Chem.
271:33515–33524. 1996.
|
|
5
|
Kannouche P, Pinon-Lataillade G, Mauffrey
P, et al: Overexpression of kin17 protein forms intranuclear foci
in mammalian cells. Biochimie. 79:599–606. 1997.
|
|
6
|
Mazin A, Milot E, Devoret R, et al: KIN17,
a mouse nuclear protein, binds to bent DNA fragments that are found
at illegitimate recombination junctions in mammalian cells. Mol Gen
Genet. 244:435–438. 1994.
|
|
7
|
Mazin A, Timchenko T, Ménissier-de Murcia
J, et al: Kin17, a mouse nuclear zinc finger protein that binds
preferentially to curved DNA. Nucleic Acids Res. 22:4335–4341.
1994.
|
|
8
|
Timchenko T, Bailone A and Devoret R:
Btcd, a mouse protein that binds to curved DNA, can substitute in
Escherichia coli for H-NS, a bacterial nucleoid protein.
EMBO J. 15:3986–3992. 1996.
|
|
9
|
Blattner C, Kannouche P, Litfin M, et al:
UV-induced stabilization of c-fos and other short-lived mRNAs. Mol
Cell Biol. 20:3616–3625. 2000.
|
|
10
|
Masson C, Menaa F, Pinon-Lataillade G, et
al: Identification of KIN (KIN17), a human gene encoding a nuclear
DNA-binding protein, as a novel component of the TP53-independent
response to ionizing radiation. Radiat Res. 156:535–544. 2001.
|
|
11
|
Biard DS, Saintigny Y, Maratrat M, et al:
Enhanced expression of the Kin17 protein immediately after low
doses of ionizing radiation. Radiat Res. 147:442–450. 1997.
|
|
12
|
Wang S, Liu H, Ren L, et al: Inhibiting
colorectal carcinoma growth and metastasis by blocking the
expression of VEGF using RNA interference. Neoplasia. 10:399–407.
2008.
|
|
13
|
Kannouche P, Mauffrey P, Pinon-Lataillade
G, et al: Molecular cloning and characterization of the human KIN17
cDNA encoding a component of the UVC response that is conserved
among metazoans. Carcinogenesis. 21:1701–1710. 2000.
|
|
14
|
Tissier A, Kannouche P, Mauffrey P, et al:
Molecular cloning and characterization of the mouse Kin17 gene
coding for a Zn-finger protein that preferentially recognizes bent
DNA. Genomics. 38:238–242. 1996.
|
|
15
|
Masson C, Menaa F, Pinon-Lataillade G, et
al: Global genome repair is required to activate KIN17, a
UVC-responsive gene involved in DNA replication. Proc Natl Acad Sci
USA. 100:616–621. 2003.
|
|
16
|
Miccoli L, Frouin I, Novac O, et al: The
human stress-activated protein kin17 belongs to the multiprotein
DNA replication complex and associates in vivo with mammalian
replication origins. Mol Cell Biol. 25:3814–3830. 2005.
|
|
17
|
Miccoli L, Biard DS, Creminon C, et al:
Human kin17 protein directly interacts with the simian virus 40
large T antigen and inhibits DNA replication. Cancer Res.
62:5425–5435. 2002.
|
|
18
|
Zeng T, Gao H, Yu P, et al: Up-regulation
of kin17 is essential for proliferation of breast cancer. PLoS One.
6:e253432011.
|
|
19
|
Kannouche P and Angulo JF: Overexpression
of kin17 protein disrupts nuclear morphology and inhibits the
growth of mammalian cells. J Cell Sci. 112:3215–3224. 1999.
|
|
20
|
Li X, Hao Z, Fan R, et al: CIAPIN1
inhibits gastric cancer cell proliferation and cell cycle
progression by downregulating CyclinD1 and upregulating P27. Cancer
Biol Ther. 6:1539–1545. 2007.
|
|
21
|
Cuadrado M, Gutierrez-Martinez P, Swat A,
et al: p27Kip1 stabilization is essential for the maintenance of
cell cycle arrest in response to DNA damage. Cancer Res.
69:8726–8732. 2009.
|