Introduction
Cancer is a multifactorial disease that results from
complex interactions between numerous genetic and environmental
factors (1). Accumulative etiologic
factors associated with cancer have been well-established by
epidemiological studies, including alcohol consumption, cigarette
smoking, obesity, occupational exposures, family history of cancer
and diet. However, only a marginal number of the individuals that
are exposed to these factors develop cancer, therefore, this
indicates that genetic susceptibility is a more significant
indication of an individual’s risk of cancer.
Proline (Pro)-directed phosphorylation, also termed
phosphorylation of proteins on serine (Ser) or threonine (Thr)
residues, is a critical intracellular signaling mechanism, which
regulates diverse cellular processes, including cell cycle
progression, transcriptional regulation, RNA processing, and cell
proliferation and differentiation (2,3). It
has been demonstrated that the deregulation of Pro-directed
phosphorylation is a prevalent and specific event in human cancers,
which results in cell transformation and oncogenesis (2).
Peptidyl-prolyl cis-trans isomerase NIMA-interacting
1 (encoded by the PIN1 gene) specifically isomerizes the
conformation of Pro-directed phosphorylation sites, revealing a
novel post-phosphorylation regulatory mechanism (4,5).
PIN1 exhibits a high specificity for substrates with
Ser/Thr-Pro motifs, and changes the conformation of phosphoproteins
by recognizing and binding to specific phospho-Ser/Thr-Pro motifs
(6). PIN1 substrates, which
contain phospho-Ser/Thr-Pro motifs, include a number of important
cell cycle regulators, as well as oncogenic and tumor suppressor
proteins. These include cyclin D1 (7), p53 (8), Cdc25 (9), c-Myc (10), c-Jun (8), β-catenin (11), glycogen synthase kinase-3β (10) and Bcl-2 (12). Therefore, by targeting these
significant substrates, which contain phospho-Ser/Thr-Pro motifs,
PIN1-induced conformational changes may function as a
critical catalyst for the potentiation of multiple oncogenic
signaling pathways during cancer development (13). It has been reported that PIN1
is aberrantly overexpressed in numerous types of cancer, including
prostate and lung cancer (14),
esophageal squamous cell carcinoma (7), and breast cancer (8,11). By
contrast, the inhibition of PIN1 in cancer cells may trigger
apoptosis or suppress the transformed phenotype (15,16).
These results indicated that the PIN1 gene may exhibit an
oncogenic role in tumorigenesis.
The human PIN1 gene, which is located at
chromosome 19p13, contains four exons within a 14-kb region,
encodes a 163-amino-acid protein and has a promoter region of 1.5
kb (17). Several putative
functional single nucleotide polymorphisms (SNPs) have been
identified in the coding and promoter regions of PIN1, and
the most important is rs2233678 G>C: c.−842 G>C (842
nucleotides upstream of the initiation transcription code ATG) in
the promoter. The PIN1 promoter polymorphism (−842 G>C)
was first identified in a study of Alzheimer’s disease, which
revealed that the variant −842C allele was associated with an
increased risk of Alzheimer’s disease (18). Recent studies have investigated the
association between the PIN1 promoter polymorphism (−842
G>C) and the risk of cancer in various organs, including the
liver (19), lungs (17) and breast (20,21),
squamous cell carcinoma of the head and neck (22), and nasopharyngeal (23) and esophageal cancer (24). However, the results of these studies
remain controversial. Considering the extensive role of PIN1
in the carcinogenic process, a meta-analysis was performed that
included all eligible case-control studies, to estimate the overall
cancer risk associated with the PIN1 promoter polymorphism
(−842 G>C) and to quantify the potential heterogeneity between
the studies.
Methods
Identification and eligibility of
relevant studies
All available case-control studies regarding the
association between the PIN1 promoter polymorphism (−842
G>C) and cancer risk were included in the present study. PubMed
(www.ncbi.nlm.nih.gov/pubmed) and EMBASE
(www.elsevier.com/online-tools/embase)
were searched using the terms ‘PIN1’, ‘polymorphism’ and
‘cancer’ (last accessed: 8 May 2013). The search was limited to
studies written in English. Additional studies were identified by
manually searching the references of the original studies
identified using the search terms. When more than one study
investigating the same type of population was included in several
publications, the most recent studies with the largest sample sizes
were selected. Furthermore, studies included in the meta-analysis
had to meet the following inclusion criteria: i) Evaluation of the
PIN1 promoter polymorphism (−842 G>C) and the cancer
risk; ii) use of a case-control design; and iii) contained an
available genotype frequency.
Data extraction
Two authors independently extracted data from the
studies according to the inclusion criteria and subsequently
reached a consensus on the data items. The following
characteristics were recorded from each study: The first author’s
surname, year of publication, cancer type, country of origin,
patient ethnicity, source of the control groups (population- or
hospital-based controls), genotyping method, the number of cases
and controls, and the P-value for the Hardy-Weinberg equilibrium
(HWE) for the controls (Table I).
Ethnicity was categorized as European or Asian and when studies
included subjects from different ethnic groups, the data were
extracted separately for each ethnic group where possible. Studies
investigating more than one sample were included as individual data
sets.
 | Table IFeatures of the studies included in
the present meta-analysis. |
Table I
Features of the studies included in
the present meta-analysis.
| First author
(year) | Tumor type | Country | Patient
ethnicity | Source of control
group | Genotyping
method | HWE | Sample size
(case/control) |
|---|
| Segat (2007) | Liver cancer | Italy | European | Hospital | PCR-RFLP | NA | 228/250 |
| Lu (2011)a | Lung cancer | China | Asian | Hospital | PCR-RFLP | >0.05 | 1056/1056 |
| Lu (2011)b | Lung cancer | China | Asian | Hospital | PCR-RFLP | >0.05 | 503/623 |
| Naidu (2011) | Breast cancer | Malaysia | Asian | Hospital | PCR-RFLP | 0.831 | 107/80 |
| Breast cancer | China | Asian | Hospital | PCR-RFLP | 0.503 | 219/111 |
| Breast cancer | India | Asian | Hospital | PCR-RFLP | 0.901 | 61/61 |
| Han (2010) | Breast cancer | USA | European | Hospital | PCR-RFLP | 0.160 | 467/488 |
| Lu (2009) | SCCHN | USA | European | Hospital | PCR-RFLP | 0.640 | 1006/1007 |
| Lu (2012) | Nasopharyngeal
cancer | China | Asian | Hospital | PCR-RFLP | 0.063 | 178/156 |
| You (2013) | Esophageal
cancer | China | Asian | Hospital | PCR-RFLP | 0.312 | 699/729 |
Statistical analysis
For the control group of each study, the allelic
frequency was calculated, and HWE was assessed using the
χ2 test. P<0.05 was considered to indicate a
statistically significant difference. The strength of the
association between the PIN1 promoter polymorphism (−842
G>C) and cancer risk was measured using odds ratios (ORs) with
95% confidence intervals (CIs). Firstly, the risks of the GC and CC
genotypes for cancers were estimated, compared with the wild-type
GG homozygote, and then the risks of GC/CC vs. GG and CC vs. GC/GG
for cancers were evaluated, assuming dominant and recessive effects
of the variant C allele, respectively. Stratified analyses were
also performed by cancer type (when one cancer type was included in
only one individual study, it was combined into the ‘other cancers’
group), ethnicity and sample size (populations >500 in the case
and control groups). To investigate the potential for heterogeneity
across the studies, a statistical test for heterogeneity was
performed based on the Q-statistic (25) whereby P>0.1 indicated a lack of
heterogeneity between the studies. The summary ORs estimate of each
study was calculated using the fixed-effects model (the
Mantel–Haenszel method) (26).
Otherwise, the random-effects model (the DerSimonian and Laird
method) (27) was used.
Furthermore, the meta-regression model was used to investigate the
possible source of heterogeneity among the different study types
(28). Sensitivity analyses were
performed to assess the stability of the results, whereby a single
study in the meta-analysis was deleted each time to reflect the
influence of the individual data set on the pooled OR. Publication
bias was evaluated using a Begg’s funnel plot and Egger’s test
(29). All analyses were performed
using Stata software version 11.0 (StataCorp LP, College Station,
TX, USA) and all tests were two-sided.
Results
Features of the studies
A total of seven articles in English regarding the
PIN1 promoter polymorphism (−842 G>C) and cancer risk
were available for the present meta-analysis. One article
investigated two individual samples, which were collected during
different time periods. Another article investigated three
individual samples that were obtained from different countries and
each sample was counted as an individual study. Finally, a total of
10 case-control studies met the inclusion criteria, including 4,524
cases and 4,561 controls. The features of the selected studies are
presented in Table I. All studies
were case-control studies, including four breast cancer studies,
two lung cancer studies, with the others categorized into an ‘other
cancer’ group. There were three studies that included patients of
European descent and seven studies including patients of Asian
descent. The cancers were identified histologically or
pathologically in the majority of studies and the polymerase chain
reaction (PCR)-restriction fragment length polymorphism (RFLP)
genotyping method was used. The distribution of genotypes in the
controls of all studies was consistent with the HWE, with the
exception of one study, which did not include data for the CC, GC
and GG genotypes (19).
Quantitative synthesis
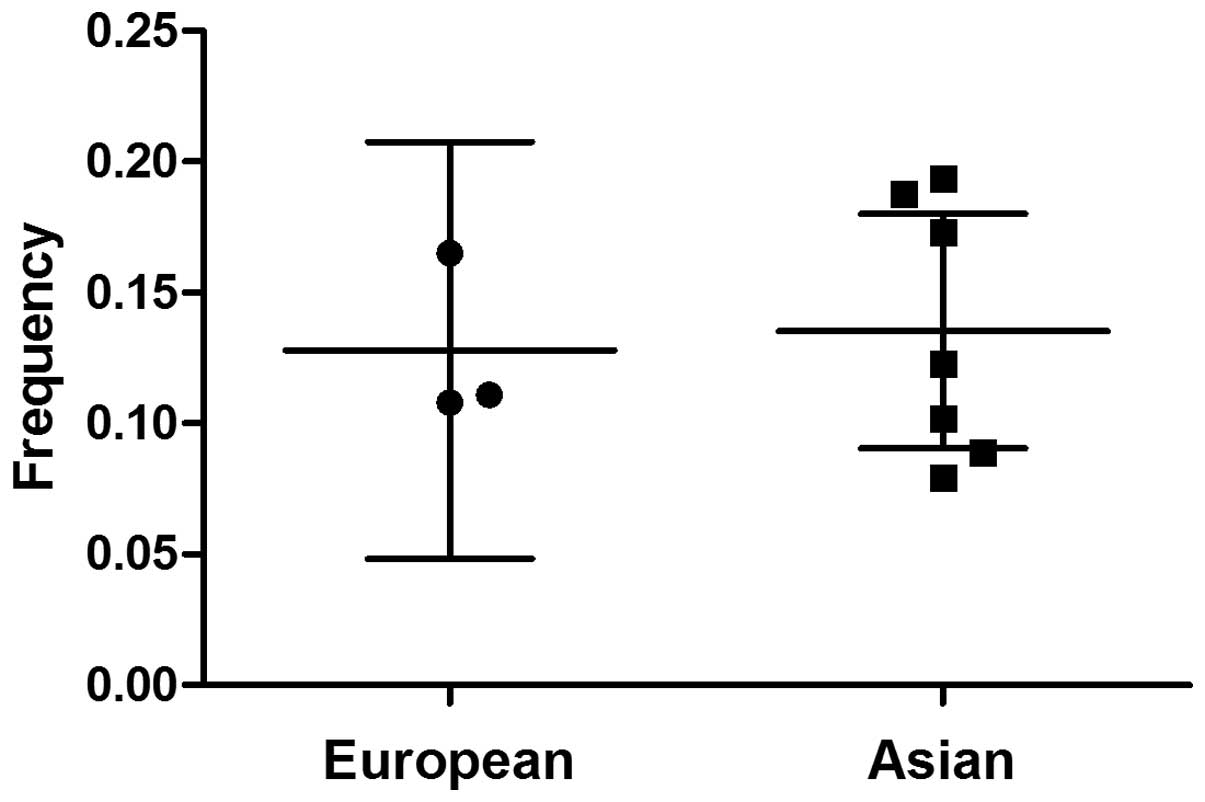
A variation in the C allele frequency was identified
across the different ethnicities. The mean C allele frequencies in
the European and Asian populations were 12.8 and 13.5%,
respectively (Fig. 1). As shown in
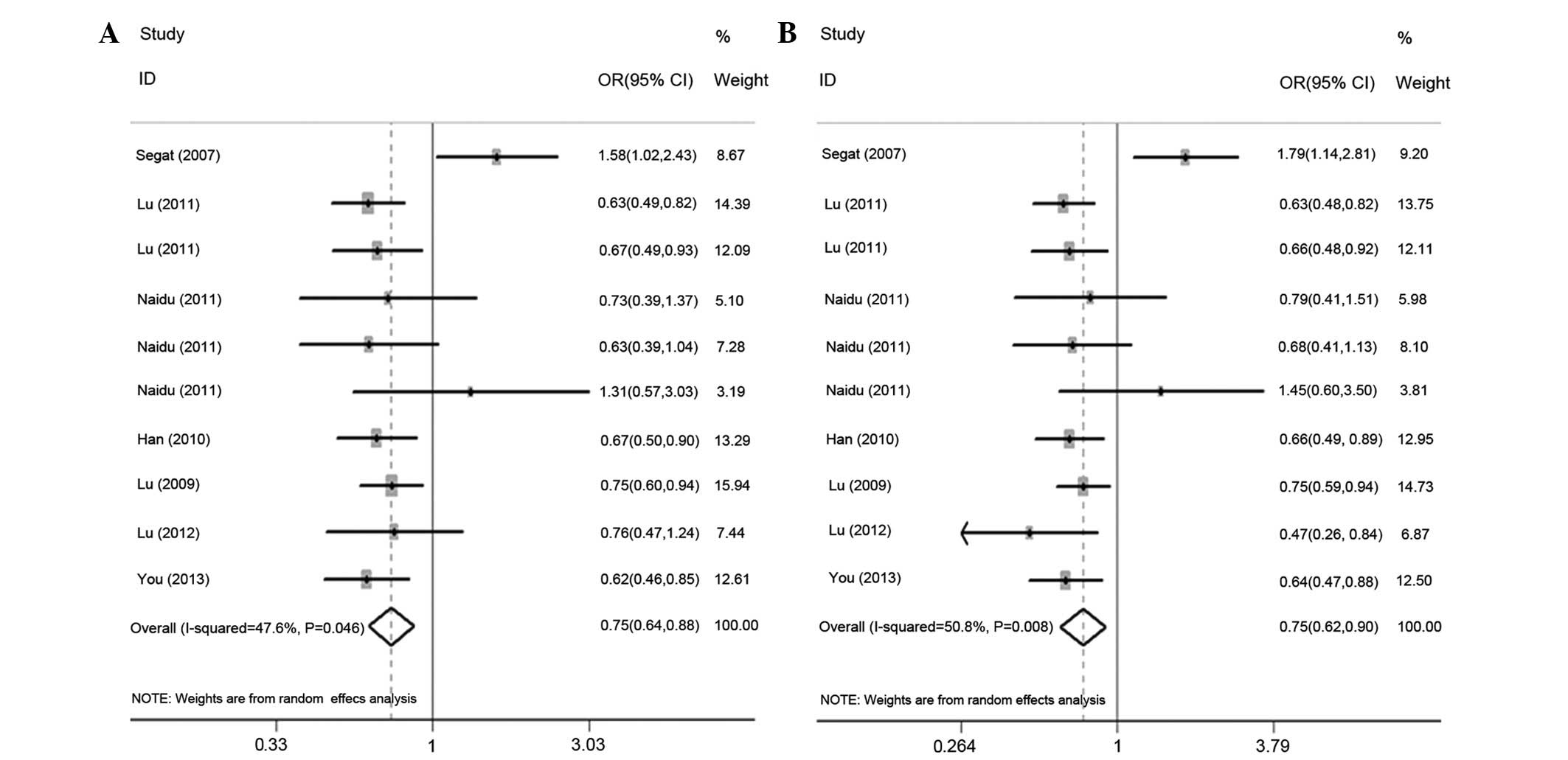
Table II, the variant genotypes
were associated with a significantly decreased cancer risk in the
dominant genetic model (OR, 0.75; 95% CI, 0.64–0.88;
Pheterogeneity =0.046; Fig.
2A). In addition, the variant GC heterozygote was associated
with a decreased cancer risk when compared with the wild-type
homozygote GG (OR, 0.75; 95% CI, 0.62–0.90;
Pheterogeneiy =0.008; Fig.
2B). Furthermore, in the stratified analysis, significantly
decreased risks of breast cancer (OR, 0.72; 95% CI, 0.57–0.90;
Pheterogeneity =0.408 for GC vs. GG; OR, 0.71; 95% CI,
0.56–0.88; Pheterogeneity =0.493 for GC/CC vs. GG
genotype) and lung cancer (OR, 0.64; 95% CI, 0.52–0.79;
Pheterogeneity =0.814 for GC vs. GG; OR, 0.65; 95% CI,
0.53–0.79, Pheterogeneity =0.762 for GC/CC vs. GG
genotype) were identified. In the subgroup analysis of ethnicity, a
significantly decreased risk of cancer was identified among Asians
for the GC vs. GG genotype (OR, 0.66; 95% CI, 0.57–0.76;
Pheterogeneity =0.564) and the GC/CC vs. GG genotype
(OR, 0.67; 95% CI, 0.58–0.77; Pheterogeneity =0.777).
According to the sample size, a markedly decreased risk was
identified in the subgroup with a sample size >500 (OR, 0.68;
95% CI, 0.59–0.78; Pheterogeneity =0.786 for GC vs. GG;
OR, 0.68; 95% CI, 0.59–0.78; Pheterogeneity =0.738 for
the GC/CC vs. GG genotype).
 | Table IISummary OR of the PIN1
(−842G>C) polymorphism with regard to cancer risk. |
Table II
Summary OR of the PIN1
(−842G>C) polymorphism with regard to cancer risk.
| | | CC vs. GG | GC vs. GG | GC/CC vs. GG
(dominant) | CC vs. GC/GG
(recessive) |
|---|
| | |
|
|
|
|
|---|
| Variable | nb | Cases/Controls | OR (95% CI)c | P-valued | OR (95% CI)c | P-valued | OR (95% CI)c | P-valued | OR (95% CI)c | P-valued |
|---|
| Cancer type |
| Breast | 4 | 854/740 | 0.53
(0.26–1.11) | 0.498 | 0.72
(0.57–0.90) | 0.408 | 0.71
(0.56–0.88) | 0.493 | 0.58
(0.28–1.20) | 0.483 |
| Lung | 2 | 1559/1679 | 0.77
(0.33–1.84) | 0.721 | 0.64
(0.52–0.79) | 0.814 | 0.65
(0.53–0.79) | 0.762 | 0.82
(0.35–1.95) | 0.713 |
| Other | 4 | 2111/2142 | 0.78
(0.33–1.85) | 0.066 | 0.80
(0.52–1.25) | 0.001 | 0.85
(0.60–1.20) | 0.006 | 0.81
(0.31–2.07) | 0.034 |
| Ethnicity |
| Europen | 3 | 1701/1745 | 0.70
(0.38–1.26) | 0.627 | 0.93
(0.57–1.50) | 0.001 | 0.89
(0.59–1.35) | 0.004 | 0.73
(0.40–1.31) | 0.483 |
| Asian | 7 | 2823/2816 | 0.83
(0.52–1.33) | 0.126 | 0.66
(0.57–0.76) | 0.564 | 0.67
(0.58–0.77) | 0.777 | 1.05
(0.64–1.74) | 0.105 |
| Sample sizea |
| ≤500 | 6 | 1260/1146 | 0.66
(0.29–1.49) | 0.091 | 0.85
(0.56–1.29) | 0.002 | 0.86
(0.63–1.19) | 0.022 | 0.68
(0.28–1.62) | 0.053 |
| >500 | 4 | 3264/3415 | 0.67
(0.38–1.17) | 0.771 | 0.68
(0.59–0.78) | 0.786 | 0.68
(0.59–0.78) | 0.738 | 0.82
(0.44–1.52) | 0.934 |
| Total | 10 | 4524/4561 | 0.78
(0.54–1.12)e | 0.273e | 0.75
(0.62–0.90)e | 0.008e | 0.75
(0.64–0.88)e | 0.046e | 0.84
(0.58–1.20)e | 0.181e |
Test of heterogeneity
Significant heterogeneity was identified in the
heterozygote comparison (GC vs. GG, Pheterogeneity
=0.008) and dominant model comparison (GC/CC vs. GG,
Pheterogeneity =0.046), although not in the homozygote
comparison (CC vs. GG, Pheterogeneity =0.273) or the
recessive model comparison (CC vs. GC/GG, Pheterogeneity
=0.181). The source of heterogeneity was also assessed in the
heterozygote comparison by the year of publication, cancer type
(breast, lung, and other types of cancer), ethnicity (European or
Asian), and sample size (>500 subjects in each of the cases and
control groups or ≤500). The year of publication (P=0.090) was
found to contribute to substantial heterogeneity, however, no
significant differences in cancer type (P=0.539), ethnicity
(P=0.341) or sample size (P=0.434) were identified.
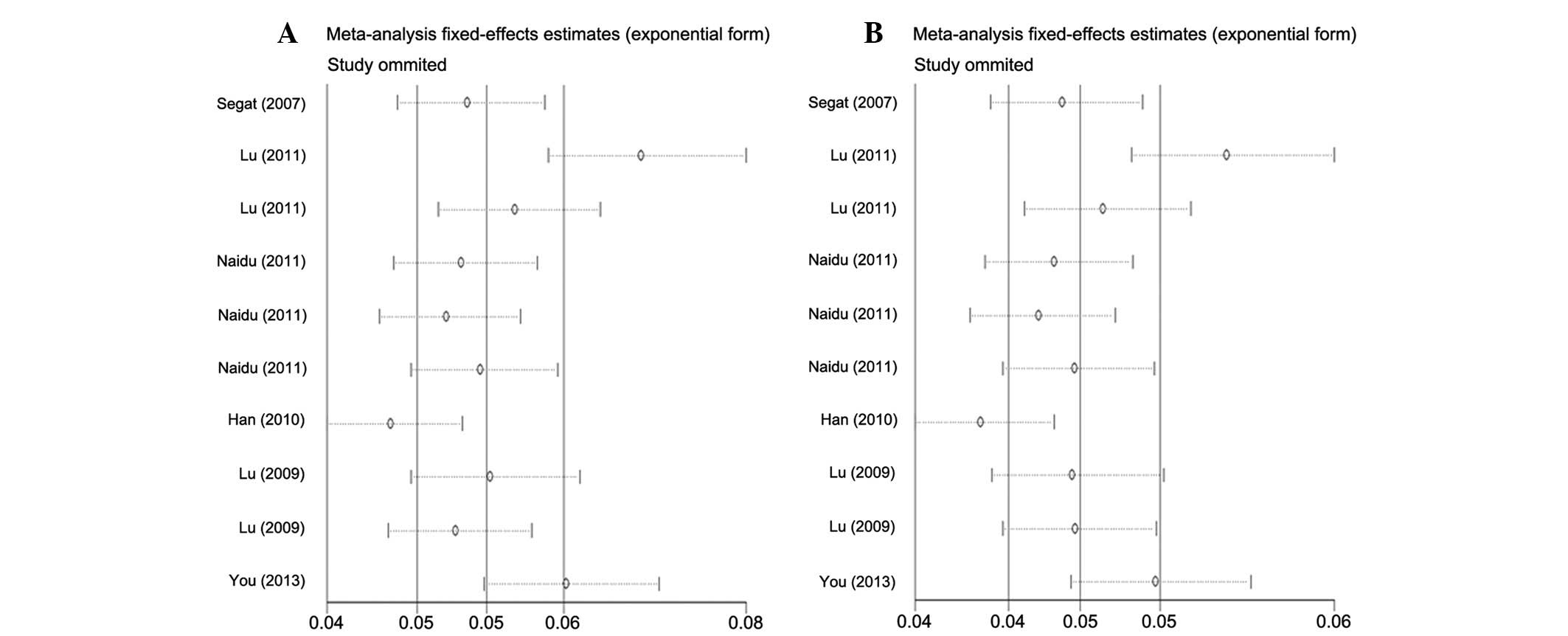
Sensitivity analysis
The influence of each study on the pooled OR was
examined by repetition of the sensitivity analysis. As shown in
Fig. 3, the sensitivity analysis
indicated that the results of the present study were reliable and
robust. Furthermore, when excluding the study by Segat et al
(19), which did not contained the
available data required for HWE, the estimated pool OR remained
unchanged.
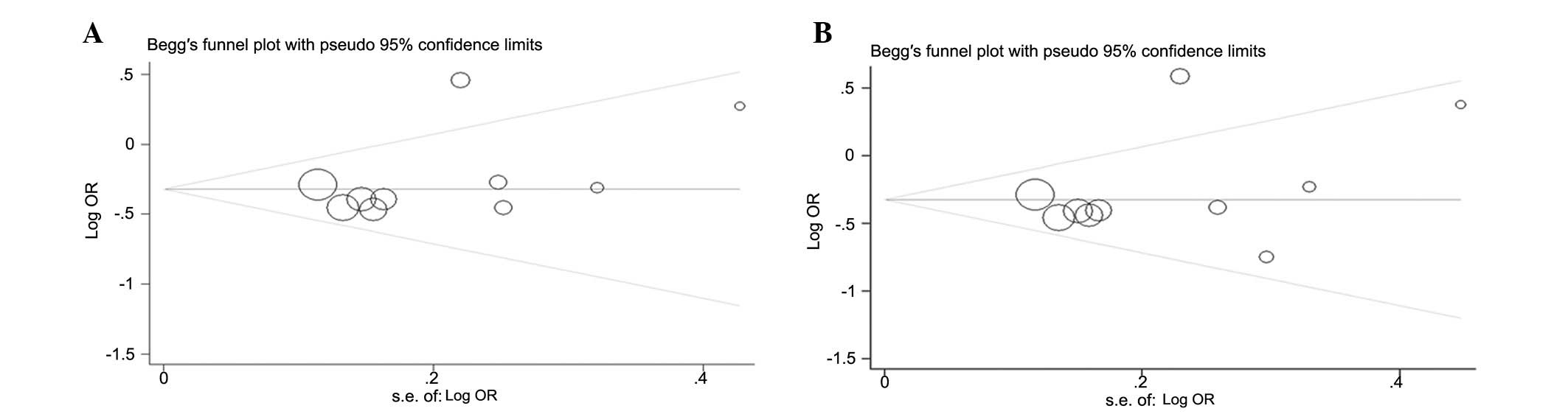
Publication bias
Begger’s funnel plot and Egger’s tests were used to
assess the publication bias of the included studies. The graphical
funnel plots appeared to be symmetrical in the dominant model
comparison (Fig. 4A) and
heterozygote comparison (Fig. 4B).
Egger’s test was then used to provide statistical evidence of the
funnel plot symmetry. As expected, the results did not reveal any
evidence of publication bias (t=0.95 and P=0.370 for GC vs. GG;
t=1.31 and P=0.228 for GC/CC vs. GG).
Discussion
The present meta-analysis, which included 4,524
cases and 4,561 controls from seven case-control studies, evaluated
the association between the PIN1 promoter polymorphism (−842
G>C) and cancer risk. To the best of our knowledge, this is the
first meta-analysis investigating this association. It was found
that individuals exhibiting the variant GC and GC/CC genotypes were
significantly associated with a decreased risk of cancer,
particularly breast and lung cancer, which was revealed by the
subgroup analysis. Furthermore, when stratifying the ethnicity and
sample size, in Asian populations and sample sizes of >500,
individuals with variant GC and GC/CC genotypes were also found to
exhibit a decreased cancer risk.
PIN1 is not an oncogene itself, however, it
may present as an indispensable translator and amplifier of
oncogenic signal transduction. PIN1 specifically recognizes
phospho-Ser/Thr-Pro motifs and regulates the conformation of
Pro-directed phosphorylation sites, which potentiates multiple
oncogenic signaling pathways during carcinogenesis (13). The overexpression of PIN1 is
a prevalent and specific event in human cancers (30). Consistently, studies have found that
high PIN1 expression correlates with poor prognosis in
patients with different types of cancer, including prostate cancer
(31) and esophageal squamous cell
carcinoma (32). Thus, it is
biologically possible that the functional polymorphisms of
PIN1 may be important in the etiology of cancer. Consistent
with the results of the present study, previous studies have
reported that the variant −842 C allele is significantly associated
with a decreased risk of cancer (17,19,20,22–24).
By contrast, no significant association between the −842 G>C SNP
and cancer risk was identified in a study of hepatocellular
carcinoma (19) and breast cancer
by Naidu et al (21).
Furthermore, previous functional analyses of the PIN1
promoter polymorphism (−842 G>C) have demonstrated that
PIN1 gene expression, induced by the −842 C allele, was
significantly lower than those induced by the variant −842 G allele
via inhibition of the transcriptional activity of PIN1
(17,22,24).
In addition, Lu et al (17)
indicated that the −842 CC genotype was associated with lower
levels of PIN1 protein expression in lung cancer samples.
Therefore, the −842 C variant genotype may lead to a reduced
expression of PIN1 and may be associated with a reduced risk of
cancer. Functional studies of the PIN1 promoter polymorphism
(−842 G>C) further support the results of the present study,
whereby individuals with variant GC and GC/CC genotypes were
significantly associated with a decreased risk of cancer.
Accounting for the tumor origin may affect the
results of the meta-analysis, therefore, stratified analyses were
performed according to the cancer type. With the exception of the
‘other cancers’ subgroup, it was found that the C allele may be a
protective factor against breast and lung cancer. Although the
reasons for these discrepancies are not completely understood, one
factor that may contribute to the discrepancies between the various
studies is that this polymorphism may exert a particular effect at
different cancer sites. Additionally, even at the same tumor site,
due to the potentially marginal effect of this genetic polymorphism
on cancer risk and the relatively small sample size of certain
studies, the discrepancies may become apparent. For example, Han
et al (20) found the
PIN1 promoter polymorphism (−842 G>C) was associated with
a decreased risk of breast cancer, however, Naidu et al
(21) did not come to this
conclusion. With regard to other types of cancer in the present
analysis, just one study was available for each specific cancer
site, which may have limited the identification of a reliable
association. For lung cancer, only two studies were included in the
analysis. Although, breast cancer was investigated in four studies,
each study had a small sample size. Therefore, the results of the
present study must be interpreted with caution and large-scale,
detailed and mechanistic studies are required to support the
association between the PIN1 promoter polymorphism (−842
G>C) and decreased cancer risk.
In the subgroup analysis of ethnicity, a decreased
risk in C allele carriers was identified among Asian individuals,
although not in European individuals (Table II). Although the exact mechanism
for this ethnic discrepancy is unclear, certain factors may account
for it. A possible reason is variations in genetic backgrounds,
and/or environmental and social factors in the different
populations. In addition, the different C allele frequency may be a
reflection of natural selection stresses or indicate that the C
allele together with other unidentified genes are involved in
cancer development. Other factors, including selection bias,
different matching criteria, misclassifications on disease status
and publication bias may also be involved. Furthermore, only three
studies included European individuals, therefore, larger-scale
studies and combined analysis are required to further support the
association between the PIN1 promoter polymorphism (−842
G>C) and decreased cancer risk.
The test of heterogeneity markedly influenced the
results of the present meta-analysis. Evident heterogeneity between
the studies was observed in the overall comparisons. Thus, the
studies were stratified by cancer type, ethnicity and sample size.
It was found that the sources of heterogeneity were the cancer site
and ethnicity, indicating that ethnicity and cancer type are
significant when an individual possesses the same polymorphisms as
another individual.
When interpreting the results of the current study,
certain limitations of the meta-analysis must be considered.
Firstly, the results were based on unadjusted estimates, however, a
more precise analysis is required when more detailed, individual
data becomes available, which would allow for an adjusted estimate
including other factors, such as age and gender. Furthermore, a
lack of information for the data analysis may result in confounding
bias. Secondly, for a number of the reviewed studies the original
data was not available, which limited further evaluation of
potential interactions, as the interactions among gene-gene,
gene-environment and different polymorphic loci of the same gene,
may modulate cancer risk. Thirdly, in the present meta-analysis,
the studies were all based on hospital patients and only contained
Asian and European individuals. Thus, validations with larger
population-based studies in different ethnic groups are required.
Finally, the number of published studies was not sufficiently large
for a comprehensive analysis, particularly for a specific cancer
type. However, this meta-analysis also had certain advantages.
Firstly, a substantial number of cases and controls were pooled
from different studies, which significantly increased the
statistical impact of the analysis. Secondly, the quality of the
case-control studies that were included in current meta-analysis
were satisfactory and no publication bias was detected, indicating
that the entire pooled result was unbiased.
In conclusion, the present meta-analysis provides
clear evidence that the PIN1 promoter polymorphism (−842
G>C) contributes to a decreased cancer risk, supporting the
hypothesis that the polymorphism may present as a biomarker for
susceptibility to cancer. However, large studies investigating
different ethnic groups using standardized unbiased methods,
involving specifically selected cancer patients and well-matched
controls, with more detailed individual data are required to
validate the results of the current study. Finally, investigations
of the gene-environment interaction may lead to an improved, more
comprehensive understanding of the roles of PIN1
polymorphisms in the etiology of cancer.
Acknowledgements
The present study was supported by grants from the
Anhui Provincial Health Administration (grant no. 09A070).
References
|
1
|
Pharoah PD, Dunning AM, Ponder BA and
Easton DF: Association studies for finding cancer-susceptibility
genetic variants. Nat Rev Cancer. 4:850–860. 2004.
|
|
2
|
Lu KP: Pinning down cell signaling, cancer
and Alzheimer’s disease. Trends Biochem Sci. 29:200–209. 2004.
|
|
3
|
Lu KP: Prolyl isomerase Pin1 as a
molecular target for cancer diagnostics and therapeutics. Cancer
Cell. 4:175–180. 2003.
|
|
4
|
Ranganathan R, Lu KP, Hunter T and Noel
JP: Structural and functional analysis of the mitotic rotamase Pin1
suggests substrate recognition is phosphorylation dependent. Cell.
89:875–886. 1997.
|
|
5
|
Lu KP, Hanes SD and Hunter T: A human
peptidyl-prolyl isomerase essential for regulation of mitosis.
Nature. 380:544–547. 1996.
|
|
6
|
Lu KP and Zhou XZ: The prolyl isomerase
Pin1: a pivotal new twist in phosphorylation signalling and
disease. Nat Rev Mol Cell Biol. 8:904–916. 2007.
|
|
7
|
Miyashita H, Mori S, Motegi K, Fukumoto M
and Uchida T: Pin1 is overexpressed in oral squamous cell carcinoma
and its levels correlate with cyclin D1 overexpression. Oncol Rep.
10:455–461. 2003.
|
|
8
|
Wulf GM, Ryo A, Wulf GG, et al: Pin1 is
overexpressed in breast cancer and cooperates with Ras signaling in
increasing the transcriptional activity of c-Jun towards cyclin D1.
EMBO J. 20:3459–3472. 2001.
|
|
9
|
Zhou XZ, Kops O, Werner A, et al:
Pin1-dependent prolyl isomerization regulates dephosphorylation of
Cdc25C and tau proteins. Mol Cell. 6:873–883. 2000.
|
|
10
|
Arnold HK, Zhang X, Daniel CJ, et al: The
Axin1 scaffold protein promotes formation of a degradation complex
for c-Myc. EMBO J. 28:500–512. 2009.
|
|
11
|
Ryo A, Nakamura M, Wulf G, Liou YC and Lu
KP: Pin1 regulates turnover and subcellular localization of
beta-catenin by inhibiting its interaction with APC. Nat Cell Biol.
3:793–801. 2001.
|
|
12
|
Pathan N, Aime-Sempe C, Kitada S, et al:
Microtubule-targeting drugs induce bcl-2 phosphorylation and
association with Pin1. Neoplasia. 3:550–559. 2001.
|
|
13
|
Ryo A, Liou YC, Lu KP and Wulf G: Prolyl
isomerase Pin1: a catalyst for oncogenesis and a potential
therapeutic target in cancer. J Cell Sci. 116:773–783. 2003.
|
|
14
|
Ayala G, Wang D, Wulf G, et al: The prolyl
isomerase Pin1 is a novel prognostic marker in human prostate
cancer. Cancer Res. 63:6244–6251. 2003.
|
|
15
|
Ryo A, Liou YC, Wulf G, et al: Pin1 is an
E2F target gene essential for Neu/Ras-induced transformation of
mammary epithelial cells. Mol Cell Biol. 22:5281–5295. 2002.
|
|
16
|
Ryo A, Uemura H, Ishiguro H, et al: Stable
suppression of tumorigenicity by Pin1-targeted RNA interference in
prostate cancer. Clin Cancer Res. 11:7523–7531. 2005.
|
|
17
|
Lu J, Yang L, Zhao H, et al: The
polymorphism and haplotypes of PIN1 gene are associated with
the risk of lung cancer in Southern and Eastern Chinese
populations. Hum Mutat. 32:1299–1308. 2011.
|
|
18
|
Segat L, Pontillo A, Annoni G, et al: Pin1
promoter polymorphisms are associated with Alzheimer’s disease.
Neurobiol Aging. 28:69–74. 2007.
|
|
19
|
Segat L, Milanese M and Crovella S: Pin1
promoter polymorphisms in hepatocellular carcinoma patients.
Gastroenterology. 132:2618–2619. 2007.
|
|
20
|
Han CH, Lu J, Wei Q, et al: The functional
promoter polymorphism (−842G>C) in the Pin1 gene is associated
with decreased risk of breast cancer in non-Hispanic white women 55
years and younger. Breast Cancer Res Treat. 122:243–249. 2010.
|
|
21
|
Naidu R, Har YC and Taib NA: Analysis of
peptidyl-propyl-cis/trans isomerase 1 (PIN1) gene −842 (G>C) and
−667 (T>C) polymorphic variants in relation to breast cancer
risk and clinico-pathological parameters. Scand J Clin Lab Invest.
71:500–506. 2011.
|
|
22
|
Lu J, Hu Z, Wei S, et al: A novel
functional variant (−842G>C) in the PIN1 promoter contributes to
decreased risk of squamous cell carcinoma of the head and neck by
diminishing the promoter activity. Carcinogenesis. 30:1717–1721.
2009.
|
|
23
|
Lu Y, Huang GL, Pu XX, et al: Association
between PIN1 promoter polymorphisms and risk of nasopharyngeal
carcinoma. Mol Biol Rep. 40:3777–3782. 2013.
|
|
24
|
You Y, Deng J, Zheng J, et al: Functional
polymorphisms in PIN1 promoter and esophageal carcinoma
susceptibility in Chinese population. Mol Biol Rep. 40:829–838.
2013.
|
|
25
|
Handoll HH: Systematic reviews on
rehabilitation interventions. Arch Phys Med Rehabil.
87:8752006.
|
|
26
|
Mantel N and Haenszel W: Statistical
aspects of the analysis of data from retrospective studies of
disease. J Natl Cancer Inst. 22:719–748. 1959.
|
|
27
|
DerSimonian R and Laird N: Meta-analysis
in clinical trials. Control Clin Trials. 7:177–188. 1986.
|
|
28
|
Whitehead A and Whitehead J: A general
parametric approach to the meta-analysis of randomized clinical
trials. Stat Med. 10:1665–1677. 1991.
|
|
29
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997.
|
|
30
|
Bao L, Kimzey A, Sauter G, et al:
Prevalent overexpression of prolyl isomerase Pin1 in human cancers.
Am J Pathol. 164:1727–1737. 2004.
|
|
31
|
Arboleda MJ, Lyons JF, Kabbinavar FF, et
al: Overexpression of AKT2/protein kinase Bbeta leads to
up-regulation of beta1 integrins, increased invasion, and
metastasis of human breast and ovarian cancer cells. Cancer Res.
63:196–206. 2003.
|
|
32
|
Fukuchi M, Fukai Y, Kimura H, et al:
Prolyl isomerase Pin1 expression predicts prognosis in patients
with esophageal squamous cell carcinoma and correlates with
cyclinD1 expression. Int J Oncol. 29:329–334. 2006.
|