Introduction
Hepatocellular carcinoma (HCC) is a common primary
liver cancer with a rising incidence globally (1). The estimated number of new cases of
HCC is ~564,000 per year worldwide (2). Human HCC development and progression
is a long-term, multi-step process that is correlated with the
sequential evolution of stages that are morphologically distinct
and result in fully developed HCC. The main risk factors for HCC
are alcoholism, hepatitis B and C, aflatoxin, cirrhosis of the
liver, hemochromatosis, Wilson’s disease and type 2 diabetes
(3). Numerous studies have
investigated the genes underlying the development and progression
of HCC, and have proposed that the pathogenesis of HCC may be
affected by multiple genetic factors (4–6).
Located on chromosome 19 (19q13.2), the X-ray repair
cross-complementing group 1 (XRCC1) gene is known to encode a vital
scaffold protein that has close associations with the base excision
repair (BER) pathway (7). The
functions of the BER pathway in the process of DNA repair require
the use of the XRCC1 protein, which has a significant role in
genome integrity and stability, and in human cancer pathogenesis
and progression (8). Although
>300 validated single nucleotide polymorphisms (SNPs) have been
identified and described in the XRCC1 gene, only three common SNPs
have been extensively studied: Argenine399glutamine (Arg399Gln;
rs25487, G/A substitution at position 28,152 on exon 10),
argenine280histidine (Arg280His; rs25489, G/A substitution at
position 27,466 on exon 9) and arginine194tryptophan (Arg194Trp;
rs1799782, C/T substitution at position 26,304 on exon 6). These
variations were shown to be able to alter the function of XRCC1,
diminish repair kinetics and lead to altered protein efficiency,
eventually inducing the development of cancer (9).
Previous studies have demonstrated that the XRCC1
Arg194Trp gene polymorphism is associated with the susceptibility
to esophageal, gastric, lung, breast and other types of cancer
(10–12). There is little known regarding the
association between the XRCC1 Arg194Trp gene polymorphism and the
susceptibility to HCC. Over the past decade, several case-control
studies have focused on the association between the Arg194Trp gene
polymorphism and the HCC risk, however, the results remain
controversial (13–18). In the present study, a meta-analysis
was performed to investigate whether the Arg194Trp gene
polymorphism was associated with the risk of HCC.
Materials and methods
Identification and eligibility of
relevant studies
Two researchers independently investigated the
titles, abstracts and full texts of relevant studies to determine
whether they could be included in the present study. The results
were compared and disagreements were resolved by consensus. All
case-control studies of the XRCC1 Arg194Trp gene polymorphism and
HCC risk published up to October 25, 2013 were identified through
systematic searches in PubMed (http://www.ncbi.nlm.nih.gov/pubmed/), Google Scholar
(http://scholar.google.co.uk/) and the
China National Knowledge Infrastructure (CNKI) databases
(http://www.cnki.net/), using English and Chinese.
The search terms used were: X-ray repair cross-complementing group
1, XRCC1, polymorphism, variation and mutation, plus all of these
terms in combination with hepatocellular carcinoma, HCC, liver
cancer, liver tumor, liver neoplasms and hepatic tumor. The
reference lists of the retrieved articles were hand-searched to
obtain other relevant publications. All associated publications
were evaluated to identify the most eligible literature. Studies
that were reported by the same authors were checked for possible
overlapping participant groups.
Inclusion and exclusion criteria
The inclusion criteria were as follows; i)
Case-controlled studies that addressed HCC cases and healthy
controls; ii) studies that evaluated the association between the
XRCC1 Arg194Trp gene polymorphism and the HCC risk; iii) all
patients with clinically diagnosed HCC; iv) studies that included
sufficient genotype data for extraction; v) the studies contained
at least two comparison groups (cancer group vs. control group);
and vi) the studies included detailed genotyping data. The
exclusion criteria were as follows: i) Not case-control studies
that evaluated the association between the XRCC1 Arg194Trp gene
polymorphism and the HCC risk; ii) animal studies; iii) studies
that were based on incomplete raw data or no usable data reported;
and iv) duplicated publications.
Data extraction
Two investigators independently performed the
extraction of data from all the eligible publications, according to
the inclusion and exclusion criteria. Any discrepancy between the
two investigators was settled by discussion until a consensus was
reached. For each study, the following data were considered: First
author’s name, year of publication, country of the study,
ethnicity, numbers of genotyped cases and controls, and deviation
from the Hardy-Weinberg Equilibrium (HWE) of the control group.
Statistical methods
The HWE in the controls was assessed for each study
using the χ2 test, and P<0.05 was considered to
indicate a statistically significant disequilibrium. In the overall
and subgroup meta-analyses, the strength of the association between
the XRCC1 Arg194Trp gene polymorphism and the HCC risk was measured
by odds ratios (ORs) and 95% confidence intervals (CIs). The
combined ORs and 95% CIs were calculated respectively for a
homozygote comparison (Trp/Trp vs. Arg/Arg), a heterozygote
comparison (Trp/Trp vs. Arg/Trp), a dominant model (Arg/Arg +
Arg/Trp vs. Trp/Trp) and a recessive model (Trp/Trp + Arg/Trp vs.
Arg/Arg) between groups. The effect of heterogeneity was quantified
using an I2 test. I2 ranged between 0 and
100% and represented the proportion of inter-study variability that
could be attributed to heterogeneity rather than chance.
I2 values of 25, 50 and 75% were defined as low,
moderate and high estimates, respectively. When
I2>50% indicated heterogeneity across studies, a
random-effects model was used for the meta-analysis, otherwise a
fixed-effects model was used (19,20).
Publication bias was examined by plotting a Begg’s funnel plot, and
P<0.05 was considered to indicate a statistically significant
publication bias. Stratified analyses were performed by sample size
(subjects >300) and P<0.05 was considered to indicate a
statistically significant difference. To assess the reliability of
the outcomes in the meta-analysis, a sensitivity analysis was
performed, excluding studies whose allele frequencies in the
controls exhibited a significant deviation from the HWE. All
statistical tests were performed using STATA v.12.0 software (Stata
Corporation, College Station, TX, USA).
Results
Study selection
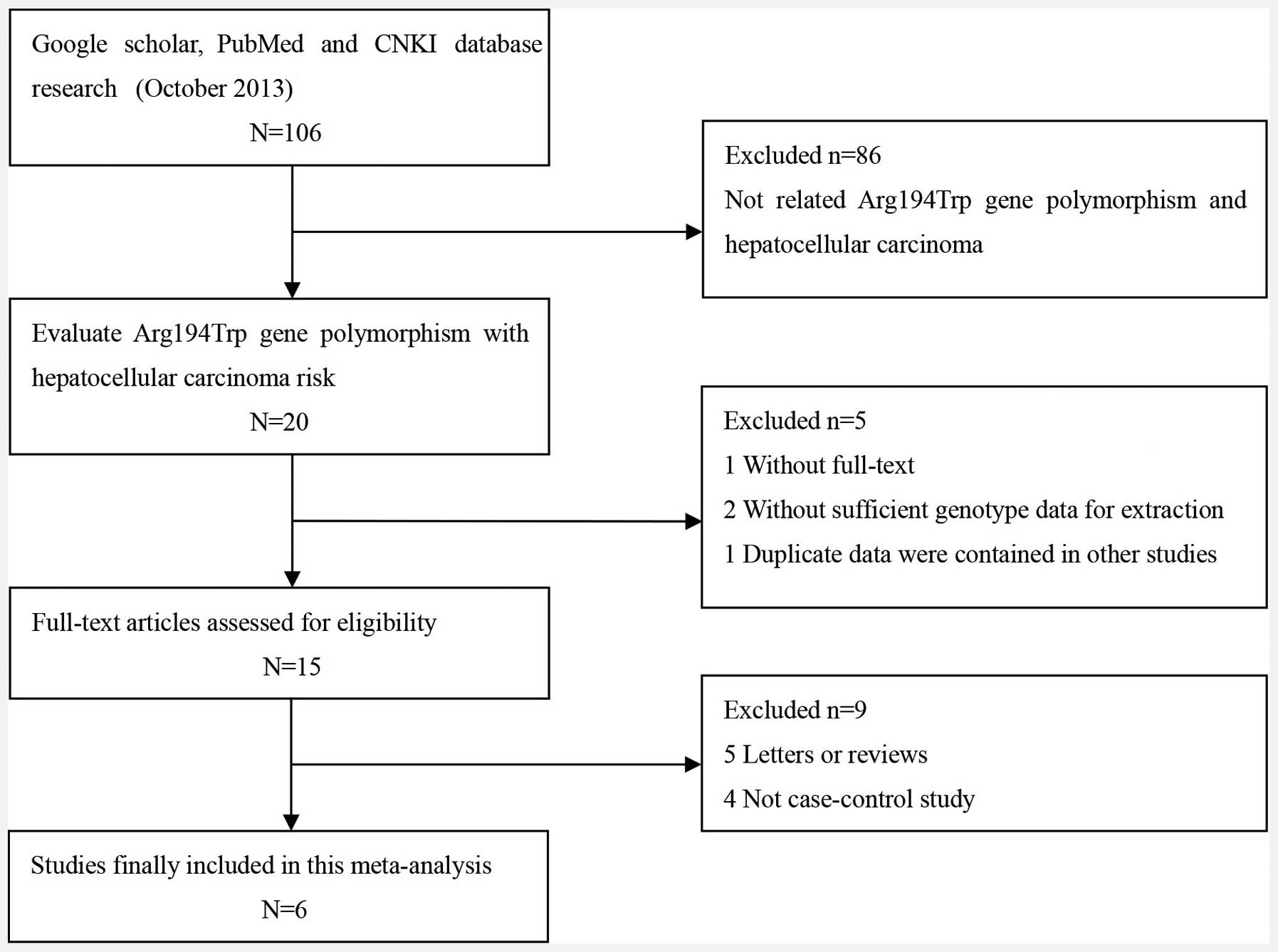
A total of 106 potentially relevant publications
were systematically identified through a search of PubMed, Google
Scholar and CNKI up to October 2013. Based on the preliminary
search criteria, 100 studies were excluded as they did not satisfy
the inclusion criteria. In total, 1,451 cases and 1,398 controls
were included in the meta-analysis. The study characteristics are
summarized in Fig. 1 and Table I. The six studies were all of
individuals of Asian descent, from which, five were from China and
one was from India (13–18). The distribution of the genotypes in
the controls was consistent with the HWE in all studies except for
that of Bo et al (16) and
Guo et al (17).
Furthermore, three studies that were conducted with >300
subjects were included in the subgroup meta-analysis (14,17,18).
Of these six studies, one used TaqMan probe methodology and five
used a polymerase chain reaction-restriction fragment length
polymorphism method to identify the XRCC1 SNPs.
 | Table ICharacteristics of literature studies
included in the meta-analysis. |
Table I
Characteristics of literature studies
included in the meta-analysis.
| | | | | Genotypes for
cases | Genotypes for
controls | |
|---|
| | | | |
|
| |
|---|
| First author
(ref.) | Year | Area | Race | Cases/controls | Arg/Arg | Arg/Trp | Trp/Trp | Arg/Arg | Arg/Trp | Trp/Trp | HWE test |
|---|
| Kiran et al
(14) | 2009 | India | Asian | 63/143 | 8 | 43 | 12 | 27 | 64 | 52 | 0.35 |
| Zeng et al
(15) | 2010 | China | Asian | 545/515 | 305 | 200 | 40 | 275 | 202 | 38 | 0.91 |
| Tang et al
(16) | 2011 | China | Asian | 147/150 | 91 | 41 | 15 | 81 | 58 | 11 | 0.89 |
| Bo et al
(17) | 2011 | China | Asian | 130/130 | 94 | 31 | 5 | 116 | 12 | 2 | 0.02 |
| Guo et al
(18) | 2012 | China | Asian | 314/210 | 264 | 109 | 37 | 292 | 96 | 23 | 0.00 |
| Yuan et al
(19) | 2012 | China | Asian | 252/250 | 119 | 115 | 18 | 128 | 101 | 21 | 0.86 |
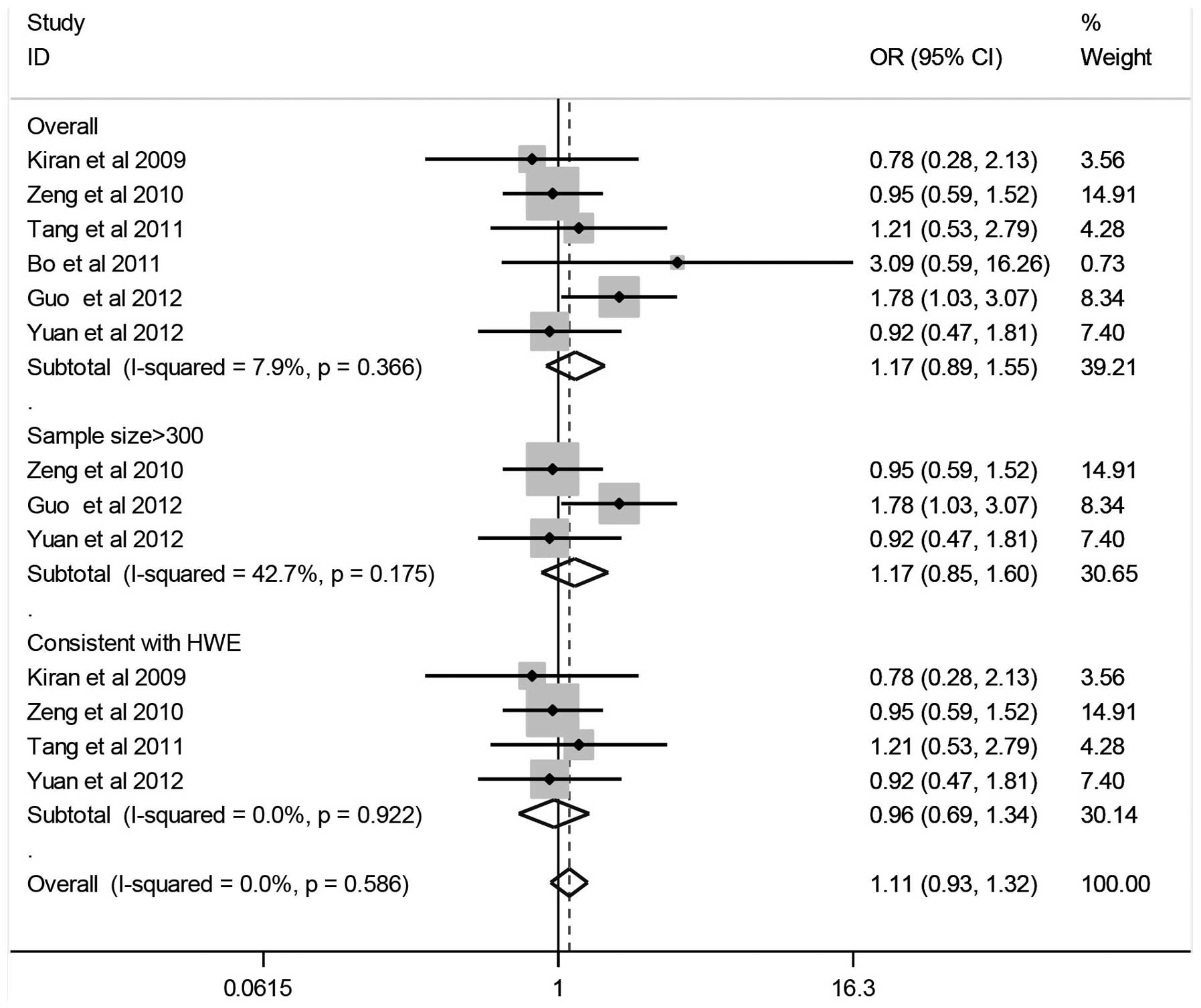
Quantitative data synthesis
The results of the associations between the
Arg194Trp gene polymorphism and the HCC risk, the heterogeneity
test and the test of publication bias are shown in Fig. 2 and Table II. The combined results based on
all the studies showed that variant genotypes were not associated
with an increased HCC risk in different genetic models (Trp/Trp vs.
Arg/Arg: OR, 1.17; 95% CI, 0.89–1.55; Trp/Trp vs. Arg/Trp: OR,
0.94; 95% CI, 0.59–1.51; dominant model: OR, 0.97; 95% CI,
0.63–1.49; recessive model: OR, 1.22; 95% CI, 0.89–1.67). In the
stratified analysis by sample size (subjects, >300), no
significant association was identified between the Arg194Trp gene
polymorphism and HCC (Trp/Trp vs. Arg/Arg: OR, 1.17; 95% CI,
0.85–1.60; Trp/Trp vs. Arg/Trp: OR, 1.07; 95% CI, 0.78–1.49;
dominant model: OR, 0.88; 95% CI, 0.65–1.19; recessive model: OR,
1.11; 95% CI, 0.86–1.44).
 | Table IISummary of ORs and 95% CIs of the
Arg194Trp gene polymorphism and HCC risk. |
Table II
Summary of ORs and 95% CIs of the
Arg194Trp gene polymorphism and HCC risk.
| | Sample size, n | | Test of
heterogeneity | Test of
association | Test of publication
bias |
|---|
| |
| |
|
|
|
|---|
| Subgroup | Genetic model | Case | Control | Type of model | I2
(%) | P-value | OR | 95% CI | z | P-value |
|---|
| Overall | Trp/Trp vs.
Arg/Arg | 1451 | 1398 | Fixed | 7.9 | 0.37 | 1.17 | 0.89–1.55 | 0.00 | 1.00 |
| Trp/Trp vs.
Arg/Trp | Random | 59.2 | 0.03 | 0.94 | 0.59–1.51 | 0.00 | 1.00 |
| Dominant model | Random | 57.0 | 0.04 | 0.97 | 0.63–1.49 | 0.00 | 1.00 |
| Recessive model | Random | 72.3 | 0.00 | 1.22 | 0.89–1.67 | 0.00 | 1.00 |
| Sample size
>300 | Trp/Trp vs.
Arg/Arg | 1111 | 975 | Fixed | 42.7 | 0.18 | 1.17 | 0.85–1.60 | 1.04 | 0.30 |
| Trp/Trp vs.
Arg/Trp | Fixed | 0.0 | 0.40 | 1.07 | 0.78–1.49 | 1.04 | 0.30 |
| Dominant model | Fixed | 36.0 | 0.21 | 0.88 | 0.65–1.19 | 1.04 | 0.30 |
| Recessive
model | Random | 57.1 | 0.10 | 1.11 | 0.86–1.44 | 1.04 | 0.30 |
| Consistent with
HWE | Trp/Trp vs.
Arg/Arg | 1007 | 1058 | Fixed | 0.0 | 0.92 | 0.96 | 0.69–1.34 | 0.34 | 1.00 |
| Trp/Trp vs.
Arg/Trp | Random | 70.4 | 0.02 | 0.84 | 0.45–1.57 | 0.34 | 1.00 |
| Dominant model | Fixed | 49.9 | 0.11 | 1.19 | 0.88–1.60 | 0.34 | 1.00 |
| Recessive
model | Fixed | 31.0 | 0.23 | 0.36 | 0.80–3.15 | 0.34 | 1.00 |
Tests of heterogeneity
Statistically significant heterogeneity was observed
between the trials of the following analyses using the
I2 test (Trp/Trp vs. Arg/Trp: I2=59.2%,
P=0.03; dominant model: I2=57.0%, P=0.04; recessive
model: I2=72.3%, P=0.00) (Table II), and a random-effects model was
employed in these studies. There was no significant heterogeneity
identified for Trp/Trp vs. Arg/Arg (I2=7.9%, P=0.37)
after performing a fixed-effects model.
Sensitivity analysis
A sensitivity analysis was performed following the
removal of the studies by Bo et al (16) and Guo et al (17) due to the genotype distribution in
the control groups deviating from the HWE. The results suggested
that no individual study significantly affected the pooled ORs,
although in certain cases, the I2 value for
heterogeneity was reduced. The sensitivity analysis therefore
confirmed that the data of this meta-analysis was statistically
robust.
Publication bias
The funnel plot and Begg’s test was used to assess
the publication bias of the selected literature. No evidence of
publication bias was detected in the study, and therefore
publication bias was low in the present meta-analysis (all
P>0.05). Information concerning the Begg’s funnel plot is shown
in Table II.
Discussion
The estimated incidence of new HCC cases each year
is >0.5 million (19), with a
wide geographic variation in incidence regions at an international
level; a high incidence can be found in Eastern and South-Eastern
Asia, while a low incidence can be observed in developed regions
(20). It is well-known that
hepatitis B virus is the predominant risk factor for the
pathogenesis of HCC. In addition, epidemiological investigations
have demonstrated that the occurrence and development of HCC has a
strong genetic predisposition (23). The XRCC1 protein is vital in the
multistep nucleotide excision repair pathway, and it is the first
mammalian gene to be isolated that affects the sensitivity of cells
to ionizing radiation (24).
Various studies have focused on the association between the
Arg194Trp gene polymorphism and HCC. However, the observed
associations of these studies were inconclusive (13–18).
The reason for the inconsistencies among these studies is most
likely to be that they were single, small-sample, case-control
studies. To help resolve these conflicting results, the present
study performed a meta-analysis to combine the study types in order
to increase the sample size and statistical power.
A meta-analysis technique was used to collect
comparable published or unpublished data, and statistical methods
were applied to synthesize the independent results of the studies
with the same research target, in order to obtain a combined
quantitative conclusion. This method could provide scientific,
repeatable and objective reasoning as to why similar studies
produced different results (25,26).
The present meta-analysis, including 1,451 cases and 1,398 controls
from six case-control studies, explored the association between the
Arg194Trp XRCC1 gene polymorphism and the HCC risk.
The results of the present meta-analysis revealed
that the XRCC1 Arg194Trp gene polymorphism was not associated with
an increased or decreased risk of HCC. However, a previous study by
Kiran et al (13), reported
that this same polymorphism increased the risk of susceptibility to
HCC in Indian patients with hepatitis (OR, 2.27; 95% CI,
1.01–5.08). This difference in result may be associated with ethnic
and regional differences. The present meta-analysis also involved
several studies with a small sample size; there may have been a
selective bias for the association between the XRCC1 Arg194Trp gene
polymorphism and HCC development, and therefore, large-sample
studies should be used to re-evaluate this association. When
stratifying by sample size (>300), the present meta-analysis
detected no significant association, indicating that there was no
evidence of a small-study bias in the meta-analysis. Further
sensitivity analysis confirmed the significant association between
the maternal XRCC1 Arg194Trp gene polymorphism and the HCC risk.
There was no evidence to suggest a publication bias in the present
meta-analysis for the XRCC1 Arg194Trp gene polymorphism
(P>0.05).
The effect of the XRCC1 Arg194Trp gene polymorphism
may have a limited impact on HCC. As with other malignant tumors,
the development of HCC is due to the combined effect of multiple
genes and gene-environment interactions (27). Previous data have suggested that for
the combination of XRCC1 Arg194Trp and Arg280His or Arg399Gln,
there is a markedly increased risk of hepatitis-related HCC
(13). Furthermore, the risk of HCC
for the XRCC1 Arg194Trp genotype is 1.29 times higher than that of
the XRCC1 194Arg genotype with exposure to alcohol. Drinking may
therefore increase the HCC risk, although there appears to be no
significant difference between the genotypes (P>0.05) (18). Further studies of gene-gene and
gene-environment interactions should be taken into consideration in
future analyses, which should lead to an improved, comprehensive
understanding of the association between the XRCC1 Arg194Trp gene
polymorphism and the HCC risk.
Certain limitations of the present meta-analysis
should be acknowledged in order to establish a complete
interpretation of the data. Firstly, the present meta-analysis was
based on unadjusted OR estimates since not all the published
studies presented adjusted ORs. In cases where the adjusted OR was
presented, they were not adjusted by the same potential
confounders, such as age, gender, ethnicity and exposures. A lack
of information for the data analysis may cause a confounding bias.
Secondly, the number of studies and the number of subjects in the
studies included in the meta-analysis by specific subgroups were
small. Thirdly, a lack of original data limited a further
evaluation of the potential gene-gene and gene-environment
interactions.
In conclusion, the present meta-analysis suggested
that the XRCC1 Arg194Trp gene polymorphism may be not associated
with the HCC risk. Further studies estimating the effects of
gene-gene and gene-environment interactions may provide an improved
comprehensive understanding of the association between XRCC1 and
the HCC risk.
References
|
1
|
Jemal A, Clegg LX, Ward E, et al: Annual
report to the nation on the status of cancer. 1975–2001, with a
special feature regarding survival. Cancer. 101:3–27. 2004.
|
|
2
|
Bosch FX, Ribes J, Díaz M and Cléries R:
Primary liver cancer: worldwide incidence and trends.
Gastroenterology. 127(5 Suppl 1): S5–S16. 2004.
|
|
3
|
Parkin DM, Bray FI and Devesa S: Cancer
burden in the year 2000. The global picture. Eur J Cancer.
37:S4–S66. 2001.
|
|
4
|
Hu Z and Zhao W: Novel insights into the
molecular mechanisms of α-fetoprotein expression and malignant
phenotypes of hepatocellular carcinoma. Cell Mol Immunol. 9:7–8.
2012.
|
|
5
|
Karabork A, Kaygusuz G and Ekinci C: The
best immunohistochemical panel for differentiating hepatocellular
carcinoma from metastatic adenocarcinoma. Pathol Res Pract.
206:572–577. 2010.
|
|
6
|
Yang M, Chen G, Dang Y and Luo D:
Significance of decoy receptor 3 in sera of hepatocellular
carcinoma patients. Ups J Med Sci. 115:232–237. 2010.
|
|
7
|
Thompson LH, Bachinski LL, Stallings RL,
et al: Complementation of repair gene mutations on the hemizygous
chromosome 9 in CHO: a third repair gene on human chromosome 19.
Genomics. 5:670–679. 1989.
|
|
8
|
Poehlmann A and Roessner A: Importance of
DNA damage checkpoints in the pathogenesis of human cancers. Pathol
Res Pract. 206:591–601. 2010.
|
|
9
|
Whitehouse CJ, Taylor RM, Thistlethwaite
A, et al: XRCC1 stimulates human polynucleotide kinase activity at
damaged DNA termini and accelerates DNA single-strand break repair.
Cell. 104:107–117. 2001.
|
|
10
|
Lee SG, Kim B, Choi J, et al: Genetic
polymorphisms of XRCC1 and risk of gastric cancer. Cancer Lett.
187:53–60. 2012.
|
|
11
|
Park JY, Lee SY, Jeon HS, et al:
Polymorphism of the DNA repair gene XRCC1 and risk of primary lung
cancer. Cancer Epidemiol Biomarkers Prev. 11:23–27. 2002.
|
|
12
|
Kim SU, Park SK, Yoo KY, et al: XRCC1
genetic polymorphism and breast cancer risk. Pharmacogenetics.
12:335–338. 2002.
|
|
13
|
Kiran M, Saxena R, Chawla YK, et al:
Polymorphism of DNA repair gene XRCC1 and hepatitis-related
hepatocellular carcinoma risk in Indian population. Mol Cell
Biochem. 7–13. 2009.
|
|
14
|
Zeng X, Yu H and Qiu X: A case-control
study of polymorphism of XRCC1 gene and the risk of hepatocellular
carcinoma. Zhongguo Jibing Kongzhi Zazhi. 14:760–763. 2010.
|
|
15
|
Tang Y, Li X, Liu T, et al: Genetic
polymorphisms of DNA repair genes in patients with hepatocellular
carcinoma. Shandong Yiyao. 51:19–20. 2011.
|
|
16
|
Bo W, Zhang G, Li D, et al: Polymorphisms
of DNA repair gene XRCC1 and susceptibility to hepatic cancer.
Xiandai Zhongliu Yixue. 19:1724–1726. 2011.
|
|
17
|
Guo LY, Jin XP, Niu W, et al: Association
of XPD and XRCC1 genetic polymorphisms with hepatocellular
carcinoma risk. Asian Pac J Cancer Prev. 13:4423–4426. 2012.
|
|
18
|
Yuan T, Deng S, Liu H, et al: Relationship
between XRCC1 and XPD polymorphisms and the risk of the development
of hepatocellular carcinoma: A case-control study. Exp Ther Med.
4:285–290. 2012.
|
|
19
|
Jemal A, Bray F, Center MM, et al: Global
cancer statistics. CA Cancer J Clin. 61:69–90. 2011.
|
|
20
|
Nordenstedt H, White DL and El-Serag HB:
The changing pattern of epidemiology in hepatocellular carcinoma.
Dig Liver Dis. 3:S206–S214. 2010.
|
|
21
|
Cochran WG: The combination of estimates
from different experiments. Biometrics. 10:101–129. 1954.
|
|
22
|
Higgins JP and Thompson SG: Quantifying
heterogeneity in a meta-analysis. Stat Med. 21:1539–1558. 2002.
|
|
23
|
Kirk GD, Lesi OA, Mendy M, et al: 249(ser)
TP53 mutation in plasma DNA, hepatitis B viral infection, and risk
of hepatocellular carcinoma. Oncogene. 24:5858–5867. 2005.
|
|
24
|
Thompson LH, Brookman KW, Jones NJ, et al:
Molecular cloning of the human XRCC1 gene, which corrects defective
DNA strand break repair and sister chromatid exchange. Mol Cell
Biol. 10:6160–6171. 1990.
|
|
25
|
Zhong H, Feng Y, Zheng GX, et al: A
meta-analysis of the association between glutathione S-transferase
P1 gene polymorphism and the risk of adenocarcinomas of lung
cancer. Cancer Biomark. 13:29–35. 2013.
|
|
26
|
Pakiz M, Potocnik U, But I and Mujezinovic
F: A CYP17A1 gene polymorphism in association with multiple uterine
leimyomas; a meta-analysis. Cancer Biomark. 8:29–34. 2011.
|
|
27
|
Sato K and Mori M: Evolving molecular
mechanism-based strategies for control of hepatocellular carcinoma.
Curr Med Chem. 18:4375–4388. 2011.
|