Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common type of cancer and the third most frequent cause of
cancer-related mortality worldwide. Over half a million new cases
are diagnosed worldwide each year (1). Treatment of HCC consists of surgery
with/without radiotherapy (RT) and chemotherapy. Ionizing radiation
(IR) is a powerful modality of cancer treatment. For its advantage
in preserving normal tissues, fractionated RT is often applied in
preoperative treatment. However, hepatoma cells are prone to
present radioresistant properties following irradiation, which are
correlated with increased recurrence and therapeutic failure in
numerous patients (2). PRF/PLC/5
HCC cells are derived from a primary hepatocellular carcinoma of an
individual with positivity for serum hepatitis B virus surface
antigen (HBsAg) (3). The PLC/PRF/5R
cells of the present study were derived from PLC/PRF/5 cells, which
show a higher radioresistance following irradiation treatment. As
chronic hepatitis B virus (HBV) is the most common cause of HCC,
this cell model is suitable for investigation of the exact
molecular mechanisms underlying the adaptive resistance of HCC
cells to IR.
Previous studies have suggested that glutathione
S-transferases (GSTs), which have well-established roles in
detoxification and clearance of ROS, have novel functions in
pivotal signaling pathways for cell proliferation or apoptosis. For
instance, GSTM1 functions as a negative regulator of MEKK1, which
act as a upstream kinase in MAPK/P38 and JNK/ERK signal pathways
(4). GSTP, another isoform of GSTs,
has functions in ERK activation and JNK repression, which may
account for the dual roles of GSTP in enhancement of growth
inhibition and inhibition of apoptosis (5–7). GSTM3
has overlapping substrates with GSTM1, and GSTM genes are
tandem-aligned on chromosome 1q13.3 (8). Thus, they may have similar functions
as the signal transducer in key signal pathways. If GSTM3 is
involved in regulating proliferation/apoptosis-associated pathways,
it may potentially affect the radioresistance property in cells. As
GSTM3 is always expressed at low levels in cancer tissues compared
with that in normal tissues, it may possess tumor suppressive
properties in tumor initiation and development (8–10).
GSTM3 was also downregulated in radioresistant HCC cells according
to the present findings. Therefore, the present study aimed to
investigate the role of GSTM3 as a tumor suppressor and a novel
target of radioresistance.
In order to identify whether GSTM3 is a potential
therapeutic target to conquer the resistance in HCC, the current
study explored the effects of GSTM3 overexpression on the
radioresistance of PLC/PRF/5R cells to fractionated RT and the
potential mechanism. The current study revealed a novel function of
GSTM3 in regulating the adaptive radioresistance in HCC cells.
Materials and methods
Cell culture and transfection
Human HCC PLC/PRF/5R cell lines were purchased from
American Type Culture Collection (Manassas, VA, USA). The
HBsAg-producing cell line, PLC/PRF/5, was chosen for analyses. The
radiation-resistant cell line, PLC/PRF/5R, was derived from
fractional irradiated PLC/PRF/5 cells (six weeks of FR with total
doses of 21 Gy). Cells were cultured in Dulbecco’s modified Eagle’s
medium (Hyclone Laboratories, Logan, UT, USA) with 10% fetal calf
serum (FCS; Gibco, Invitrogen, Grand Island, NY, USA), 100 units/ml
penicillin and 100 μg/ml streptomycin at 37°C, 5% CO2
and 95% humidity. GSTM3 cDNA was inserted into a pcDNA3.1 vector
(Invitrogen Life Technologies, Carlsbad, CA, USA) resulting in
pcDNA3.1-GSTM3. The plasmids were transfected into PLC/PRF/5R cells
using Lipofectamine 2000 (Invitrogen Life Technologies) to generate
PLC/PRF/5R-con (pcDNA3.1/His-TOPO) cells and PLC/PRF/5R-GSTM3
(pcDNA3.1/His-TOPO-GSTM3) cells. The wild-type vector of
pcDNA3.1/His-TOPO was used as a control. Stable cell lines were
established by G418 screening. For the irradiation experiments,
cells were treated with 2, 4, 6, 8 or 0 Gy (control) of X-ray using
a linear accelerator (VARIAN 21EX 6 MV; Varian Medical Systems,
Inc., Palo Alto, CA USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Cells were washed with phosphate-buffered saline
(PBS) and homogenized in TRIzol reagent (Invitrogen Life
Technologies). Total RNA was extracted and the concentration was
determined on a Nanodrop ND-1000 spectrophotometer (Nanodrop,
Wilmington, DE, USA). RNA (2 μg) was reverse-transcribed using a
PrimeScript™ RT reagent kit (Takara Shuzo Co. Ltd., Dalian, China)
according to the manufacturer’s instructions. GAPDH was used as a
housekeeping gene, and the relative expression of the apoptosis and
proliferation-related genes was detected using qPCR on a
LightCycler 480 real-time instrument (Roche Diagnostics, Mannheim,
Germany). The reactions were performed in a final volume of 20 μl
containing 10 μl 2X SYBR Green Master mix (Takara Shuzo Co. Ltd.),
1 μl cDNA template and 0.25 μM each primer. The qPCR cycling
parameters were as follows: 95°C for 30 sec; and 40 cycles of 95°C
for 10 sec, 56°C for 25 sec and 72°C for 25 sec. At the end of the
PCR, melting curve analyses of amplification products were carried
out to confirm that only one product was amplified. The relative
mRNA level of each gene was calculated as 2−ΔΔCt
(11). Differences between the
PLC/PRF/5, PLC/PRF/5R-con and PLC/PRF/5R-GSTM3 groups were assessed
by one-way analysis of variance. The primer sequences for GAPDH and
proliferation/apoptosis-related molecules are listed in Table I.
 | Table IPrimers used in reverse
transcription-polymerase chain reaction. |
Table I
Primers used in reverse
transcription-polymerase chain reaction.
| Gene name | Forward primers
(5′-3′) | Reverse primers
(5′-3′) | Fragments (bp) |
|---|
| GAPDH |
TGCCGTCTAGAAAAACCTGC |
ACCCTGTTGCTGTAGCCAAA | 485 |
| Bcl-2 |
GTGGAGGAGCTCTTCAGGGA |
AGGCACCCAGGGTGATGCAA | 368 |
| Bax |
AAGAAGCTGAGCGAGTGT |
GGAGGAAGTCCAATGTC | 462 |
| p21 |
CACCCTAGTTCTACCTCAGGCA |
ACTCCCCCATCATATACCCCT | 412 |
| p27 |
ACGGGAGCCCTAGCCTGGAGC |
TGCCCTTCTCCACCTCTTGCC | 500 |
| p53 |
TGGCCATCTACAAGCAGTCACA |
GCAAATTTCCTTCCACTCGGAT | 375 |
Western blot analysis
Cells were lysed in RIPA buffer plus protease
inhibitors one day after irradiation, and protein was quantified by
the bradford assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
In total, 20 μg protein of each group was separated by SDS-PAGE and
transferred to a PVDF membrane. The protein bands were
immunoblotted with the primary antibodies [rabbit polyclonal
anti-human B-cell chronic lymphocytic leukemia/lymphoma 2 (Bcl-2),
rabbit polyclonal anti-human Bax, mouse monoclonal anti-human p21,
mouse monoclonal anti-human p27, mouse monoclonal anti-human p53
and rabbit polyclonal anti-human β-actin; 1:500; Cell Signaling
Technology, Inc., Beverly, MA, USA] and secondary antibody
(polyclonal goat anti-rabbit-HRP IgG; 1:1000; Pierce Biotechnology,
Rockford, IL, USA). Following this, the bands were visualized by
enhanced chemiluminescence substrate solution (GE Healthcare,
Little Chalfont, UK) on a SyngeneTM gel imaging analysis
system (Syngene, Cambridge, UK). β-actin was considered as a
loading control.
Cell proliferation assay
Cell viability was examined by Cell Counting Kit-8
(CCK-8; Dojindo, Kumamoto, Japan) according to the manufacturer’s
instructions. Briefly, cells were plated at a density of
0.5×104 cells/well in 96-well plates. Following
incubation, cells were treated with 10 μl CCK-8 solution for 3 h.
Absorbance was measured at 450 nm using a multiwell plate reader
(Synergy HT, Bio-Tek, Winooski, VT, USA).
Clonogenic assay
Twenty-four hours after irradiation, cells were
plated on 9-cm dishes with 1,000 cells/well and maintained in
culture for 10 days to alow colony formation. When visible colonies
(at least 50 cells) emerged, they were fixed with 95% methanol,
stained with 0.5% crystal violet and counted. Plating efficiency
(PE) and surviving fraction (SF) were calculated as follows: PE =
(colony number/seeded cell number) ×100%, SF = (PEirradiated
group/PEnon-irradiated group) ×100%. The cell
survival curve was fitted using Origin 8.0 software (OriginLab
Corporation, Northampton, MA, USA).
Hoechst 33258 staining
Cells were washed with PBS and fixed using 4%
paraformaldehyde for 30 min. Cells were then washed again and
incubated in Hoechst 33258 solution for 10 min in the dark, at
37°C. Finally, the solution was removed, and cells were washed and
observed with an inverted fluorescence microscope (Leica DMI300B,
Leica, Wetzlar, Germany).
Flow cytometric analysis of the cell
cycle
A total of 1×106 cells were trypsinized,
washed and resuspended in 1 ml PBS with 5% FCS. Subsequently, 3 ml
ice-cold ethanol was added to fix the samples. Prior to FCS
analysis, samples were centrifuged at 1,500 × g, the supernatant
was discarded and the cell pellet was resuspended in 1 ml PBS with
propidium iodide (Sigma-Aldrich, St. Louis, MO, USA) and RNAse
(Huamei, Shanghai, China) and incubated overnight at 4°C.
Flow cytometric analysis of
apoptosis
Cells were washed twice with ice-cold PBS, and then
resuspended in binding buffer. Following this, cells were stained
in Annexin V-fluorescein isothiocyanate (BD Biosciences, San Jose,
CA, USA) and propidium iodide (Sigma-Aldrich), according to the
manufacturer’s instructions. After a 30-min incubation in the dark
at 37°C, cells were immediately examined on a FACSCalibur (BD
Biosciences) flow cytometer and the data was analyzed with
CellQuest software (BD Biosciences). All of the samples were tested
in triplicate. The cell apoptosis rate was calculated following the
formula: (Napoptotic cells/Ntotal cells)
×100%.
Statistical analyses
All experiments were independently repeated two or
three times. The data are presented as the mean ± standard
deviation. Statistical analyses were performed using SPSS 16.0
software (SPSS, Inc., Chicago, IL, USA). Values of P<0.05 were
considered to indicate a statistically significant difference.
Results
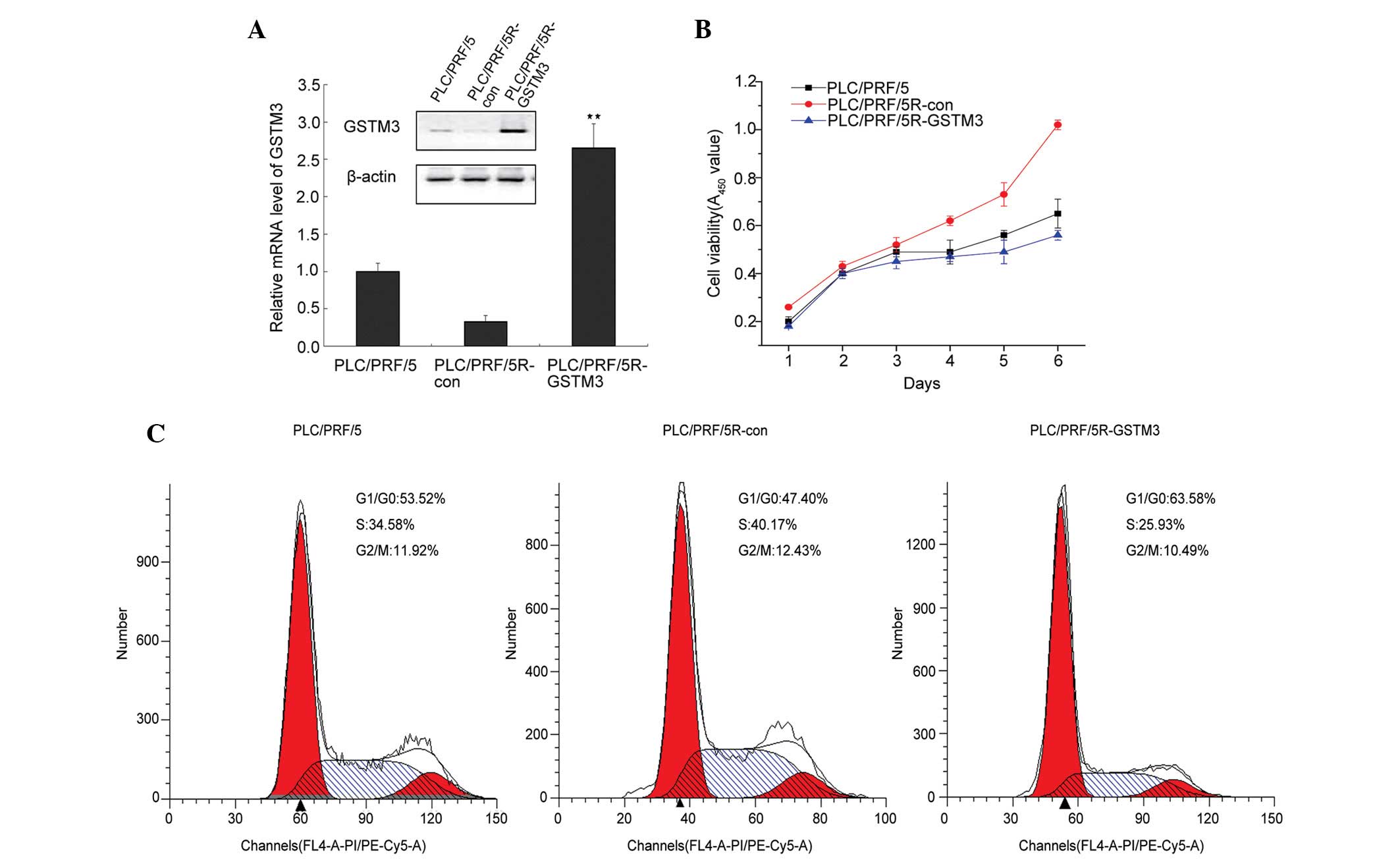
GSTM3 is expressed at low levels in
adaptive radioresistant HCC cells
The expression levels of GSTM3 were detected in the
radiation-resistant cell line, PRF/PLC/5R, and the parental cell
line, PRF/PLC/5. GSTM3 expression levels in the PRF/PLC/5 cells
were three-fold greater than those in the PRF/PLC/5R cells.
Additionally, GSTM3 protein expression levels, examined by western
blotting, confirmed the RT-qPCR results (Fig. 1A). GSTM3 expression was also
measured in the HepG3BR and HepG3B radioresistant and
radiosensitive HCC cell lines: GSTM3 expression levels were
significantly lower in the radioresistant types (data not shown).
These results suggest that low levels of GSTM3 expression may be
associated with the adaptive radioresistance of HCC.
GSTM3 overexpression inhibits the growth
of PRF/PLC/5R cells
To investigate the role of GSTM3 in the
radiosensitivity of PRF/PLC/5R cells, a GSTM3-overepressing vector
was transfected into PRF/PLC/5R cells to construct a cell line that
stably expressed GSTM3 (PRF/PLC/5R-GSTM3). This showed a nine-fold
increase in GSTM3 transcript levels in contrast to PRF/PLC/5R-con
cells, which was confirmed by immunoblotting (Fig. 1A). As GSTM1 plays a role in
inhibiting the activity of proliferation regulator MEKK (4), we proposed that it may act as a
transducer in tumor growth signaling pathways. Studies have
reported low GSTM3 expression in several types of tumors (8–10), and
this has been found to be associated with cisplatin resistance in
breast cancer (10). Therefore, we
hypothesized that GSTM3 may be similar to GSTM1, and act as a
signal transductor in proliferation-related signaling pathways. We
first examined the effect of GSTM3 on cell proliferation. The CCK-8
assay showed that GSTM3 dramatically inhibited the cell
proliferation of PRF/PLC/5R cells. The cell viability of
PLC/PRF-5R-GSTM3 was less than that of the PLC/PRF/5R-con at day
four (P<0.05; Fig. 1B). Cell
cycle analysis indicated that GSTM3 induced G0/G1-phase arrest and
subsequently a decreased number of cells in the S-phase compared to
the control PRF/PLC/5R-con and PRF/PLC/5 cells (Fig. 1C). Meanwhile, the apoptotic cell
number increased to 11.4% in the PRF/PLC/5R-GSTM3 cells (Fig. 2A and C).
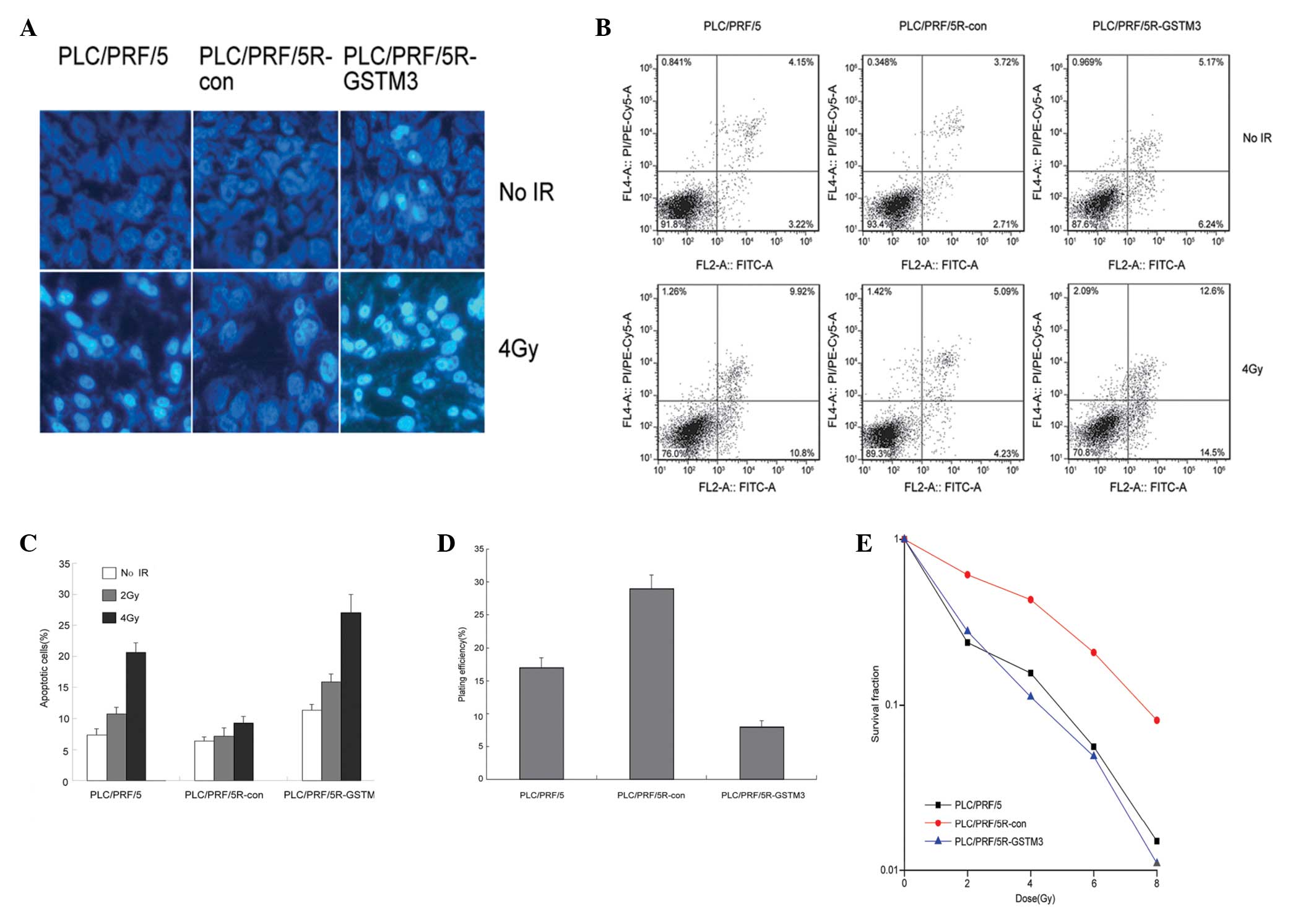
GSTM3 overexpression sensitizes
PLC/PRF/5R cells to irradiation
To assess whether GSTM3 overexpression could reverse
the radioresistance of the PLC/PRF/5R cell line, we assessed the
role of GSTM3 on cell apoptosis in radiated PLC/PRF/5R cells. As
cell death by apoptosis may occur following IR, we first used
Hoechst33258 staining to measure the morphological changes of
apoptotic HCC cells induced by IR (Fig.
2B). Cells with characteristic bright blue fluorescent nuclei
were defined as apoptotic cells, as the condensed chromatin of
apoptosis cells was strongly stained. As shown in Fig. 2B, apoptotic cells in the PLC/PRF/5
and PLC/PRF/5R-GSTM3 groups were dramatically increased compared
with those of the radioresistant PLC/PRF/5R-con group at 4 Gy IR.
The Annexin V/PI staining assay was conducted to determine the
apoptotic cell number induced by irradiation. PLC/PRF/5 cells were
sensitive to irradiation; the apoptotic rate increased from 7.37 to
20.7% post-IR. The PLC/PRF/5R-con cells were resistant to
irradiation; the apoptotic rate increased from 6.43 to 9.32% after
4 Gy IR. Although it increased, this difference was not
significant, and therefore, no significant difference was
identified in the levels of apoptosis post-IR. GSTM3 overexpression
significantly increased the apoptotic rate from 11.41 to 27.1% in
the PLC/PRF/5R-GSTM3 group (Fig.
2A). Similar data were observed in the 2-Gy irradiated HCC
cells (Fig. 2C). The effect of
GSTM3 on the radiation response of PLC/PRF/5R cells was also
evaluated using a clonogenic assay. GSTM3 overexpression decreased
the PE of PLC/PRF/5R cells to 8% in contrast to the control
PLC/PRF/5R-con cells to 29% (Fig.
2D), and increased the radiosensitivity of PLC/PRF/5R cells, as
shown by the notable reduction in SF of clonogenic cells at 2, 4, 6
and 8 Gy (SF of PLC/PRF-5R-GSTM3 vs. PLC/PRF-5R-con; P<0.05).
The sensitization enhancement ratio was 1.5 (Fig. 2E).
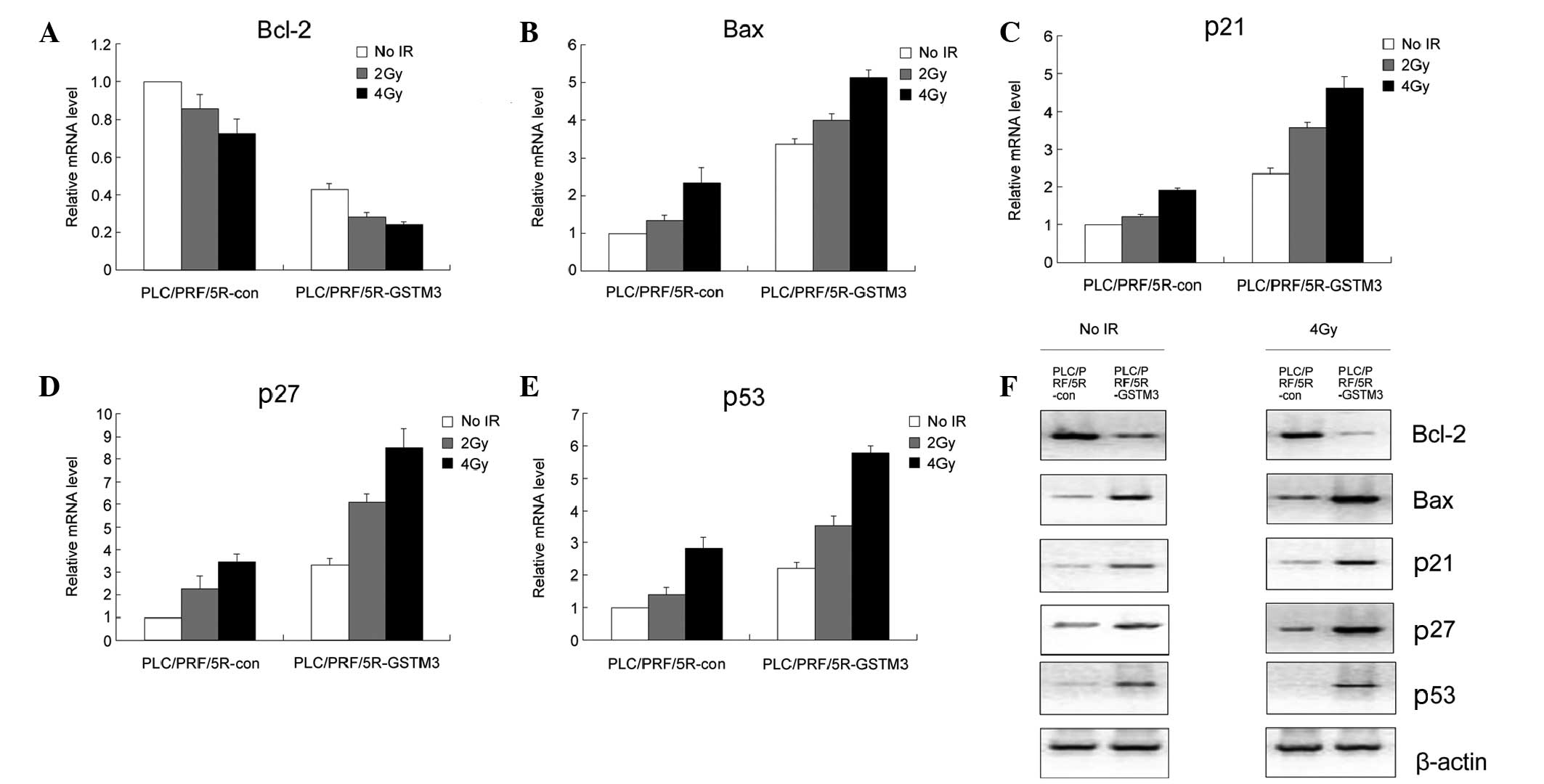
GSTM3 overexpression affects the
expression of cell cycle/apoptosis-related proteins
As abovementioned, GSTM3 overexpression may be
correlated with the arrest of cell cycle progression and
potentiation of apoptosis in PLC/PRF/5R cells. Therefore, the
expression of a number of molecules involved in these two processes
was analyzed. First, RT-qPCR was used to compare the mRNA
expression levels of Bcl-2, Bax, p21, p27 and p53. GSTM3
overexpression markedly decreased the Bcl-2 expression and
increased the Bax, p21, p27 and p53 expression in contrast to that
in the PLC/PRF/5R-con cells. Following irradiation, the expression
of Bax, p21, p27 and p53 were elevated, but GSTM3 overexpression
induced markedly higher expression levels of these molecules
compared with those in the control cells (Fig. 3A–E). Data regarding the protein
expression levels of Bcl-2, Bax, p21, p27 and p53 at 0 and 4 Gy
also confirmed the results of the RT-qPCR (Fig. 3F).
Discussion
While RT provides useful palliation on HCC, advanced
HCC remains incurable, as the previously sensitive tumor rapidly
becomes resistant to IR. The acquired resistance may arise from
tolerance of stimuli, decreased transducer activity, inactivation
of normal cell death, decreased pro-apoptotic factors, altered cell
cycling factors and proliferation signals (12–14).
Therefore, there is an urgent requirement for the identification of
novel biomarkers which may predict the prognosis and help to select
the patients that will benefit from RT. Furthermore, elucidating
the underlying molecular mechanisms of the radioresistance of HCC
cells is important for the development of novel target agents for
radiation sensitization, and for exploring optimal radiotherapeutic
regimens.
GSTs are members in gene superfamilies of the phase
II detoxification enzymes. GSTs can be classified into four
categories on the basis of sequence homology and chromosomal
localization, namely α, μ, π and θ (15). GSTM3 is a GST-Mu class member, which
is known to be involved in regulating the susceptibility to cancer
(8,16–19).
However, little is known regarding the role of GSTM3 in
antitumorigenesis or IR sensitization.
To the best of our knowledge, this is the first
study to anlayze the novel effect of GSTM3 on RT sensitization. The
results showed that GSTM3 overexpression significantly increased
the cell apoptosis rate and apoptosis-related gene expression in
PLC/PRF/5R cells, at the base level and after IR. Bcl-2 and Bax
both are members of Bcl-2 family. The former is an anti-apoptosis
protein and the latter is a pro-apoptosis protein (20). In the present study, overexpression
of GSTM3 downregulated the Bcl-2 expression and upregulated the Bax
expression. This was consistent with the effects of GSTM3 in
promoting the apoptosis of radioresistant PLC/PRF/5R cells. In
addition, p21, p27 and p53 expression was increased in
GSTM3-overexpressing cells. p21 and p27 are famous cyclin-dependent
kinase inhibitors. They are members in the Cip/Kip family, which
can arrest the cell cycle and serve as tumor suppressors (21). Cell cycle arrest contributes to
proliferation inhibition and apoptosis induction. In the present
flow cytometric assay, GSTM3 overexpression was shown to cause
G0/G1 arrest in PLC/PRF/5R cells. Lamore and Wondrak (22) and Cabello et al (23) reported that GSTM3 upregulation is
coordinated with an increase in p21 and p53 expression. p53 is a
pivotal molecule in HBV-caused HCC for its transcriptional
suppression on HBx (24). GSTM3 may
act as a tumor suppressor and overcome the effect of HBx in
inhibiting p53 transcription. According to the present clonogenic
assay results, GSTM3 overexpression increased the radiosensitivity
of PLC/PRF/5R cells and decreased the SF of clonogenic cells at 2,
4, 6 and 8 Gy. In addition, the expression of Bax, p21, p27 and p53
were markedly higher following irradiation in the
GSTM3-overexpressing cells. Overall, GSTM3 regulation of apoptosis-
or proliferation-related pathways may contribute to the effects of
GSTM3 in reversing the radioresistance of HCC cells.
In conclusion, the results of the present study
suggest that GSTM3 is not only a enzyme in phase II
biotransformation, but also has novel functions in irradiation
sensitization of HCC cells. GSTM3 regulates the expression of cell
cycle/apoptosis-related proteins and, thus, inhibits the
proliferation and reverses the radioresistance of PLC/PRF/5R cells.
Therefore, previously discovered enzymes may exert effects other
than their superficial functions. This study provides promising
prospects for novel radiotherapeutic regimens acheiving more
benefits from fractionated RT. However, the exact molecular
mechanisms of GSTM3 in sensitizing cells to IR and the optimal
target sensitizer in HCC require further investigation.
References
|
1
|
El-Serag HB: Hepatocellular carcinoma. N
Engl J Med. 365:1118–1127. 2011.
|
|
2
|
Kim JJ and Tannock IF: Repopulation of
cancer cells during therapy: an important cause of treatment
failure. Nat Rev Cancer. 5:516–525. 2005.
|
|
3
|
Leveille-Webster CR and Arias IA:
Establishment and serial quantification of intrahepatic xenografts
of human hepatocellular carcinoma in severe combined
immunodeficiency mice, and development of therapeutic strategies to
overcome multidrug resistance. Clin Cancer Res. 2:695–706.
1996.
|
|
4
|
Ryoo K, Huh SH, Lee YH, et al: Negative
regulation of MEKK1-induced signaling by glutathione S-transferase
Mu. J Biol Chem. 279:43589–43594. 2004.
|
|
5
|
Lu M, Xia L, Luo D, Waxman S and Jing Y:
Dual effects of glutathione-S-transferase pi on
As2O3 action in prostate cancer cells:
enhancement of growth inhibition and inhibition of apoptosis.
Oncogene. 23:3945–3952. 2004.
|
|
6
|
Adler V, Yin Z, Fuchs SY, et al:
Regulation of JNK signaling by GSTp. EMBO J. 18:1321–1334.
1999.
|
|
7
|
Yin Z, Ivanov VN, Habelhah H, Tew K and
Ronai Z: Glutathione S-transferase p elicits protection against
H2O2-induced cell death via coordinated
regulation of stress kinases. Cancer Res. 60:4053–4057. 2000.
|
|
8
|
Yu KD, Fan L, Di GH, et al: Genetic
variants in GSTM3 gene within GSTM4-GSTM2-GSTM1-GSTM5-GSTM3 cluster
influence breast cancer susceptibility depending on GSTM1. Breast
Cancer Res Treat. 121:485–496. 2010.
|
|
9
|
Tan X, Zhai Y, Chang W, et al: Global
analysis of metastasis-associated gene expression in primary
cultures from clinical specimens of clear-cell renal-cell
carcinoma. Int J Cancer. 123:1080–1088. 2008.
|
|
10
|
Smith L, Welham KJ, Watson MB, et al: The
proteomic analysis of cisplatin resistance in breast cancer cells.
Oncol Res. 16:497–506. 2007.
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCt method. Methods. 25:402–408. 2001.
|
|
12
|
Chen YJ, Lin CP, Hsu ML, et al: Sonic
hedgehog signaling protects human hepatocellular carcinoma cells
against ionizing radiation in an autocrine manner. Int J Radiat
Oncol Biol Phys. 80:851–859. 2011.
|
|
13
|
Urick ME, Chung EJ, Shield WP III, et al:
Enhancement of 5-fluorouracil-induced in vitro and in vivo
radiosensitization with MEK inhibition. Clin Cancer Res.
17:5038–5047. 2011.
|
|
14
|
Piao LS, Hur W, Kim TK, et al:
CD133+ liver cancer stem cells modulate radioresistance
in human hepatocellular carcinoma. Cancer Lett. 315:129–137.
2012.
|
|
15
|
Mannervik B, Awasthi YC, Board PG, et al:
Nomenclature for human glutathione transferases. Biochem J.
282:305–306. 1992.
|
|
16
|
Salinas-Souza C, Petrilli AS and de Toledo
SR: Glutathione S-transferase polymorphisms in osteosarcoma
patients. Pharmacogenet Genomics. 20:507–515. 2010.
|
|
17
|
Chatzimichalis M, Xenellis J,
Tzagaroulakis A, et al: GSTT1, GSTM1, GSTM3 and NAT2 polymorphisms
in laryngeal squamous cell carcinoma in a Greek population. J
Laryngol Otol. 124:318–323. 2010.
|
|
18
|
Golka K, Schmidt T, Seidel T, et al: The
influence of polymorphisms of glutathione S-transferases M1 and M3
on the development of human urothelial cancer. J Toxicol Environ
Health A. 71:881–886. 2008.
|
|
19
|
Jain M, Kumar S, Lal P, et al: Role of
GSTM3 polymorphism in the risk of developing esophageal cancer.
Cancer Epidemiol Biomarkers Prev. 16:178–181. 2007.
|
|
20
|
Hao YX, Zhong H, Yu PW, et al: Effects of
HIF-1alpha on human gastric cancer cell apoptosis at different
CO(2) pressures. Clin Exp Med. 9:139–147. 2009.
|
|
21
|
Liu F, Wei YG, Luo LM, et al: Genetic
variants of p21 and p27 and hepatocellular cancer risk in a Chinese
han population: a case-control study. Int J Cancer. 132:2056–2064.
2013.
|
|
22
|
Lamore SD and Wondrak GT: Zinc pyrithione
impairs zinc homeostasis and upregulates stress response gene
expression in reconstructed human epidermis. Biometals. 24:875–890.
2011.
|
|
23
|
Cabello CM, Lamore SD, Bair WB III, et al:
DCPIP (2,6-dichlorophenolindophenol) as a genotype-directed redox
chemotherapeutic targeting NQO1*2 breast carcinoma. Free
Radic Res. 45:276–292. 2011.
|
|
24
|
He HB, Wu XL, Yu B, et al: The effect of
desacetyluvaricin on the expression of TLR4 and P53 protein in Hepg
2.2.15. Hepat Mon. 11:364–367. 2011.
|