Introduction
The liver kinase B1 (LKB1)/serine-threonine kinase
11 tumor suppressor gene encodes a ubiquitously expressed and
evolutionarily conserved serine-threonine kinase. LKB1 was
originally identified as a tumor suppressor gene due to its
association with an increased risk of malignancy and Peutz-Jeghers
syndrome (PJS; a rare autosomal dominant syndrome characterized by
benign polyps of the gastrointestinal tract) (1). An increased incidence of carcinomas of
the gastrointestinal tract, as well as breast, ovarian, uterine,
cervical, lung and testicular cancer, has been observed in PJS
patients and their relatives (2,3).
Somatic mutations in LKB1 have also been observed in sporadic
pulmonary, breast, pancreatic and biliary cancers and melanomas
(4–8). Although LKB1 has been identified as a
tumor suppressor, its function in chemoresistance remains
unclear.
Gemcitabine, also known as
2′,2′-difluorodeoxycytidine (dFdC) or Gemzar, is an analogue of
deoxycytidine, with two fluorine atoms substituted at the
2′-position of the ribose ring. It has been widely used in the
treatment of several types of cancer, including non-small cell
lung, pancreatic and metastatic breast cancer (9–11).
After entering the cell, gemcitabine is phosphorylated to
gemcitabine monophosphate (dFdCMP), diphosphate (dFdCDP) and
triphosphate (dFdCTP) in a stepwise manner (12,13).
dFdCTP may be incorporated into DNA, leading to strand termination
and cellular apoptosis. Other mechanisms associated with the
anticancer effects of gemcitabine include ribonucleotide reductase
(RNR) inhibition, RNA incorporation and thymidylate synthase
inhibition (12). However, one of
the main factors hindering gemcitabine application is its
chemoresistance. Resistance to gemcitabine may be attributed to
cellular events during drug uptake and metabolism, and a number of
molecular markers have been found to correlate with gemcitabine
sensitivity (14).
To the best of our knowledge, the present study is
the first to investigate the effect of forced LKB1 expression on
chemosensitivity to gemcitabine in breast cancer cells.
Materials and methods
Cell lines and cell culture
The human breast cancer cell line, MDA-MB-231, was
obtained from the American Type Culture Collection (Manassas, VA,
USA). The MDA-MB-231 gemcitabine resistance subline was developed
by over one year of exposure to gemcitabine, beginning with 1 μM
and increasing stepwise to 840 μM. Cells were maintained in
Leibovitz’s L-15 medium (Sigma-Aldrich, St. Louis, MO, USA)
containing 10% fetal bovine serum (Gibco-BRL, Carlsbad, CA, USA),
100 units/ml penicillin and 100 μg/ml streptomycin (Gibco-BRL) at
37°C in a humidified atmosphere with 5% CO2.
Adenovirus production and
transfection
The coding sequence of LKB1 was amplified by
polymerase chain reaction (PCR) and then cloned into the adenoviral
vector plasmid with the flag tag, using the Gateway Cloning system
(Invitrogen Life Technologies, Carlsbad, CA, USA). The LKB1
expression plasmids and corresponding vector plasmids were
transfected into HEK-293T cells with the gag-pol packaging and
VSV-G envelope plasmids using polyethylenimine reagent. The medium
containing virus particles was harvested. Cells were seeded into a
25-cm2 cell culture flask 1 day prior to transfection,
and 2 ml of collected medium containing virus particles, 2 ml L15
medium and 4 μl 8 mg/ml polybrene mixture was added to transfect
the cells. Medium containing 1 μg/ml puromycin was used to screen
for stable transfected cells.
RNA isolation, reverse transcription and
quantitative PCR (qPCR)
Total RNA was extracted from cells with TRIzol
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) and the
reverse transcription reaction was performed using the Reverse
Transcription system (Promega Corporation, Shanghai, China). qPCR
was performed using SYBR Premix Ex Taq (Takara Biotechnology
(Dalian) Co., Ltd., Dalian, China), and GAPDH was used as an
internal control. The primer sequences used were as follows:
Forward, 5′-GAGAAGCGTTTCCCAGTGTG-3′ and reverse,
5′-CCCAGGTCGGAGATTTTGA-3′ for LKB1; and forward,
5′-GGATTTGGTCGTATTGGG-3′ and reverse, 5′-GGATTTGGTCGTATTGGG-3′ for
GAPDH.
Western blot analysis
Cells were washed twice with phosphate-buffered
saline (PBS) followed by T-PER tissue extraction buffer (Thermo
Fisher Scientific, Rockford, IL, USA) supplemented with protease
and phosphatase inhibitor tablets (Roche Diagnostics, Indianapolis,
IN, USA) on ice for 30 min. The lysates were then centrifuged at
12,000 g for 30 min at 4°C. Next, a total of 20 μg protein was
resolved by sodium dodecyl sulfate-polyacrylamide gel
electrophoresis and transferred to polyvinylidene fluoride film.
The film was then incubated with blocking solution containing 5%
bovine serum albumin [BSA; Sangon Biotech (Shanghai) Co., Ltd.,
Shanghai, China) in Tris-buffered saline with Tween 20 [Sangon
Biotech (Shanghai) Co., Ltd.] at room temperature for 1 h.
Subsequently, the film was immunoblotted with polyclonal rabbit
anti-human LKB1 (Calbiochem, Dormstadt, Gemany), monoclonal mouse
anti-human ribonucleotide reductase M1 (RRM1; Abcam, Hong Kong,
China), monoclonal rabbit anti-human phosphorylated (p)-ATR,
monoclonal rabbit anti-human p-CHK1, monoclonal rabbit anti-human
p-ATR, monoclonal rabbit anti-human p-CHK2 and monoclonal mouse
anti-human γH2AX (Cell Signaling Technology, Inc., Danvers, MA,
USA) antibodies, or monoclonal mouse anti-human GAPDH antibody
(Sigma-Aldrich). Polyclonal goat anti-rabbit and goat anti-mouse
IgG (H+L) (Jackson ImmunoResearch Laboratories, Inc., West Grove,
MA, USA) secondary antibodies were used. The signals were detected
using chemiluminescent horseradish peroxidase substrate (Millipore,
Billerica, MA, USA) according to the manufacturer’s
instructions.
Cytotoxicity and cell proliferation
assays
Cells at the logarithmic growth phase were plated at
a density of 2×103 cells/well in 96-well plates.
Following overnight adherence, complete medium was replaced with
medium containing 16 different concentrations of gemcitabine
ranging between 0.00001 and 900 μmol/l. Following gemcitabine
treatment for six days, cell cytotoxicity was measured by the Cell
Counting Kit-8 (CCK-8) kit (Dojindo Laboratories, Kumamoto, Japan).
The IC50 value of gemcitabine was estimated from
semilogarithmic dose-response curves generated using GraphPad Prism
(GraphPad Software, Inc., La Jolla, CA, USA). Each experiment was
performed in triplicate. Cell proliferation rate was also measured
using the CCK-8 kit every 24 hours for seven days and a
proliferation curve was generated using Prism 5 software (GraphPad
Software, Inc.).
Colony formation assay
The cells were seeded at a density of 500 cells per
60-mm dish. Following 24 h, complete medium was replaced with
medium containing different concentrations of gemcitabine (0.0001,
0.0003 and 0.0006 μmol/l). In addition, 100 cells of each type were
seeded as standard controls, without gemcitabine treatment. After
14 days, clones were fixed and stained with crystal violet
containing 40% methanol for ~30 min. Stained clones with a diameter
of >1 mm were counted and standardized. The cloning efficiency
was calculated using the following formula: Cloning efficiency (%)
= (clone number/total cell number)/(control clone number/control
total cell number) × 100. Each independent experiment was performed
in triplicate. For colony formation assays without gemcitabine
treatment, 300 cells were seeded in each dish.
Immunofluorescence assay
Cells were seeded at a density of 2×104
cells/well onto coverslips in a 24-well plate. Following 30 min of
fixation in 4% paraformaldehyde, 10 min of permeabilization with
0.5% Triton X-100 solution and 1 h of blockage with 5% BSA in PBS,
cells were incubated with primary monoclonal mouse anti-human γH2AX
antibody (Millipore) for 2 h, and Alexa Fluor 555 polyclonal goat
anti-rabbit IgG (H+L) secondary antibody (red; Invitrogen Life
Technologies) for 1 h. DAPI was used to stain the nuclei. All
images were captured using a confocal laser microscope (Leica TCS
SP5; Leica, Mannheim, Germany).
Statistical analysis
Statistical analysis was performed using SPSS
software, version 17.0 (SPSS, Inc., Chicago, IL, USA). One-way
analysis of variance was used to determine the statistical
significance of differences between experimental groups. P<0.05
was considered to indicate a statistically significant
difference.
Results
LKB1 suppresses the proliferation of
breast cancer cells
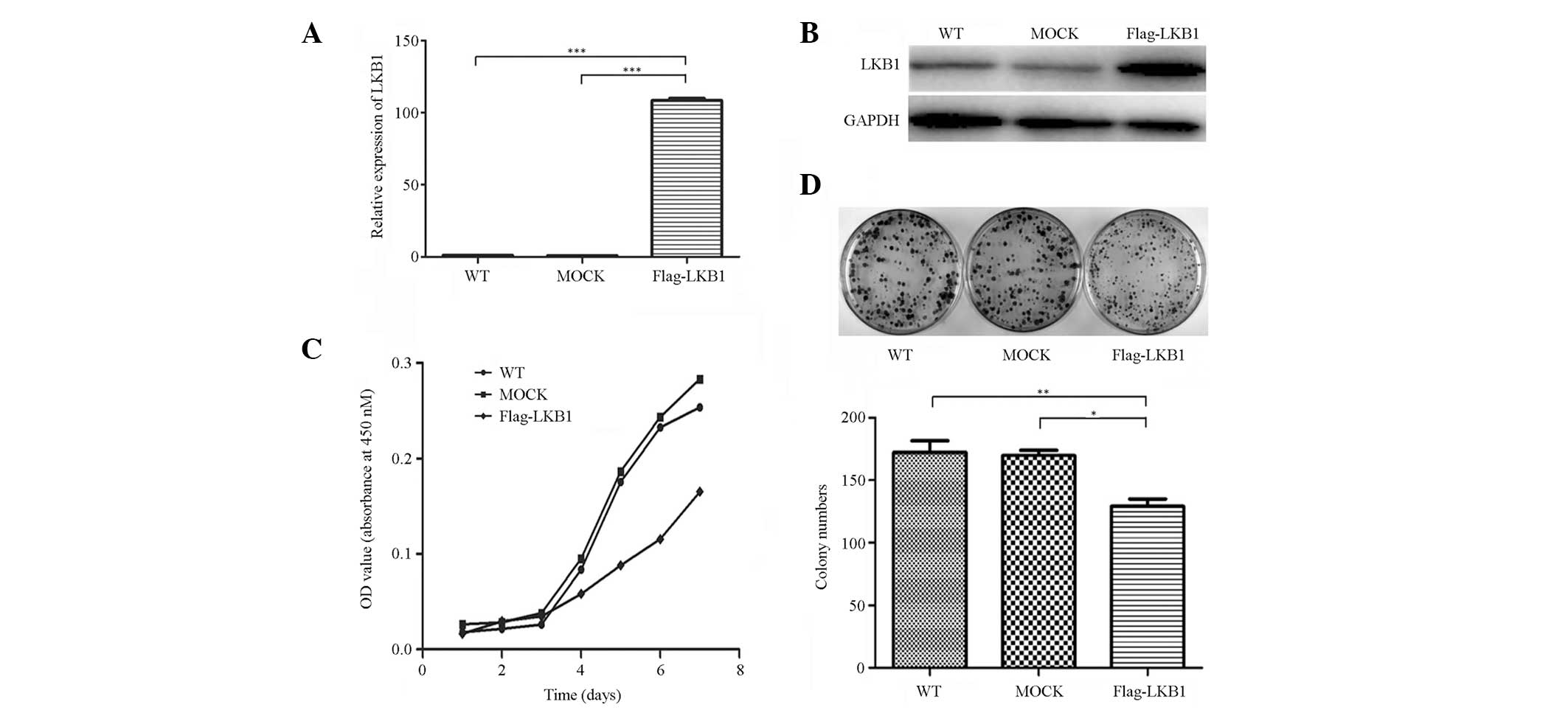
Successful construction of LKB1 stably transfected
cells and the mock-transfected controls was determined by qPCR, and
further confirmed by western blot analysis (Fig. 1A and B). To investigate the function
of LKB1 in breast cancer cells, cell proliferation and colony
formation assays were performed. The results indicated that LKB1
markedly decreased cell proliferation rate and clonogenicity
(Fig. 1C and D). These results were
consistent with previous studies, which have identified LKB1 as a
tumor suppressor gene (15,16).
Forced expression of LKB1 is associated
with increased gemcitabine chemoresistance
To further explore the effect of LKB1 on gemcitabine
sensitivity in breast cancer cells, a cytotoxicity assay was
conducted. Semilogarithmic dose-response curves from the
cytotoxicity assays of wild-type, mock-transfected,
LKB1-transfected and gemcitabine-resistant MDA-MB-231 cells are
shown in Fig. 2A. In addition, the
estimated IC50 values of these cells are shown in
Fig. 2B. The IC50 value
of LKB1-transfected cells was 46.07 nM, which was more than
seven-fold greater that of MDA-MB-231 cells (5.85 nM) (P<0.001),
more than five-fold greater than that of mock-transfected cells
(7.93 nM) (P<0.001), and less than that of gemcitabine-resistant
cells (61.27 nM) (P<0.001).
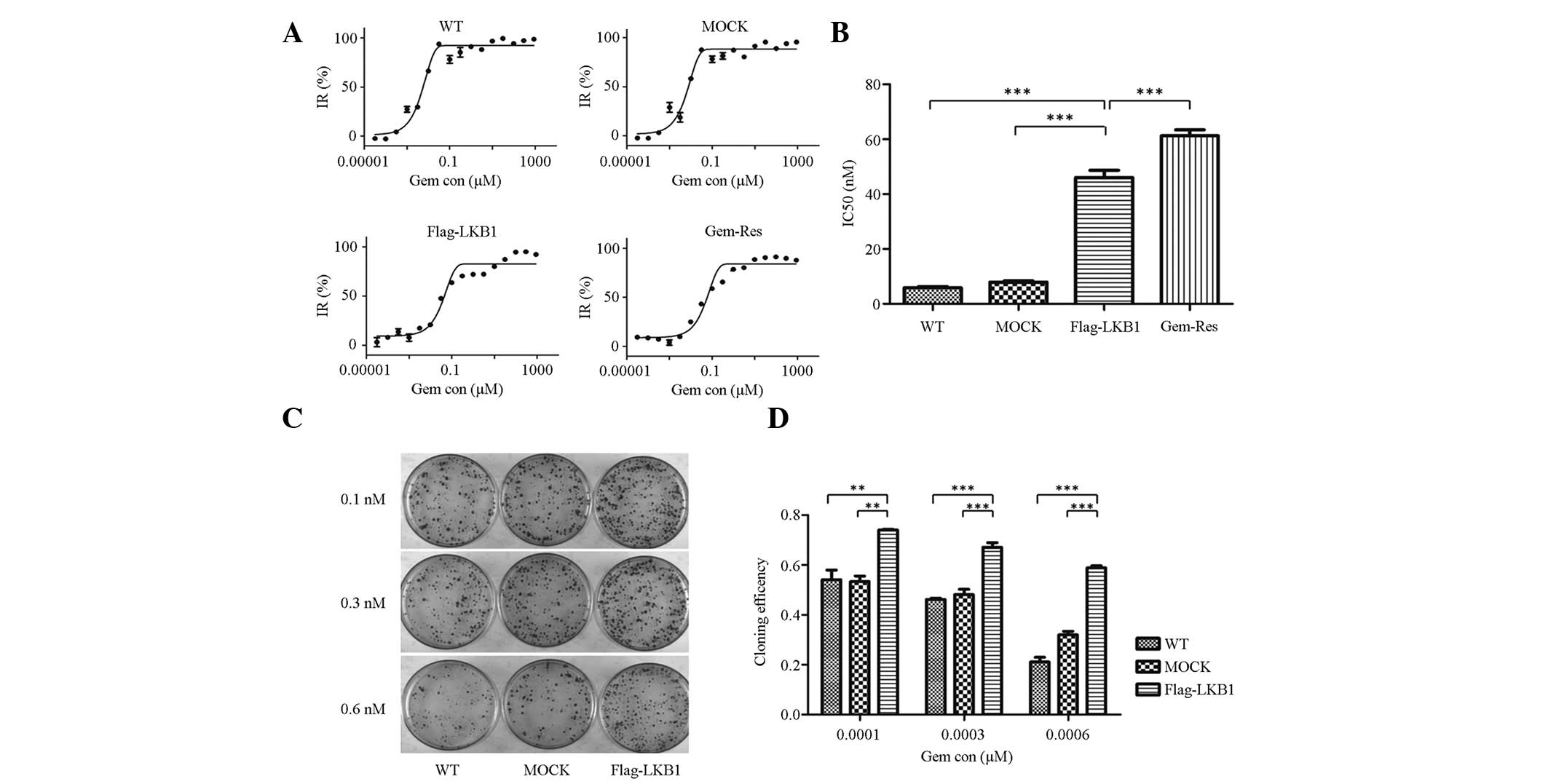 | Figure 2Forced LKB1 expression is associated
with increased gemcitabine chemoresistance. (A) Semilogarithmic
dose-response curves of cytotoxicity assays as measured by Cell
Counting Kit-8 assay. (B) Estimated IC50 value of
wild-type, mock-transfected, LKB1-transfected and
gemcitabine-resistant sublines of MDA-MB-231 cells. The
IC50 value of LKB1-transfected cells was higher than
that of wild-type and mock-transfected cells; however, it was lower
than that of the gemcitabine-resistant subline. Each independent
experiment was performed in triplicate and data are presented as
the mean ± SD. (C) Cloning efficiency of the three cell lines at
three different concentrations (0.1, 0.3 and 0.6 nM) of
gemcitabine. The control group contained 100 cells/dish without
gemcitabine. Cloning efficiency was calculated using the following
formula: Cloning efficiency (%) = (clone number/total cell
number)/(control clone number/control total cell number) × 100.
Each independent experiment was performed in triplicate and the
data are presented as the mean ±SD. (D) Colony formation assays
revealed higher clonogenicity in LKB1-transfected cells when
compared with wild-type and mock-transfected cells, in the presence
of gemcitabine. (*P<0.05, **P<0.01 and
***P< 0.001). IR, inhibition rate; Gem con,
gemcitabine concentration; LKB1, liver kinase B1; WT, wild
type. |
In the colony formation assay, the colony number
decreased with increasing gemcitabine concentration in each cell
line. Furthermore, in each gemcitabine concentration group, LKB1
increased the colony number when compared with wild-type
(P<0.01) or mock-transfected (P<0.01) cells (Fig. 2C and D). Cloning efficiency was
standardized by control group (100 cells/dish, without gemcitabine)
to exclude the inaccuracy of manual operation and culturing
conditions. Overall, the cytotoxicity and colony formation assays
indicated that LKB1 enhances chemoresistance to gemcitabine.
LKB1 overexpression alleviates
gemcitabine-induced DNA damage
γH2AX expression was analyzed using
immunofluorescence to assess the effect of LKB1 on the DNA damage
caused by gemcitabine. Cells were fixed at various time points (0,
12 and 24 h) following treatment with 1 μM gemcitabine for 24 h,
followed by an immunofluorescence assay to detect γH2AX foci, the
marker of DNA double-strand breaks (DSBs), in cell nuclei. Cells
with clear red foci in the nucleus were considered to be DNA
damage-positive, while those without were considered as negative
(Fig. 3A). Three fields were
randomly selected for each coverslip to calculate positive rates
(Fig. 3B). In each cell line,
positive rates decreased in a time-dependent manner. At each time
point, the positive rate for LKB1-transfected cells was lower than
that for wild-type and mock-transfected cells, and the differences
were statistically significant.
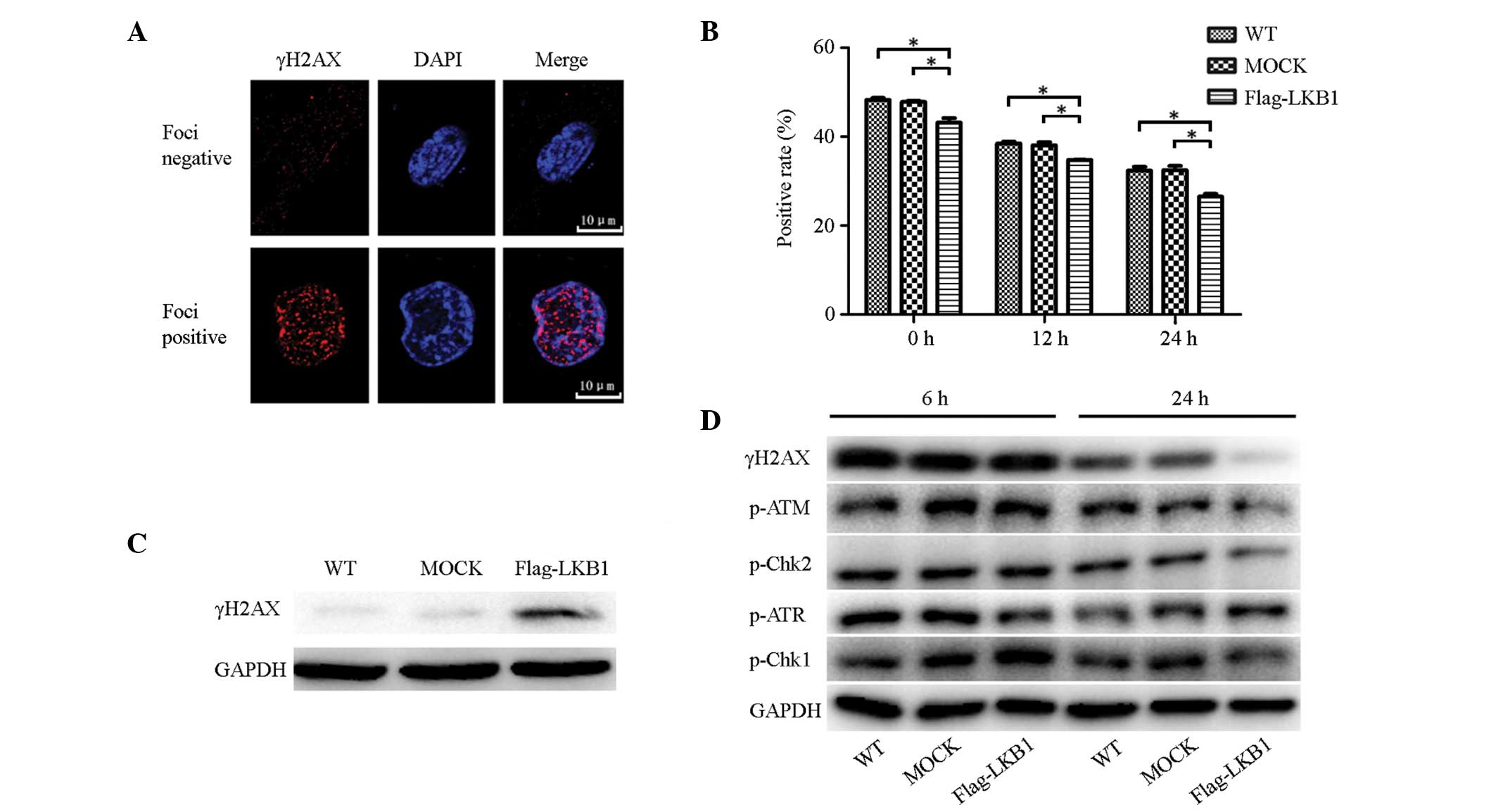 | Figure 3LKB1 overexpression alleviates
gemcitabine-induced DNA damage possibly by affecting the ATM-CHK2
pathway. (A) Negative and positive examples of γH2AX foci in the
immunofluorescence assay. (B) Positive rates of γH2AX foci in
cells. Following gemcitabine treatment (1 μM; 24 h), three time
points (0, 12 and 24 h) were selected to perform the
immunofluorescence assay. Three fields were randomly selected for
each coverslip and positive rates were calculated. LKB1 decreased
positive rates of γH2AX foci. Each independent experiment was
performed three times and data are presented as the mean ± SD. (C)
γH2AX expression in normal conditions was analyzed by western blot
analysis. LKB1 increased γH2AX expression. (D) Expression of DNA
damage-associated proteins (p-ATR, -CHK1, -ATR, -CHK2 and γH2AX) 6
and 24 h following gemcitabine treatment (1 μM; 24 h),
respectively. LKB1 affected the expression of p-ATR, p-CHK2 and
γH2AX in the 24 h group. (*P< 0.05,
**P<0.01 and ***P<0.001). LKB1, liver
kinase B1; p, phosphorylated; WT, wild-type. |
In addition, western blot analysis was used to
compare γH2AX expression levels prior to and following treatment
with 1 μM gemcitabine for 24 h. In the absence of gemcitabine, low
levels of γH2AX were expressed in wild-type and mock-transfected
cells, while LKB1-transfected cells expressed significantly more
γH2AX (Fig. 3C). Notably, this
trend was gradually reversed following gemcitabine treatment and, 6
h following gemcitabine withdrawal, γH2AX expression in the
wild-type and mock-transfected cells increased to the same level as
that in the LKB1-transfected cells. Furthermore, 24 h following
gemcitabine withdrawal, as DNA repair progressed, γH2AX expression
began to decline, decreasing most significantly in LKB1-transfected
cells (Fig. 3D). This result
indicated that LKB1 expression activates a process of resistance to
the DNA damage caused by gemcitabine. This protective function was
not evident directly following exposure to gemcitabine; however, it
was identified several hours following the withdrawal of
gemcitabine treatment.
In addition to γH2AX, other proteins associated with
DNA damage (p-ATR, -CHK1, -ATM and -CHK2) were examined by western
blot analysis following gemcitabine treatment (1 μM for 24 h). The
protein expression 24 h following gemcitabine withdrawal was
generally lower than that of the 6-h group, indicating that less
DNA damage had occurred with time. In the 6-h group, p-ATM and
-CHK2 expression was almost equivalent in the three cell lines;
whereas in the 24 h group, the expression was markedly lower in the
LKB1-transfected cells, exhibiting the same trend as γH2AX.
However, p-ATR and -CHK1 did not exhibit a similar trend (Fig. 3D). This indicated that LKB1 only
regulates the ATM-CHK2 pathway, and not the ATR-CHK1 pathway, in
response to gemcitabine.
Ectopic expression of LKB1 increases the
expression of cytidine deaminase (CDA)
Following on from the previous results, the
mechanism whereby LKB1 enhances gemcitabine resistance was further
investigated. CDA is an enzyme that catabolizes gemcitabine to
dFdU, thus abolishing the cytotoxicity of gemcitabine. RRM1 is a
component of RNR, which antagonizes the DNA-damaging effect of
gemcitabine. The expression of the two enzymes was detected by
western blot analysis in different cells. As shown in Fig. 4A, CDA and RRM1 expression was higher
in gemcitabine-resistant sublines than in wild-type MDA-MB-231
cells. However, only CDA expression was upregulated in
LKB1-transfected cells (Fig. 4B).
Therefore, we hypothesized that the upregulation of CDA expression
is one mechanism by which LKB1 reduces sensitivity to gemcitabine
in breast cancer cells.
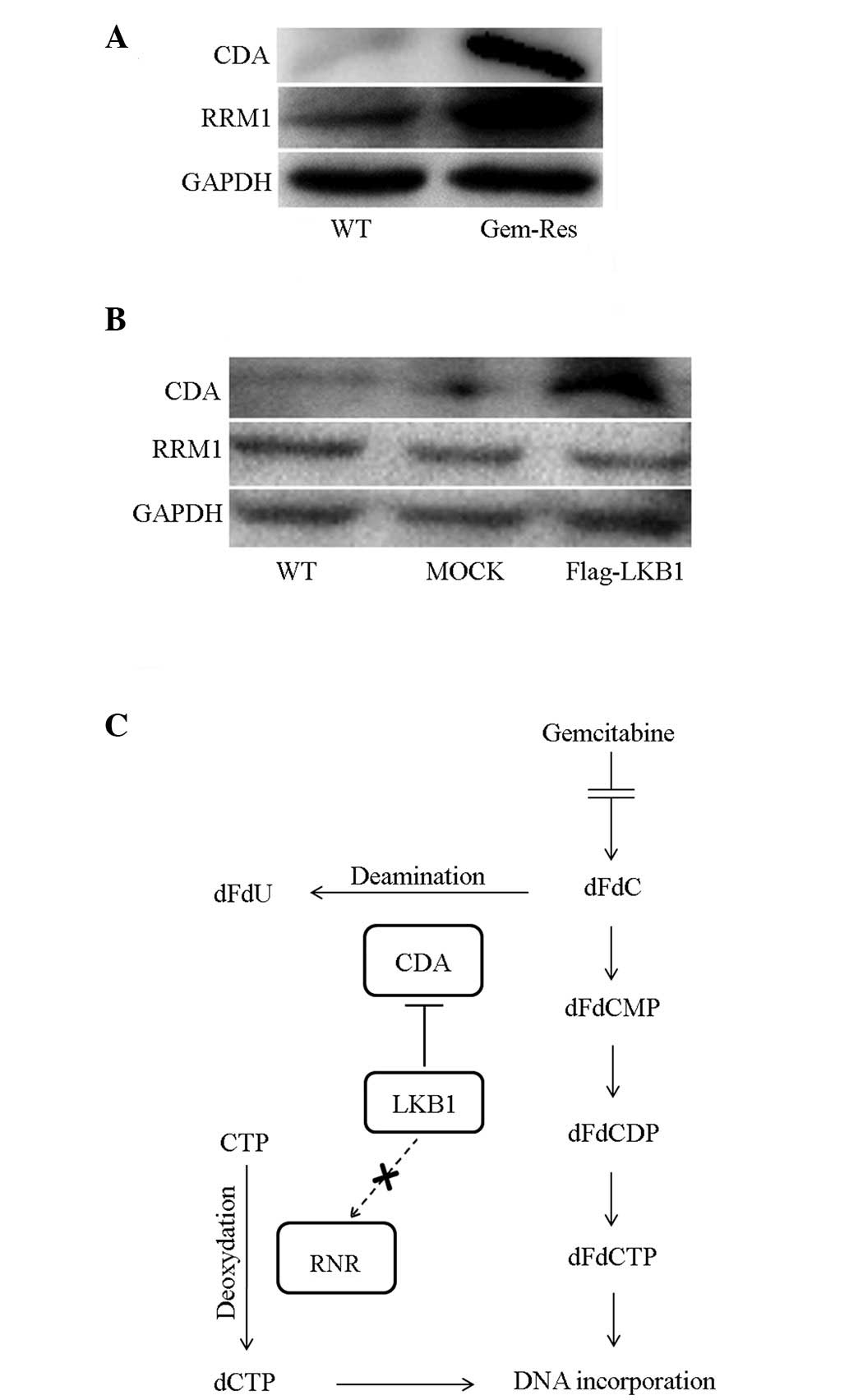 | Figure 4Ectopic expression of LKB1 increases
the expression of CDA. (A) CDA and RRM1 expression in wild-type and
gemcitabine-resistant MDA-MB-231 cells was analyzed by western blot
analysis. The two enzymes were upregulated in the
gemcitabine-resistant subline. (B) Western blot analysis of CDA and
RRM1 expression in cells. LKB1 increased CDA expression. (C)
Metabolism and mechanism of gemcitabine in the cytoplasm. CDA is
the enzyme that deaminizes gemcitabine to dFdU, eliminating the
anticancer effect of gemcitabine. RRM1 is a subunit of RNR, which
is the key enzyme involved in the production of the
deoxyribonucleotide pool and DNA synthesis (12). LKB1, liver kinase B1; CDA, cytidine
deaminase; RNR, ribonucleotide reductase; RRM1,
ribonucleoside-diphosphate reductase; dFdCMP, difluoro-deoxyuridine
monophosphate; dFdCDP, difluoro-deoxyuridine diphosphate; dFdCTP,
difluoro-deoxyuridine triphosphate; WT, wild type. |
Discussion
LKB1 has been identified as a tumor suppressor gene
in numerous studies (15); however,
its function in chemoresistance remains unclear. To the best of our
knowledge, the present study was the first to identify that LKB1
enhances gemcitabine resistance in the breast cancer MDA-MB-231
cell line, possibly by accelerating gemcitabine degradation and
protecting cells from DNA damage.
The cytotoxicity and colony formation assays
supplemented with gemcitabine revealed that LKB1-transfected cells
grew faster and formed more colonies than wild-type and
mock-transfected MDA-MB-231 cells, indicating that LKB1 enhanced
the chemosensitivity to gemcitabine. Notably, in the proliferation
and colony formation assays without gemcitabine treatment, LKB1
suppressed cell proliferation and reduced cell clonogenicity, which
was consistent with the results of previous studies (16). Overall, the protective role of LKB1
in breast cancer cells was specific to the gemcitabine
environment.
To further investigate the association between LKB1
and gemcitabine, and as gemcitabine inhibited DNA synthesis and
caused DNA damage, the extent of DNA damage was evaluated in
LKB1-transfected, mock-transfected and wild-type MDA-MB-231 cells
following exposure to gemcitabine. γH2AX is a generally accepted
sensitive marker of DNA damage, and in particular DSBs. γH2AX is
the phosphorylated form of H2AX, a member of the H2A family. The
H2A family is a histone family that facilitate the organization of
chromatin. The detection of γH2AX by immunofluorescence is a widely
used and convenient method of assessing DNA damage, which enables
the visualization of the γH2AX foci, and the quantification of
γH2AX foci indicates the degree of DNA damage. A number of
genotoxic insults may lead to DSBs, including ionizing radiation,
ultraviolet light exposure, drugs and chemicals, among others, of
which gemcitabine is one (17,18).
Previous studies have shown that gemcitabine treatment may induce
γH2AX foci in the nucleus, indicating damage to the DNA (19,20).
In the present study, according to the immunofluorescence assay and
calculated foci positive rates, in each cell line, foci positive
rates generally decreased in a time-dependent manner following
gemcitabine withdrawal. This may have been a result of gemcitabine
consumption or DNA repair. At each time period, LKB1 transfected
cells exhibited a lower foci positive rate than wild-type or
mock-transfected cells (P<0.01).
Additionally, γH2AX expression was investigated by
western blot analysis prior to and following gemcitabine treatment.
On the one hand, γH2AX expression was the highest in
LKB1-transfected cells in the absence of gemcitabine; however, on
the other hand, it was the lowest following exposure to
gemcitabine. Therefore, we hypothesized that LKB1 acts as a tumor
suppressor by increasing cell vulnerability to DSBs in the normal
environment; however, in the presence of gemcitabine, other
mechanisms were activated in LKB1-transfected cells to eliminate
this vulnerability to DSBs. Overall, we hypothesized that LKB1
prevented breast cancer cells from DSBs caused by gemcitabine, and
enhanced the chemoresistance to gemcitabine.
In addition to γH2AX, other proteins involved in DNA
damage pathways (p-ATR, -CHK1, -ATM and -CHK2) were examined
following exposure to gemcitabine. The ATR-CHK1 and ATM-CHK2
protein kinase pathways are the two predominant signaling pathways
activated by DNA damage (21). ATM
is activated primarily by radiation and genotoxins that induce DNA
DSBs (22), while ATR is activated
via recruitment to the single-stranded DNA (23). The activation of ATM at DSB sites
may affect multiple local substrates, including CHK2 and H2AX,
inducing their phosphorylation (24,25).
As shown in Fig. 3D, among p-ATR,
-CHK1, -ATM and -CHK2, only p-ATM and -CHK2 exhibited the same
trend as γH2AX, supporting the hypothesis that LKB1 prevents DSB
caused by gemcitabine via the ATM-CHK2 pathway.
The enhancement of resistance to gemcitabine is
possibly due to the involvement of LKB1 in gemcitabine metabolism;
therefore, the expression of CDA and RRM1 in wild-type and
transfected cells was assessed. The metabolism and mechanism of
gemcitabine following entrance to the cytoplasm is shown in
Fig. 4C. CDA is the enzyme that
catabolizes gemcitabine to dFdU, preventing the stepwise conversion
of gemcitabine into dFdCMP, dFdCDP and dFdCTP and inhibiting DNA
synthesis. Previous study has revealed the function of CDA in
gemcitabine resistance (26).
Another important enzyme involved in gemcitabine metabolism is RNR.
It converts ribonucleotide 5′-diphosphates to
2′-deoxyribonucleotide-5′-diphosphates, which is the rate limiting
step for deoxyribonucelotide production and DNA synthesis. RRM1 is
a subunit of RNR, which has been reported to contribute to
gemcitabine chemoresistance in vivo and in vitro
(27,28). In the present study, CDA levels were
highest in LKB1-transfected cells; however, no significant
differences in RRM1 levels were identified between wild-type,
mock-transfected and LKB1-transfected cells. The higher levels of
CDA were partly responsible for the decreased number of DSBs and
increased resistance to gemcitabine in LKB1-transfected cells.
However, other mechanisms may account for LKB1 enhancing
gemcitabine resistance.
To the best of our knowledge, the current study is
the first to indicate that LKB1 is a tumor suppressor and
gemcitabine desensitizer, simultaneously. As a result, when
patients exhibit LKB1 expression in breast cancer, the application
of gemcitabine may not achieve the expected outcome. Since LKB1 is
a potential treatment target for malignant tumors, its ability to
enhance chemoresistance to gemcitabine must be considered during
subsequent oncological management. In conclusion, the results of
the current study provide a novel insight into the antitumor
activity of gemcitabine and indicate a distinct mechanism for
improving the efficacy of gemcitabine, which is feasible for
clinical application in breast cancer patients.
Acknowledgements
This study was supported by the Shanghai Municipal
Health Bureau (grant no. 20124164) and the Shanghai Municipal
Science and Technology Commission (grant no. 124119a4700). The
study was performed at the Cancer Center and Cancer Institute,
Shanghai Medical College, Fudan University. The authors would like
to thank Mr. Jianmin Luo for supplying the gemcitabine-resistant
subline of MDA-MB-231 cells and all coworkers at the Cancer Center
and Cancer Institute.
References
|
1
|
Mehenni H, Gehrig C, Nezu J, et al: Loss
of LKB1 kinase activity in Peutz-Jeghers syndrome, and evidence for
allelic and locus heterogeneity. Am J Hum Genet. 63:1641–1650.
1998.
|
|
2
|
Lim W, Olschwang S, Keller JJ, et al:
Relative frequency and morphology of cancers in STK11 mutation
carriers. Gastroenterology. 126:1788–1794. 2004.
|
|
3
|
Hearle N, Schumacher V, Menko FH, et al:
Frequency and spectrum of cancers in the Peutz-Jeghers syndrome.
Clin Cancer Res. 12:3209–3215. 2006.
|
|
4
|
Fenton H, Carlile B, Montgomery EA, et al:
LKB1 protein expression in human breast cancer. Appl
Immunohistochem Mol Morphol. 14:146–153. 2006.
|
|
5
|
Ji H, Ramsey MR, Hayes DN, et al: LKB1
modulates lung cancer differentiation and metastasis. Nature.
448:807–810. 2007.
|
|
6
|
Sanchez-Cespedes M, Parrella P, Esteller
M, et al: Inactivation of LKB1/STK11 is a common event in
adenocarcinomas of the lung. Cancer Res. 62:3659–3662. 2002.
|
|
7
|
Wang ZJ, Churchman M, Campbell IG, et al:
Allele loss and mutation screen at the Peutz-Jeghers (LKB1) locus
(19p13.3) in sporadic ovarian tumours. Br J Cancer. 80:70–72.
1999.
|
|
8
|
Ylikorkala A, Avizienyte E, Tomlinson IP,
et al: Mutations and impaired function of LKB1 in familial and
non-familial Peutz-Jeghers syndrome and a sporadic testicular
cancer. Hum Mol Genet. 8:45–51. 1999.
|
|
9
|
Reck M, von Pawel J, Zatloukal P, et al:
Phase III trial of cisplatin plus gemcitabine with either placebo
or bevacizumab as first-line therapy for nonsquamous non-small-cell
lung cancer: AVAil. J Clin Oncol. 27:1227–1234. 2009.
|
|
10
|
Cunningham D, Chau I, Stocken DD, et al:
Phase III randomized comparison of gemcitabine versus gemcitabine
plus capecitabine in patients with advanced pancreatic cancer. J
Clin Oncol. 27:5513–5518. 2009.
|
|
11
|
Chan S, Romieu G, Huober J, et al: Phase
III study of gemcitabine plus docetaxel compared with capecitabine
plus docetaxel for anthracycline-pretreated patients with
metastatic breast cancer. J Clin Oncol. 27:1753–1760. 2009.
|
|
12
|
Mini E, Nobili S, Caciagli B, Landini I
and Mazzei T: Cellular pharmacology of gemcitabine. Ann Oncol.
17v7–v12. (Suppl 5)2006.
|
|
13
|
Wong A, Soo RA, Yong WP and Innocenti F:
Clinical pharmacology and pharmacogenetics of gemcitabine. Drug
Metab Rev. 41:77–88. 2009.
|
|
14
|
Bergman AM, Pinedo HM and Peters GJ:
Determinants of resistance to 2′,2′-difluorodeoxycytidine
(gemcitabine). Drug Resist Update. 5:19–33. 2002.
|
|
15
|
Katajisto P, Vallenius T, Vaahtomeri K, et
al: The LKB1 tumor suppressor kinase in human disease. Biochim
Biophys Acta. 1775:63–75. 2007.
|
|
16
|
Zhuang ZG, Di GH, Shen ZZ, Ding J and Shao
ZM: Enhanced expression of LKB1 in breast cancer cells attenuates
angiogenesis, invasion, and metastatic potential. Mol Cancer Res.
4:843–849. 2006.
|
|
17
|
Bonner WM, Redon CE, Dickey JS, et al:
GammaH2AX and cancer. Nature Rev Cancer. 8:957–967. 2008.
|
|
18
|
Mah LJ, El-Osta A and Karagiannis TC:
gammaH2AX: a sensitive molecular marker of DNA damage and repair.
Leukemia. 24:679–686. 2010.
|
|
19
|
Al-Ejeh F, Darby JM and Brown MP: The La
autoantigen is a malignancy-associated cell death target that is
induced by DNA-damaging drugs. Clin Cancer Res. 13:5509s–5518s.
2007.
|
|
20
|
Dufau I, Frongia C, Sicard F, et al:
Multicellular tumor spheroid model to evaluate spatio-temporal
dynamics effect of chemotherapeutics: application to the
gemcitabine/CHK1 inhibitor combination in pancreatic cancer. BMC
Cancer. 12:152012.
|
|
21
|
Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K
and Linn S: Molecular mechanisms of mammalian DNA repair and the
DNA damage checkpoints. Ann Rev Biochem. 73:39–85. 2004.
|
|
22
|
Lee JH and Paull TT: ATM activation by DNA
double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science.
308:551–554. 2005.
|
|
23
|
Zou L and Elledge SJ: Sensing DNA damage
through ATRIP recognition of RPA-ssDNA complexes. Science.
300:1542–1548. 2003.
|
|
24
|
Smith J, Tho LM, Xu N and Gillespie DA:
The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and
cancer. Adv Cancer Res. 108:73–112. 2010.
|
|
25
|
Lee JH and Paull TT: Direct activation of
the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science.
304:93–96. 2004.
|
|
26
|
Bardenheuer W, Lehmberg K, Rattmann I, et
al: Resistance to cytarabine and gemcitabine and in vitro selection
of transduced cells after retroviral expression of cytidine
deaminase in human hematopoietic progenitor cells. Leukemia.
19:2281–2288. 2005.
|
|
27
|
Bergman AM, Eijk PP, Ruiz van Haperen VW,
et al: In vivo induction of resistance to gemcitabine results in
increased expression of ribonucleotide reductase subunit M1 as the
major determinant. Cancer Res. 65:9510–9516. 2005.
|
|
28
|
Nakahira S, Nakamori S, Tsujie M, et al:
Involvement of ribonucleotide reductase M1 subunit overexpression
in gemcitabine resistance of human pancreatic cancer. Int J Cancer.
120:1355–1363. 2007.
|