Introduction
The proliferation and invasion of the first
trimester human trophoblast cell is important in embryonic
development. During the first trimester of human pregnancy,
extravillous trophoblasts (EVTs) grow out from anchoring villi,
invade the maternal decidua and remodel the uterine spiral
arteries. Furthermore, deficient EVT invasion is associated with
complications during pregnancy, including intrauterine growth
restriction and pre-eclampsia (1).
Abnormal proliferation and invasion of trophoblast cells may lead
to gestational trophoblastic diseases, which encompass a spectrum
of associated diseases, including hydatidiform mole, invasive mole,
choriocarcinoma and placental-site trophoblastic tumor (2).
The proliferation and invasion of the first
trimester human trophoblast cell is influenced by multiple
regulatory factors, including growth factors, cytokines, adhesion
molecules, proteases, matrix-derived components and oxygen tension
(3,4). Transforming growth factor-β1 (TGF-β1)
is involved in trophoblast proliferation and invasion (5–7).
Previous studies have revealed that on the one hand, the
proliferation and invasion of choriocarcinoma may be inhibited by
TGF-β1 (5–12); however, on the other hand, TGF-β1
may enhance normal trophoblast functions (9,11–12).
TGF-β receptor I (TβRI) and SMADs are the key downstream mediators
of transcriptional responses to TGF-β. TGF-β activates TβRI and
TβRII, which results in the phosphorylation of receptor-regulated
SMAD2/3 proteins, which are associated with the common mediator,
SMAD4. The SMAD2/3/4 complex translocates to the nucleus, binds DNA
and regulates the transcription of a number of genes (13). TGF-β signaling is known to be
involved in the regulation of proliferation, differentiation and
apoptosis of numerous cells (14).
It is difficult to study the proliferation and
invasion of human trophoblast cells in vivo, therefore the
analysis of the growth and invasion of trophoblast cells in culture
is essential for understanding the functions of the placenta. Two
types of trophoblast cell models have been identified: Primary cell
models and trophoblast cell lines. However, studies using primary
cell culture are hindered by practical issues. For example, it is
difficult to obtain the normal first trimester human chorionic
tissues, and the number and purity of primary trophoblasts
available are limited. Furthermore, cultures become contaminated
with other placental cell types, including fibroblasts (15). In addition, trophoblast primary
cells cannot be maintained for long time periods in culture.
Therefore, these cells are not suitable for studies involving
genetic manipulations that often require long-term cell culture
(12). The HTR-8/SVneo cell line
was established by introducing the gene encoding simian virus 40
large T antigen into first trimester human trophoblasts. It is an
immortalized trophoblast cell line, which exhibits a number of
similar characteristics to those of parental trophoblast cells
(12). Graham et al
(12) revealed that HTR-8/SVneo
cells were inhibited by recombinant TGF-β1, which is identical to
that of the parental trophoblast cells (12). Therefore, in the present study, the
HTR-8/SVneo cell line was selected to investigate the TGF-β/SMAD
signaling pathway and the involvement of such in the proliferation
and invasion of trophoblast cells.
The proliferation of HTR-8/SVneo cells was
investigated using MTT assays and the invasion ability was
determined by Transwell assay, following the incubation of cells
with various concentrations of TGF-β1. In addition, the mRNA
expression levels of TβRI, SMAD4, SMAD3, SMAD7 and tissue inhibitor
of metalloproteinases-1 (TIMP-1) were examined to elucidate which
factor leads to the abnormal regulation exhibited by TGF-β1. The
implications of the results and comparison with previous data have
been discussed.
Materials and methods
Cell culture
HTR-8/SVneo cells (the 90th passage) were provided
by Queen’s University at Kingston (Kingston, Canada). The cells
were cultured in an incubator with an atmosphere of 5%
CO2 at 37°C in RPMI-1640 medium (Hyclone, Waltham, MA,
USA) supplemented with 10% fetal bovine serum (FBS; Hangzhou
Sijiqing Biological Engineering Materials Co, Ltd, Hangzhou,
China), 1 mM pyruvic acid sodium salt, 2 mM glutathione, 100 U/ml
penicillin and 100 μg/ml streptomycin. The cells were then
subcultured with 0.25% trypsin and 0.02% EDTA (Sigma-Aldrich, St.
Louis, MO, USA) when the cell growth reached 70–80%, and the
density of subcultured cells was 1:3.
Analysis of cell viability by MTT
assay
A total of 1×105 cells/ml in 200-μl
aliquots were plated in 96-well plates and allowed to adhere
overnight. Next, the cells were incubated for 24, 48 and 72 h with
or without various concentrations of TGF-β1 (200 μl for a final
concentration of 0, 0.05, 0.5, 5, 10, 12.5, 25, 50, 100 and 200
μg/l; six wells for each concentration; PeproTech Inc., Rocky Hill,
NJ, USA). The cell viability was determined using MTT reagent
(Gibco-BRL, Carlsbad, CA, USA) and the absorbance was determined at
a wavelength of 492 nm using a microplate reader (Multiskan MK3;
Thermo Fisher Scientific, Waltham, MA, USA). The experiment was
repeated five times.
Transwell invasion assay
A thin layer of growth factor-reduced diluted
Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) was added to the
upper chambers of 6.5-mm Transwell inserts with polycarbonate
membrane filters containing 8-μm pores (Corning Inc, Acton, MA,
USA). Inserts were placed into 24-well culture plates and incubated
at 37°C for 4 h. Next, 500 μl aliquots of RPMI-1640 supplemented
with 20% FBS were added to the lower chambers. Concurrently,
1×105 cells/ml in 200 μl aliquots of serum-free
RPMI-1640 containing 0, 1, 10 and 100 μg/l TGF-β1, respectively,
were added to the upper chambers of the inserts and cultured for 48
h. Cells remaining on the upper surface of the Matrigel layer were
removed using a cotton swab and dried, and the cells that had
invaded the bottom of the membrane were fixed in 4%
paraformaldehyde for 10 min and stained with hematoxylin. The
invasive cells were observed under a light microscope (Nikon 80i;
Nikon, Tokyo, Japan) at ×100 magnification. Images of five random
fields were captured for each membrane and the average number of
cell numbers detected in each field was calculated. The total
number of transmigrated cells was counted by selecting 10 random
fields and observing the number of apoptotic cells using a light
microscope (magnification, ×200; BH-2; Olympus Corporation, Tokyo,
Japan).
Reverse transcription-polymerase chain
reaction (RT-PCR)
A total of 3×105 cells were plated in
six-well plates and incubated for 24 h, followed by culturing the
cells with serum-free RPMI-1640 for 24 h. Following cell
synchronization, the medium was changed and added to TGF-β1 with 0,
1, 10 and 100 μg/l, respectively, as the final concentrations to
incubate the cells for an additional 48 h. Total RNA from each
group of cells was isolated using TRIzol reagent (Invitrogen Life
Technologies, Carlsbad, CA, USA) according to the manufacturer’s
instructions. Next, 1% agarose gel electrophoresis was performed to
separate the RNA and 28S, 18S and 5S bands were detected. The M-MLV
first-strand synthesis system kit (Invitrogen Life Technologies)
and 1 μg RNA with oligo(dt)20 primers were used to synthesize the
cDNA. The Taq kit [Takara Biotechnology (Dalian) Co., Ltd., Dalian,
China] was used for PCR amplification. The primers used are shown
in Table I. β-actin was used as the
internal control (16). The
amplification of cDNA was performed over varying cycles: 94°C for 2
min, 30 cycles at 94°C for 30 sec, followed by the indicated
annealing temperature (Table I) for
30 sec, and 72°C for 1 min. The PCR products were electrophoresed
in 2% agarose gel, images were captured using the ZF ultraviolet
transmission reflection analyzer (Shanghai Jiapeng Technology Co.,
Ltd., Shanghai, China) and gray values were measured using Quantity
One-4.6.2 software (Bio-Rad, Hercules, CA, USA). The relative level
of the target mRNA expression was defined as the ratio of the
absorbance of the target band to the β-actin band.
 | Table IPrimers used in the polymerase chain
reaction. |
Table I
Primers used in the polymerase chain
reaction.
| Target | Primer sequences
(5′-3′) | Product size
(bp) | Annealing temperature
(°C) | Cycles, n |
|---|
| TβRI | F:GGC CAA ATA TCC CAA
ACA GAT
R: AAT CCA ACT CCT TTG CCC TTA | 509 | 60 | 35 |
| Smad3 | F: ACA GCT GTG TCT
GCC AAA CAC
R: ATG TTC TGA GAG GGG AGG GAG | 428 | 59 | 30 |
| Smad4 | F: CAG CAT CCA CCA
AGT AAT CGT
R: CTC TCA ATG GCT TCT GTC CTG | 587 | 60 | 30 |
| Smad7 | F:CCT CCT TAC TCC AGA
TAC CCA
R:ACC AGC TGA CTC TTG TTG TCC GAA T | 304 | 55 | 30 |
| TIMP-1 | F:GTT GTT GCT GTG GCT
GAT AG
R:TGT GGG ACC TGT GGA AGT A | 265 | 58 | 30 |
| β-actin | F: AGC GGG AAA TCG
TGC GTG AC
R: ACA TCT GCT GGA AGG TGG AC | 453 | 58 | 30 |
Statistical analysis
Statistical analysis was performed using SPSS
software, version 19.0 (IBM Corporation, Armonk, NY, USA). All data
are expressed as the mean ± standard deviation. Multifactorial
analysis was used to analyze the results of the MTT assay, and
one-way analysis of variance was used for other comparisons. The
pairwise comparisons of several means between groups was performed
using the Student-Newman-Keuls method. P<0.05 was considered to
indicate a statistically significant difference.
Results
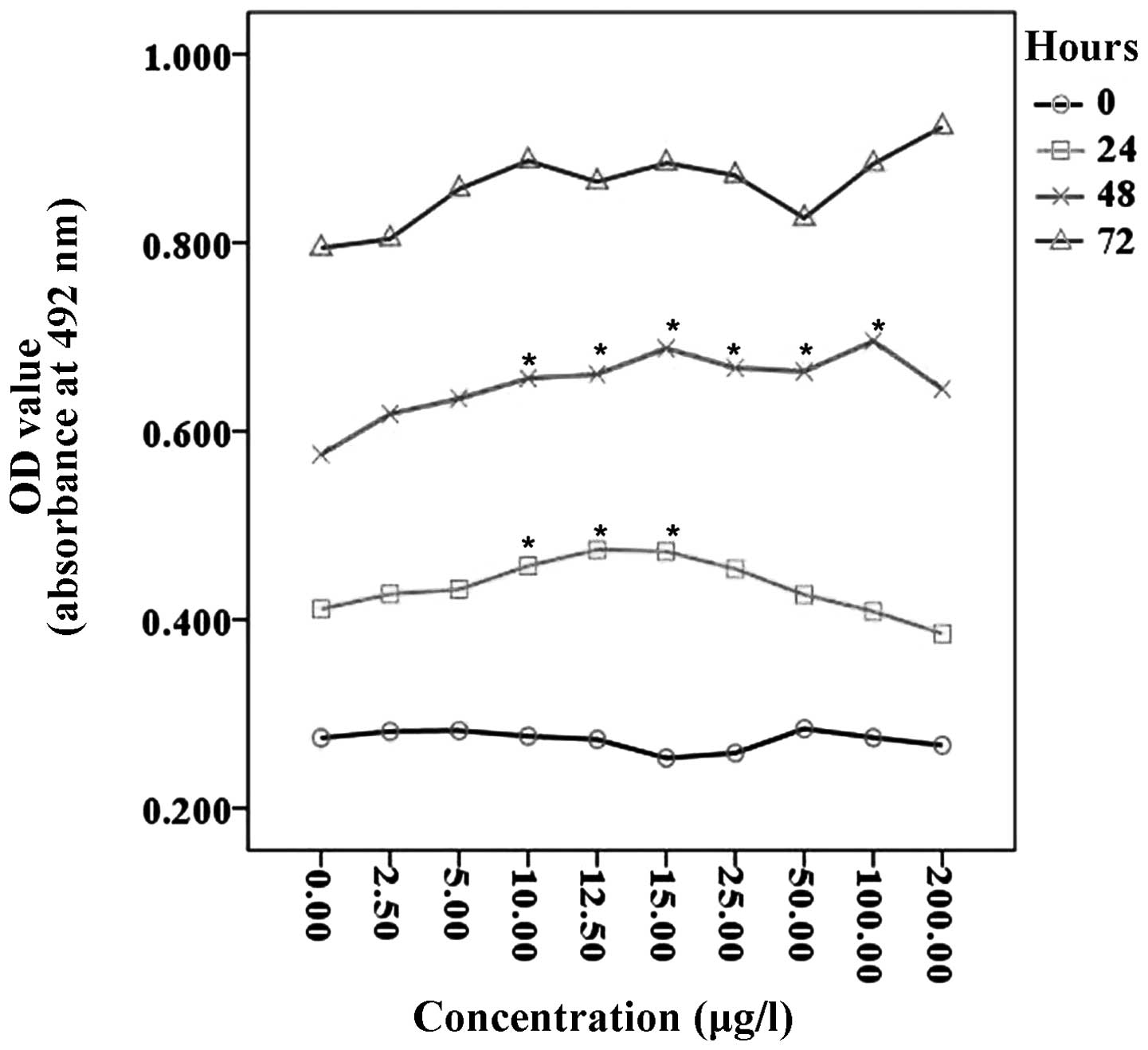
Effects of TGF-β1 on the proliferation of
HTR-8/SVneo cells
An MTT assay of the HTR-8/SVneo cells was performed
to examine cell proliferation in the presence of various
concentrations of recombinant TGF-β1. No proliferation was
identified when the HTR-8/SVneo cells were incubated for 0 h with
or without various concentrations of TGF-β1. Following incubation
with TGF-β1 for 24 h, a significant increase in HTR-8/SVneo cells
was identified, when compared with control cells, in particular for
cells grown with 10–15 μg/l TGF-β1 (P<0.05). Following
incubation with TGF-β1 for 48 h, the proliferation of HTR-8/SVneo
cells was significantly increased with 10–100 μg/l of TGF-β1
(P<0.05). In addition, an increased level of proliferation was
observed in cells incubated with TGF-β1 for 72 h; however, the
increase was not statistically significant (P>0.05; Fig. 1).
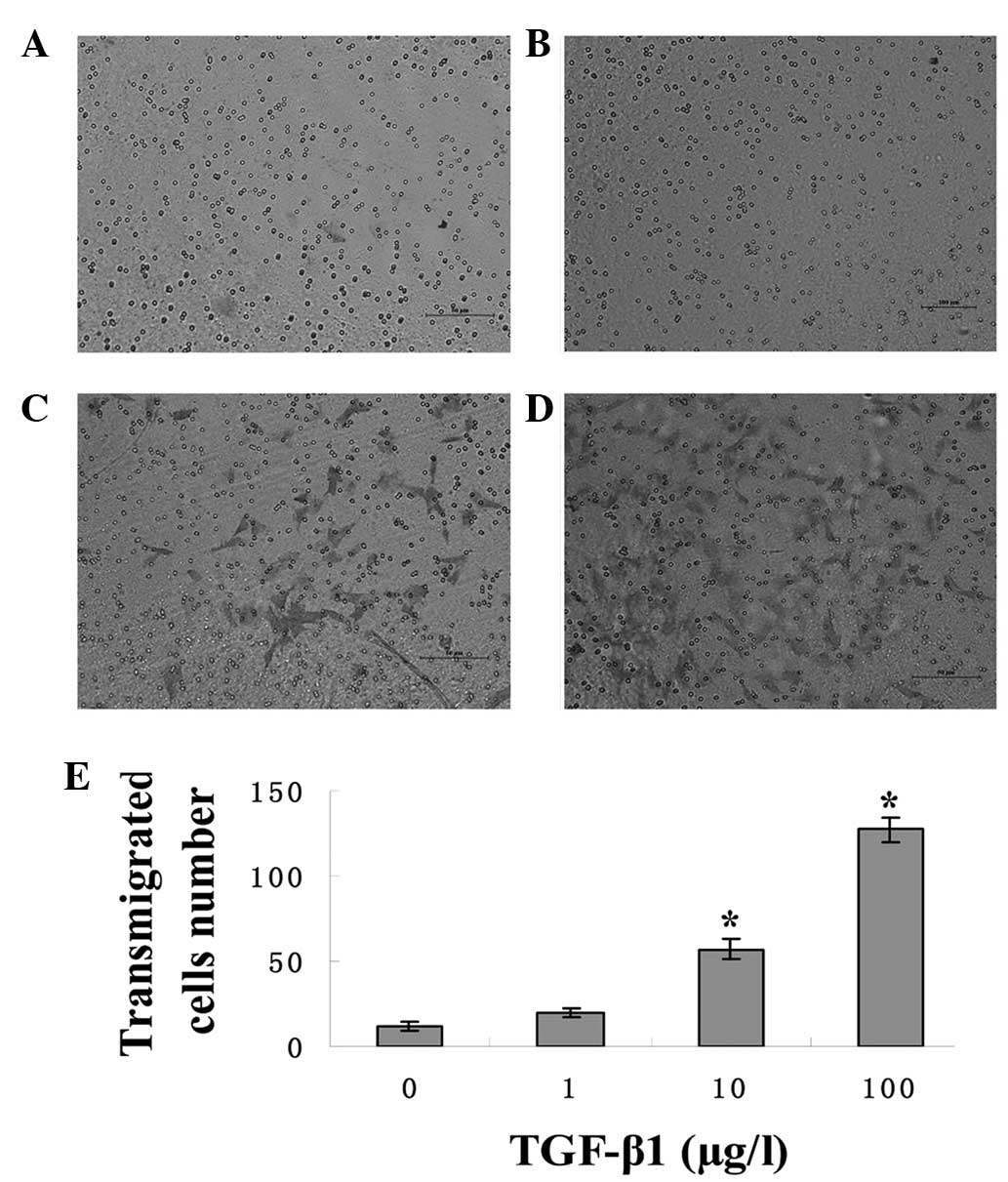
Effects of TGF-β1 on the invasion of
HTR-8/SVneo cells
The effect of TGF-β1 on the invasion ability of the
HTR-8/SVneo Cells was examined using Transwell invasion assays. The
invasion ability of the cells was compared by treating the cells
with 0, 10 or 100 μg/l of TGF-β1 for 48 h. The results demonstrated
that cells grown in the presence of 10 and 100 μg/l TGF-β1
transmigrated more than the cells grown in the absence of TGF-β1,
which indicated an increased invasion (P<0.05). The most
efficacious concentration of TGF-β1 with regard to increased cell
invasion was 100 μg/l (P<0.05) (Fig.
2).
Regulation of TβR1, SMAD4, SMAD3, SMAD7
and TIMP-1 mRNA expression by TGF-β1 in HTR-8/SVneo cells
Subsequently, the mRNA expression of the known
TGF-β1 downstream mediators was compared using RT-PCR in the
HTR-8/SVneo cells by treating these cells with various TGF-β1
concentrations for 48 h. The results indicated that the expression
of TβRI, SMAD4 and SMAD3 mRNA was decreased and that of SMAD7 mRNA
was increased in cells grown in the presence of 10 μg/l of TGF-β1
(P<0.05). However, no significant difference in the mRNA
expression of the four genes was identified in cells grown in the
presence of 1 or 100 μg/l TGF-β1 (P>0.05). In addition, no
significant differences were identified in the expression of TIMP-1
mRNA in the presence of any of the doses of TGF-β1 applied
(P>0.05; Fig. 3).
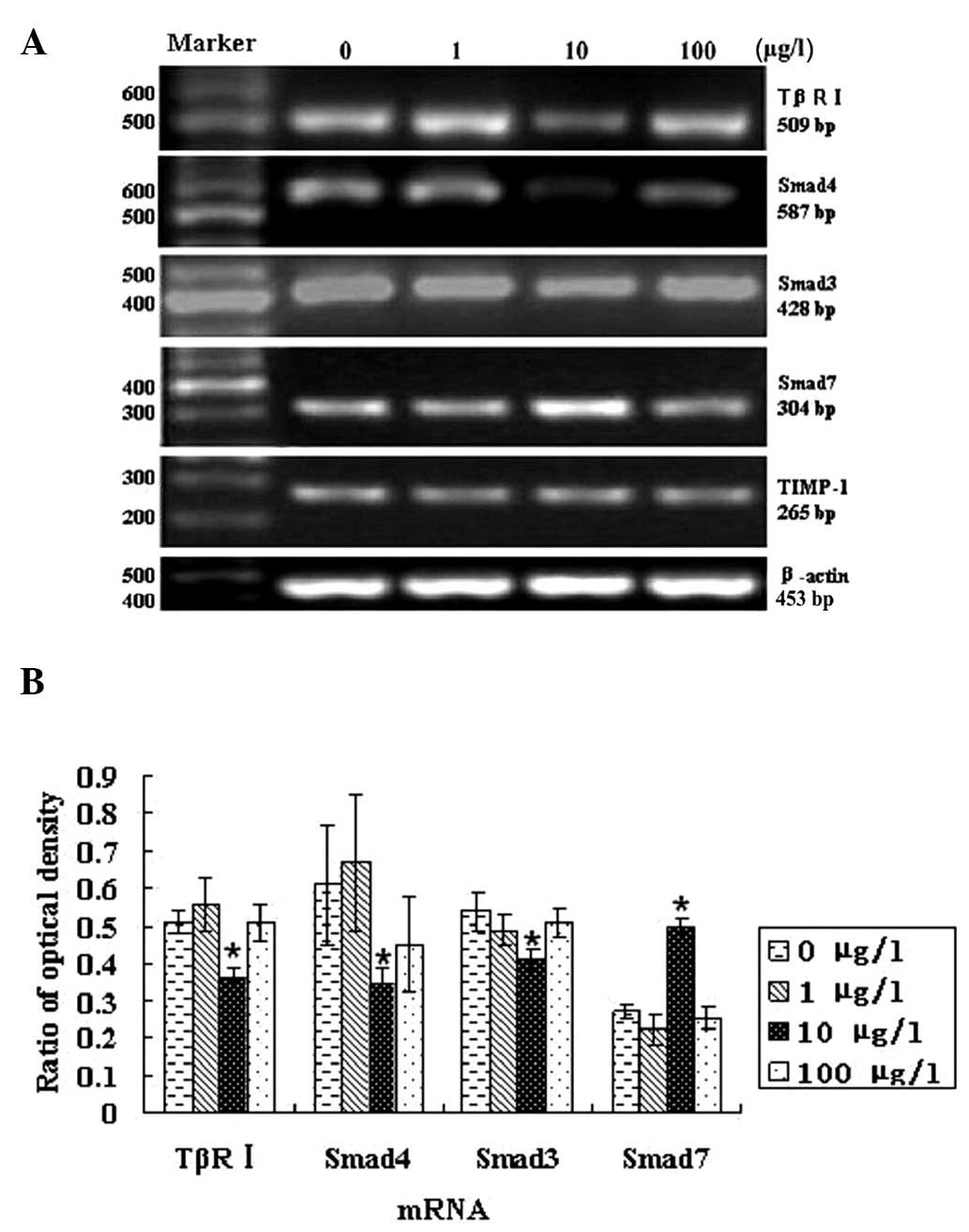 | Figure 3Regulatory effects of varying doses of
TGF β1 for 48 h on TβR1, SMAD4, SMAD3, SMAD7 and TIMP-1 mRNA in HTR
8/SVneo cells. (A) TβR1, SMAD4, SMAD3, SMAD7 and TIMP-1 mRNA
expression by TGF-β1 in HTR 8/SVneo cells. (B) RT-PCR analysis of
TβR1, SMAD4, SMAD3, SMAD7 and TIMP-1 mRNA levels by TGF-β1 in HTR
8/SVneo cells (mean ± SD; n=5). The expression of TβRI, SMAD4 and
SMAD3 mRNA was decreased, whereas that of SMAD7 mRNA increased in
cells grown with 10 μg/l of TGF-β1 (P<0.05). However, no
significant differences were identified between the mRNA expression
in the four genes in cells grown in the presence of 1 or 100 μg/l
TGF-β1 (P>0.05). In addition, the expression of TIMP-1 mRNA
remained unchanged in the presence of all TGF-β1 doses applied in
this study (P>0.05). *P<0.05, vs. the control
group. TβR1, transforming growth factor β receptor I; TIMP-1,
tissue inhibitor of metalloproteinases-1; TGF-β1, transforming
growth factor-β1. |
Discussion
It is generally accepted that the proliferation and
invasion of the first trimester human trophoblast cells is
important in embryonic development. Primary cell and transformed
trophoblast cell models have been developed to investigate the
proliferation and invasion of the placenta and placental tumors,
respectively. In particular, the HTR-8/SVneo cell line has been
established to investigate the biology of normal trophoblast cells
as they have been reported to share certain characteristics with
their parental cells.
TGF-β is a family of cytokines, which are
multifunctional peptides that regulate proliferation,
differentiation, adhesion, migration and other functions of
numerous cell types. TGF-β comprises of three isoforms: TGF-β1, β2
and β3. The normal trophoblast proliferation and invasiveness of
the uterus are strictly regulated processes that are inhibited by
TGF-β1 produced locally (6–9). TGF-β1 may inhibit the growth of
epithelial cells and induce apoptosis, thus acting as a tumor
suppressor (17). However, the
TGF-β1 gene is also frequently upregulated in tumor cells, and
mutations in this gene may result in Camurati-Engelmann disease
(18). TGF-β1 is overexpressed in
various types of malignancies. For example, Lv et al
(19) demonstrated that TGF-β1
induced the epithelial-to-mesenchymal transition of breast cancer
cells and promoted breast cancer cell metastasis (19). TGF-β1 has also been found to be
overexpressed in invasive types of hepatocellular carcinoma and may
be involved in the rapid progression of hepatocellular carcinoma
(20). Dave et al (21) found that higher TGF-β1 levels were
exhibited in the serum of breast cancer patients. Furthermore,
Dehaghani et al (22)
revealed that the TGF-β1 serum levels were significantly higher in
gestational trophoblastic disease patients when compared with those
in pregnant and non-pregnant controls.
In the current study, the proliferation of the
HTR-8/SVneo cells was found to increase following incubation with
TGF-β1 (10–100 μg/l) for 24 or 48 h. An increased invasion was also
observed when the cells were treated with 10 or 100 μg/l TGF-β1 in
the Transwell assays. Notably, previous studies have reported that
TGF-β1 may significantly inhibit the cell invasion (11). In addition, Graham et al
(12) demonstrated that HTR-8/SVneo
cells were inhibited by recombinant TGF-β1, which is the same as
that of parental trophoblast cells (12). Considering that the HTR-8/SVneo cell
line used were over 90 passages in cell culture, the results of the
present study indicated that the proliferation and invasion ability
of the HTR-8/SVneo cell line has been changed over 90 passages.
This observation is similar to the results of Khoo et al
(23), which demonstrated that the
immortalized trophoblast RSVT-2 and RSVT2/C cell lines were
hyper-proliferative and -invasive when compared with their parental
HTR8 cell line. The RSVT-2 and RSVT2/C cell lines were also
resistant to the anti-proliferative and -invasive effects of TGF-β1
to different extents. Khoo et al (23) also revealed that the downregulation
of connexins, and the resultant impairment in gap junctional
intercellular communication, caused cells to escape from the
inhibition of TGF-β1, which may be an early event in tumor
progression, as observed in the premalignant SV40 Tag transformants
(24). We hypothesized that the
changes in cytokines may cause the HTR-8/SVneo cell line to escape
from the inhibition of TGF-β1, and to exhibit a hyper-proliferative
and -invasive phenotype following numerous passages, as observed in
the current study. Therefore, further studies investigating the
expression of the downstream mediators of TGF-β1 were
performed.
TGF-β1 is normally inactive in cells and exerts its
biological effects depending on its downstream molecules. Any
change occurring to its downstream mediators leads to the
dysregulation of TGF-β1, which is illustrated by the following
examples. The expression levels of TGF-β1 and TGF-βR-1 genes have
been demonstrated to be higher in the gastric cancer tissues
(25). Exogenous glucosamine
promotes the osteogenic differentiation of human dental pulp stem
cells via increasing the levels of TGF-βRI and phosphorylated SMAD2
(26). The expression of SMAD3,
SMAD4 and phosphorylated SMAD3, as well as TGF-βR type I and type
II, were all higher in leiomyoma when compared with those in
myometrium (27).
SMADs are important intracellular proteins, which
transfer the information of TGF-β to the nucleus. In mammals, four
types of SMADs have been identified: i) receptor-regulated SMADs
(R-SMADs) comprising SMAD2 and SMAD3, which transduce TGF-β
signaling; ii) SMAD1, SMAD5 and SMAD8, which transduce the bone
morphogenetic protein signaling; iii) a common SMAD called
co-SMAD4, which is a key mesomerism that associates with R-SMADs
and translocates to the nucleus; and iv) inhibitory SMADs, SMAD6
and SMAD7, which compete with R-SMADs for receptor binding, thereby
inhibiting the R-SMAD phosphorylation (28,29).
Previous studies have demonstrated that the decreased expression
levels of TβRI, SMAD3 and SMAD4 may cause cancer cells to escape
the growth inhibition of TGF-β1, and the increased expression of
SMAD7 may block the inhibitory effects of TGF-β1. Thus TβRI, SMAD3,
SMAD4 and SMAD7 are closely associated with the development of
tumors. For example, the expression of SMAD4 was lower, and that of
SMAD7 was significantly higher, in the gastric cancer tissues than
in the peri-tumoral tissues (30).
Similarly, the expression of SMAD4 was lower, whereas SMAD7 was
found to positively correlate with tumor grading in human glioma
and gastric cancer (31,32). SMAD3 may mediate in vivo
signaling that is inhibitory to epithelial wound healing and thus,
during the physiological process of wound healing, the suppression
of SMAD3 levels may engage (33).
In the physiological tissue repair as well as the pathological
fibrosis, SMAD3 mRNA was markedly downregulated and the
antagonistic SMAD7 was rapidly and transiently induced by TGF-β1 to
stimulate collagen synthesis (34).
These results indicate that SMAD3 is involved in the growth
inhibition by TGF-β. The aberrant expression of SMAD7 has been
shown to be involved in inflammatory bowel disease and scleroderma
(35). In addition, SMAD7 mRNA
levels are increased in human pancreatic cancer. Furthermore, SMAD7
transfected colo-357 cells exhibit enhanced anchorage-independent
growth and accelerated growth in nude mice (36).
The results of the present study, obtained from
RT-PCR assays, revealed that when compared with that of the control
cells, the mRNA levels of TβRI, SMAD3 and SMAD4 were significantly
decreased in the HTR-8/SVneo cells treated with 10 μg/l TGFβ1
(P<0.05), while the mRNA level of SMAD7 was significantly
increased. In addition, no evident changes in the expression of
TβRI, SMAD3 and SMAD4 mRNAs were identified in cells treated with 1
and 100 μg/l TGFβ1 (P>0.05). These results indicated that the
decreased mRNA expression of TβRI, SMAD3 and SMAD4 may reduce the
inhibitory effect of TGF-β1 on cell proliferation, while the
enhanced mRNA expression of SMAD7 further blocks the inhibitory
effect of TGF-β1 on cell proliferation. This may explain the
significant increase in proliferation identified when cells were
treated with TGF-β1 at this concentration.
The changes in SMAD7 expression observed in the
HTR-8/SVneo cell line over several passages were similar to those
in the immortalized bronchial BEP2D epithelial cells. A previous
study on the BEP2D cells and the radiation-induced malignant
transformation of bronchial BERP35T2 epithelial cells revealed that
the SMAD7 gene was highly expressed, and the cells that exhibited a
high expression of SMAD7 demonstrated an active proliferative
capacity. In addition, the inhibitory effect of TGF-β1 on the cell
growth of these cells was weak. Furthermore, the changes in SMAD7
expression were associated with the malignant transformation of
bronchial epithelial cells and the development of lung cancer
(37).
In the present study, the mRNA expression of the
TIMP-1 gene that encodes a natural inhibitor of the matrix
metalloproteinases (MMPs) was also examined by RT-PCR. MMPs are a
family of >23 zinc-binding enzymes, which are involved in the
proteolytic degradation of the extracellular matrix. MMP-9 and -2
have been shown able to mediate the invasion of trophoblast cells,
and the natural tissue inhibitors of MMP, including the TIMP
family, inhibit their invasiveness (38). The results of the present study
indicated that the mRNA levels of TIMP-1 were not altered. However,
the mechanism underlying the increased invasion of the HTR-8/SVneo
cells regulated by TGF-β1 requires further investigation.
In conclusion, the cell lines may be convenient for
studies; however, the cell lines expressed a number of novel
characteristics (39–42). The results of this study indicate
that the effects of TGF-β1 on the proliferation and invasion of the
HTR-8/SVneo cell line at passage 90 were different from those of
the parental trophoblasts, which is in contrast to the results of
previous studies. We hypothesized that HTR-8/SVneo cell lines,
which have been grown for over 90 passages do not accurately
represent parental trophoblast cells for studies of the TGF-β/SMAD
signaling pathway.
Acknowledgements
The authors would like to thank Professor Charles H.
Graham (Queen’s University at Kingston, Kingston, Canada) for
providing the HTR-8/SVneo cell line. This study was supported by
the Key Subjects in Universities and Colleges of Hebei Province of
China (pathology and pathophysiology).
References
|
1
|
Prossler J, Chen Q, Chamley L and James
JL: The relationship between TGFβ, low oxygen and the outgrowth of
extravillous trophoblasts from anchoring villi during the first
trimester of pregnancy. Cytokine. 68:9–15. 2014.
|
|
2
|
Genest DR, Berkowitz RS and Fisher RA:
Gestational trophoblastic disease. World Health Organization
Classification of Tumours, Pathology and Genetics Tumours of the
Breast and Female Genital Organs. Tavassoli FA and Devilee P: 4.
3rd edition. IARC Press; Lyon: pp. 250–254. 2003
|
|
3
|
Knöfler M: Critical growth factors and
signalling pathways controlling human trophoblast invasion. Int J
Dev Biol. 54:269–280. 2010.
|
|
4
|
Lunghi L, Ferretti ME, Medici S, et al:
Control of human trophoblast function. Reprod Biol Endocrinol.
5:62007.
|
|
5
|
Simpson H, Robson SC, Bulmer JN, et al:
Transforming growth factor beta expression in human placenta and
placental bed during early pregnancy. Placenta. 23:44–58. 2002.
|
|
6
|
Graham CH, Lysiak JJ, McCrae KR and Lala
PK: Localization of transforming growth factor-beta at the human
fetal-maternal interface: role in trophoblast growth and
differentiation. Biol Reprod. 46:561–572. 1992.
|
|
7
|
Lysiak JJ, Hunt J, Pringle GA and Lala PK:
Location of transformationg growth factor beta and its natural
inhibitor decorin in the human placenta and deciduas throughout
gestation. Placenta. 16:221–231. 1995.
|
|
8
|
Graham CH and Lala PK: Mechanism of
control of trophoblast invasion in situ. J Cell Physiol.
148:228–234. 1991.
|
|
9
|
Zhen FA, Ying-Yong L, Guang CY and Jun LY:
Effect and significance of TGF-β1 on cell proliferation of human
first trimester cytotrophoblasts and JAR cells. MCHCC.
18:2582–2585. 2008.(In Chinese).
|
|
10
|
Lala PK and Chakraborty C: Factors
regulating trophoblast migration and invasiveness: possible
derangements contributing to pre-eclampsia and fetal injury.
Placenta. 24:575–587. 2003.
|
|
11
|
Zhao MR, Qiu W, Li YX, et al: Dual effect
of transforming growth factor beta1 on cell adhesion and invasion
in human placenta trophoblast cells. Reproduction. 132:333–341.
2006.
|
|
12
|
Graham CH, Hawley TS, Hawley RG, et al:
Establishment and characterization of first trimester human
trophoblast cells with extended lifespan. Exp Cell Res.
206:204–211. 1993.
|
|
13
|
Shi Y and Massagué J: Mechanisms of
TGF-beta signaling from cell membrane to the nucleus. Cell.
113:685–700. 2003.
|
|
14
|
Kaminska B, Kocyk M and Kijewska M: TGF
beta signaling and its role in glioma pathogenesis. Adv Exp Med
Biol. 986:171–187. 2013.
|
|
15
|
Petroff MG, Phillips TA, Ka H, et al:
Isolation and culture of term human trophoblast cells. Methods Mol
Med. 121:203–217. 2006.
|
|
16
|
Suzuki T, Higgins PJ and Crawford DR:
Control selection for RNA quantitation. Biotechniques. 29:332–337.
2000.
|
|
17
|
Sundqvist A, Ten Dijke P and van Dam H:
Key signaling nodes in mammary gland development and cancer: Smad
signal integration in epithelial cell plasticity. Breast Cancer
Res. 14:2042012.
|
|
18
|
Collet C, Laplanche JL and de Vernejoul
MC: Camurati-Engelmann disease with obesity in a newly identified
family carrying a missense p.Arg156Cys mutation in the TGFB1 gene.
Am J Med Genet A. 161A:2074–2077. 2013.
|
|
19
|
Lv ZD, Kong B, Li JG, et al: Transforming
growth factor-β 1 enhances the invasiveness of breast cancer cells
by inducing a Smad2-dependent epithelial-to-mesenchymal transition.
Oncol Rep. 29:219–225. 2013.
|
|
20
|
Lee D, Chung YH, Kim JA, et al:
Transforming growth factor beta 1 overexpression is closely related
to invasiveness of hepatocellular carcinoma. Oncology. 82:11–18.
2012.
|
|
21
|
Dave H, Shah M, Trivedi S and Shukla S:
Prognostic utility of circulating transforming growth factor beta 1
in breast cancer patients. Int J Biol Markers. 27:53–59. 2012.
|
|
22
|
Dehaghani AS, Rad NR, Fattahi MJ, et al:
Investigation of soluble HER2 and transforming growth factor Beta-1
serum levels in gestational trophoblastic disease. Pathol Oncol
Res. 15:37–40. 2009.
|
|
23
|
Khoo NK, Bechberger JF, Shepherd T, et al:
SV40 Tag transformation of the normal invasive trophoblast results
in a premalignant phenotype. I. Mechanisms responsible for
hyperinvasiveness and resistance to anti-invasive action of
TGFbeta. Int J Cancer. 77:429–439. 1998.
|
|
24
|
Khoo NK, Zhang Y, Bechberger JF, Bond SL,
Hum K and Lala PK: SV40 Tag transformation of the normal invasive
trophoblast results in a premalignant phenotype. II. Changes in gap
junctional intercellular communication. Int J Cancer. 77:440–448.
1998.
|
|
25
|
Guo RF, Zang SZ, Zhang L, et al: Relations
of transforming growth factor-beta1 expression to differentiation
and prognosis of advanced gastric cancer. Zhonghua Yi Xue Za Zhi.
86:3249–3254. 2006.(In Chinese).
|
|
26
|
Huang CH, Tseng WY, Yao CC, et al:
Glucosamine promotes osteogenic differentiation of dental pulp stem
cells through modulating the level of the transforming growth
factor-beta type I receptor. J Cell Physiol. 225:140–151. 2010.
|
|
27
|
Chegini N, Luo X, Ding L and Ripley D: The
expression of Smads and transforming growth factor beta receptors
in leiomyoma and myometrium and the effect of gonadotropin
releasing hormone analogue therapy. Mol Cell Endocrinol. 209:9–16.
2003.
|
|
28
|
Yang G and Yang X: Smad4-mediated TGF-beta
signaling in tumorigenesis. Int J Biol Sci. 6:1–8. 2010.
|
|
29
|
Brown KA, Pietenpol JA and Moses HL: A
tale of two proteins: differential roles and regulation of Smad2
and Smad3 in TGF-beta signaling. J Cell Biochem. 101:9–33.
2007.
|
|
30
|
Leng A, Liu T, He Y, et al: Smad4/Smad7
balance: a role of tumorigenesis in gastric cancer. Exp Mol Pathol.
87:48–53. 2009.
|
|
31
|
Chen QX, Shao BY, Zheng H, et al:
Expression and significance of TGF-B1 and its intracellular
signaling molecules SMAD4, SMAD7 in human glioma. Chin J Exp Surg.
1:90–92. 2005.
|
|
32
|
Lu B, Zhou YN, Li Q, et al: Correlations
of TGF-betaRII, Smad4 and Smad7 expression to clinicopathologic
characteristics and prognosis of gastric cancer. Ai Zheng.
28:538–542. 2009.
|
|
33
|
Schiller M, Javelaud D and Mauviel A:
TGF-beta-induced SMAD signaling and gene regulation: consequences
for extracellular matrix remodeling and wound healing. J Dermatol
Sci. 35:83–92. 2004.
|
|
34
|
Mori Y, Chen SJ and Varga J: Modulation of
endogenous Smad expression in normal skin fibroblasts by
transforming growth factor-beta. Exp Cell Res. 258:374–383.
2000.
|
|
35
|
Nakao A, Okumura K and Ogawa H: Smad7: a
new key player in TGF-beta-associated disease. Trends Mol Med.
8:361–363. 2002.
|
|
36
|
Kleeff J, Ishiwata T, Maruyama H, et al:
The TGF-beta signaling inhibitor Smad7 enhances tumorigenicity in
pancreatic cancer. Oncogene. 18:5363–5372. 1999.
|
|
37
|
Huo YY, Zhang KT, Li BY, et al: Regulation
of Smad7 gene by TGF-beta 1 in the process of malignant
transformation. Ai Zheng. 21:117–121. 2002.(In Chinese).
|
|
38
|
Bischof P, Meisser A and Campana A:
Paracrine and autocrine regulators of trophoblast invasion - a
review. Placenta. 21(Suppl A): S55–S60. 2000.
|
|
39
|
Burns JS, Abdallah BM, Guldberg P, et al:
Tumorigenic heterogeneity in cancer stem cells evolved form
long-term cultures of telomerase-immortalized human mesenchymal
stem cells. Cancer Res. 65:3126–3135. 2005.
|
|
40
|
Hai C, Jun D, Xi-Chuan Y, et al: Genetic
stability of immortal melanocytes transfected by SV40T antigen gene
[J]. Practical Journal of Clinical Medicine. 5:24–27. 2008.
|
|
41
|
Ahuja D, Sáenz-Robles MT and Pipas JM:
SV40 large T antigen targets multiple cellular pathways to elicit
cellular transformation. Oncogene. 24:7729–7745. 2005.
|
|
42
|
Bosticardo M, Ghosh A, Du Y, et al:
Self-inactivating retroviral vector-mediated gene transfer induces
oncogene activation and immortalization of primary murine bone
marrow cells. Mol Ther. 17:1910–1918. 2009.
|