Introduction
Female breast cancer accounts for one in 10 of all
new cancer cases diagnosed each year worldwide. As the most
prevalent cancer amongst females in developing and developed
countries, breast cancer is the leading global cause of female
cancer-related mortality (1,2).
Although improvements in the early detection and treatment of
breast cancer have decreased mortality rates in recent years, the
survival rates for patients with late-stage or metastatic breast
cancer remain poor (3). Thus, it is
important to identify novel genes or pathways involved in breast
cancer to develop faster diagnoses and safer treatments.
Hepatoma upregulated protein (HURP) was initially
classified as an upregulated protein in human hepatocellular
carcinoma and was demonstrated to be an integral part of the
spindle apparatus (4,5). Further studies have revealed HURP to
be a novel component of the Ran-importin β-regulated spindle
assembly pathway, which forms a complex with
guanosine-5′-triphosphate (RanGTP) and localizes predominantly to
the kinetochore microtubules (K-MTs) supporting kinetochore fiber
(k-fiber) stabilization (6,7). HURP is also a mitotic phosphoprotein
substrate for Aurora-A, a mitotic serine/threonine kinase with
oncogenic properties (8,9). Spindle assembly and function are
controlled by the phosphorylation of HURP by Aurora-A, which acts
as a regulatory mechanism (10).
HURP abundance is tightly regulated during the cell cycle, with the
levels of HURP fluctuating during the cycle and reaching a peak at
G2/M (5). This fact
suggests that HURP is a potential cell cycle regulator. HURP
promotes chromosome congression and controls spindle stability by
combining with k-fibers. HURP activity is necessary for correct
kinetochore capture, effective chromosome congression and prompt
mitotic progression. Defects in these regulatory process can lead
to mitotic delay, misaligned chromosomes and genomic instability
(6,9–11). As
genomic instability is a noteworthy feature of human cancer
(12,13), we hypothesized that HURP may have a
role in the progression of breast carcinogenesis.
The overexpression of HURP has thus far been
identified in hepatocellular carcinoma, adrenocortical tumors and
urogenital carcinoma (14,15). However, there is no information
about HURP in human breast carcinogenesis progression and the
clinical relevance of HURP in cancer patients. In the present
study, the clinicopathological and functional activities of HURP in
human breast carcinogenesis were investigated. The present findings
demonstrate the significance of the overexpression of HURP in human
breast carcinogenesis and its functional role in vitro.
Material and methods
Patients and specimen collection
In total, 43 breast cancer tumor samples and paired
normal tissues were obtained from patients who underwent surgery in
West China Hospital (Sichuan University, Chengdu, China) between
2011 and 2012. The normal tissue was extracted at least 5 cm distal
from the primary breast cancer and was identified as normal by a
pathologist. All specimens were immediately frozen in liquid
nitrogen and stored at −80°C until RNA was extracted. All patients
provided written informed consent and the procedures were approved
by the Human Ethics Review Board. No patients received chemotherapy
or radiotherapy prior to surgery. All demographic and pathological
data, including the patient age, tumor size and stage, number of
tumors, presence of lymph node metastasis, immunohistochemical
results and histological classification were obtained from clinical
and pathological database records. All specimens were graded using
a modification of the World Health Organization classification
system (16), and the pathological
staging was performed according to the pathological
tumor-node-metastasis (TNM) staging system (17).
RNA preparation, reverse transcription
(RT) and quantitative polymerase chain reaction (qPCR)
Total RNA was extracted from frozen tissue specimens
and cells using TRIzol reagent (Life Technologies, Gaithersburg,
MD, USA). cDNA was then synthesized with a PrimeScript RT reagent
kit with gDNA Eraser (Takara Biotechnology (Dalian) Co., Ltd.,
Dalian, China). The sequences of the HURP primers were designed as
follows: Sense, 5′-CAT GTGAAGAAGACTTTGTTTTTGA-3′; and antisense,
5′-GGTAATCCAGGACACTGAGCA-3′. The glyceraldehyde-3 phosphate
dehydrogenase (GAPDH) gene served as an internal quality RNA
reference control. The sequences of the GAPDH primers were as
follows: Sense, 5′-ACCACAGTCCATGCCATCAC-3′; and antisense,
5′-TCCACCACCCTGTTGCTGTA-3′. qPCR was performed in MyiQTM and iQTM5
Real-Time PCR Detection Systems (Bio-Rad Laboratories, Hercules,
CA, USA) using the SsoFast™EvaGreen®Supermix (Bio-Rad
Laboratories). qPCR was performed as follows: Enzyme activation at
95°C for 30 sec, followed by 40 cycles of denaturation at 95°C for
5 sec and annealing at 55°C for 5 sec. All mRNA copy numbers were
calculated relative to the concentration of cDNA from Human
Universal Reference total RNA (Takara Bio, Inc., Shiga, Japan). The
HURP copy number was then divided by the copy number of the
endogenous reference (GAPDH) to obtain normalized expression
values.
HURP protein expression analysis
Total protein was extracted from the specimens and
cells using RIPA Lysis Buffer (YuanPingHao Bio, Beijing, China).
Aliquots of total protein were separated on 10% acrylamide gradient
gels. Following electrophoresis, the samples were electroblotted
(45 mA, 90 min) onto a polypropylene difluoride membrane
(Millipore, Billerica, MA, USA). Anti-HURP rabbit polyclonal
antibody (Ab; Santa Cruz Biotechnology, Inc., CA, USA) detected
HURP protein at a 1:200 dilution. The protein level of HURP was
normalized to the level of GAPDH protein, which was detected by a
1:10,000 dilution of GAPDH rabbit monoclonal (Mc)Ab (Epitomics,
Inc., Burlingame, CA, USA). Following incubation with a secondary
Ab, peroxidase-conjugated Affinipure goat anti-rabbit
immunoglobulin G (ZSGB-Bio, Beijing, China) at a dilution of
1:5,000, the protein signals were visualized by chemiluminescence
(Pierce, Rockford, IL, USA). The intensity of the bands was
measured by Quantity One Software (Bio-Rad) and normalized using
the intensity of GAPDH.
Cell lines and cell culture
In total, four human breast cancer cell lines,
MCF-7, MDA-MB-231, MDA-MB-435S and ZR-75-30, were obtained from
Zhengzhou Jinrong Biotechnology Co., Ltd., (Zhengzhou, Henan,
China) and were routinely maintained in RPMI-1640 with 10%
(vol/vol) fetal bovine serum at 37°C, in a humidified atmosphere of
95% air and 5% CO2.
Small interfering (si)RNA
Gene-specific 27mer siRNA duplexes (DsiRNAs)
designed to target the HURP gene (DsiRNA1,
rArGrArCrCrArGrUrArCrArGrGrArT; DsiRNA2,
rCrCrUrArUrCrArArGrUrArArCrArCrCrUrArUrGrArCrUCC; and DsiRNA3,
rArCrCrUrArArGrUrCrUrGrUrCrArArCrArArArGrCrUrGTA) were obtained
from a Trilencer-27 siRNA kit (OriGene, Rockville, MD, USA), and
universal scrambled negative control siRNA duplex (OriGene) was
used as a negative control. The cells (1.2×105) were
transfected with a 10-nM final concentration of the respective
siRNAs, using a siTRAN (OriGene) for 24 h to harvest the cells and
detect the mRNA levels in the parental, negative and HURP siRNA
cells, according to the manufacturer’s instructions.
Protein transfection
The Xfect Protein Transfection Reagent (Takara
Biotechnology (Dalian) Co., Ltd.) was used to bind and transport
active anti-HURP rabbit polyclonal Ab (Santa Cruz Biotechnology,
Inc.) directly into the breast cancer cells. The cells
(1.2×105) were transfected with concentration gradients
of the anti-HURP Ab (4 and 5 μg, respectively)using the Xfect
Protein Transfection Reagent, according to the manufacturer’s
instructions. Following incubation at 37°C for 60 minutes, the
cells were harvested for the next cell viability assays. In
addition, β-galactosidase was used as the control. The cells were
stained with X-gal to determine the efficiency of β-galactosidase
transfection using the β-Galactosidase Staining kit (Takara Bio,
Inc.).
Cell proliferation assays
Cell proliferation assays were performed using Cell
Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). The cells
were plated in 96-well plates, at 1×105 cells per well,
and cultured in the growth medium. At the indicated time-points,
the cell numbers in triplicate wells were measured at the
absorbance (450 nm) of reduced WST-8
[2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium,
monosodium salt].
Statistical analysis
The statistical analysis was conducted using SPSS
software (version 19.0; SPSS, Inc., Chicago, IL, USA). The HURP
normalized expression values were expressed as the mean ± standard
deviation (SD) and compared using Student’s t-test. Differences
between the groups were determined using the Mann-Whitney-Wilcoxon
U test and Kruskal-Wallis test. P<0.05 was considered to
indicate a statistically significant difference.
Results
HURP overexpression in human breast
cancer
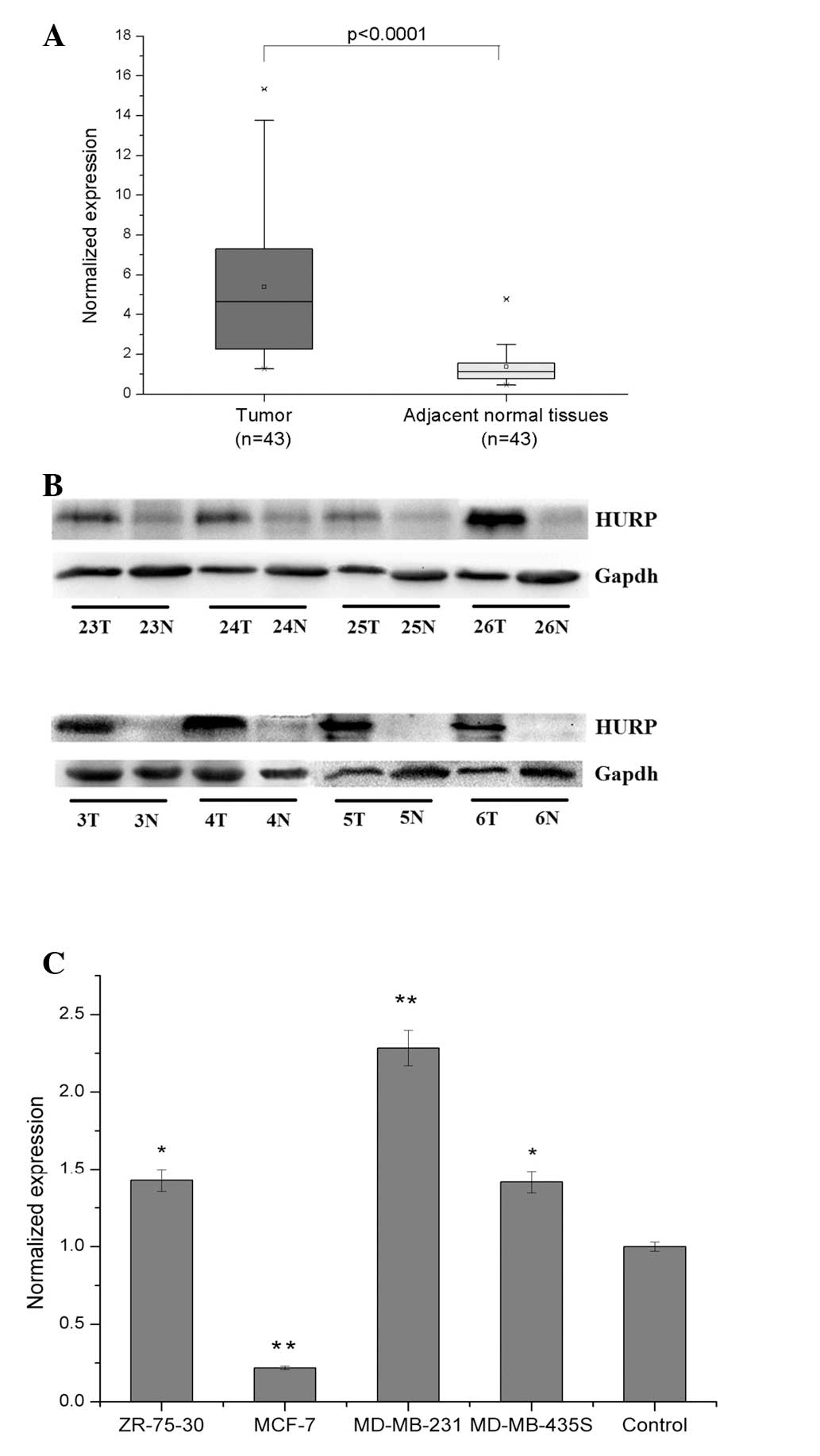
qPCR analysis of HURP mRNA expression in 43 pairs of
primary breast tumors and adjacent histologically normal tissues
revealed a significantly (P<0.0001) higher expression level of
HURP in the tumor tissue (n=43; 90%) compared with the normal
tissue. The mean expression level of the HURP mRNA in the tumor
tissues was 5.38±3.71 (mean ± SD), which was significantly higher
than the mean of 1.37±0.87 in the corresponding paired adjacent
normal tissues (Fig. 1A). Western
blot analysis with anti-HURP Ab verified that HURP protein levels
were unregulated in the human breast tumor tissues compared with
the normal tissues. Representative images of western blots are
shown in Fig. 1B. Significantly
high levels of HURP mRNA expression were also detected in three,
MDA-MB-231, MDA-MB-435S and ZR-75-30, of the four, MDA-MB-231,
MDA-MD-435S, ZR-75-30 and MCF-7, human breast cancer cell lines
examined (Fig. 1C). Together, these
results indicate that HURP is overexpressed in human breast cancer
cells that have a high proliferative and invasive ability.
Clinicopathological significance of HURP
expression in human breast cancer
To further investigate the association between HURP
and breast cancer, 43 malignant tumors were analyzed. The
clinicopathological factors analyzed in relation to HURP normalized
expression are shown in Table I.
The expression of HURP was positively correlated with the TNM
staging (P=0.003). Conversely, no significant differences were
observed between the age, size, estrogen receptor response,
progesterone receptor response, human epidermal receptor presence,
extent of lymph node metastasis or differentiation and HURP
expression.
 | Table IHURP mRNA expression and
clinocopathological factors |
Table I
HURP mRNA expression and
clinocopathological factors
| Prognostic
factors | No. of patients
(n=43) | Expression of HURP,
mean ± SD | P-value |
|---|
| Age, years |
| >50 | 13 | 4.91±2.70 | 0.881 |
| ≤50 | 30 | 5.31±3.86 | |
| Size, mm |
| <20 | 19 | 5.67±3.77 | 0.490 |
| 20–50 | 20 | 4.97±3.46 | |
| >50 | 4 | 3.56+2.07 | |
| Lymph node
metastasis |
| Absent | 27 | 4.72±3.39 | 0.125 |
| Present | 16 | 6.51±4.06 | |
| ER |
| Positive | 34 | 5.22±3.72 | 0.798 |
| Negative | 9 | 5.03±2.62 | |
| PR |
| Positive | 33 | 5.32±3.74 | 0.890 |
| Negative | 10 | 4.69±2.67 | |
| HER |
| Positive | 39 | 5.18±3.62 | 0.643 |
| Negative | 4 | 5.17±1.96 | |
| Differentiation |
| G1 | 2 | 2.28±0.82 | 0.253 |
| G2 | 17 | 5.15±4.36 | |
| G3 | 19 | 5.75±3.65 | |
| pTNM |
| I | 5 | 2.32±0.58 | 0.017a |
| II | 29 | 5.09±3.36 | |
| III | 9 | 8.05±4.29 | |
Suppression of HURP expression in breast
cancer cells inhibits proliferation
To investigate the clinical findings of HURP in
breast cancer and to understand its role in carcinogenesis, in
vitro functional studies of HURP were performed. The primary
focus was whether HURP overexpression is associated with the
proliferative potency of breast cancer cells, as HURP has been
reported to be associated with proliferation activity in other
cancers (18). The highest mRNA
expression level of HURP among the MDA-MB-231, MDA-MD-435S,
ZR-75-30 and MCF-7 cell lines was exhibited by MDA-MB-231 (Fig. 1C). MDA-MB-231 was also the most
sensitive to HURP-specific DsiRNA transfection and had consistent
stability with DsiRNA transfection. Therefore, MDA-MB-231 was
selected as the representative cell line for study.
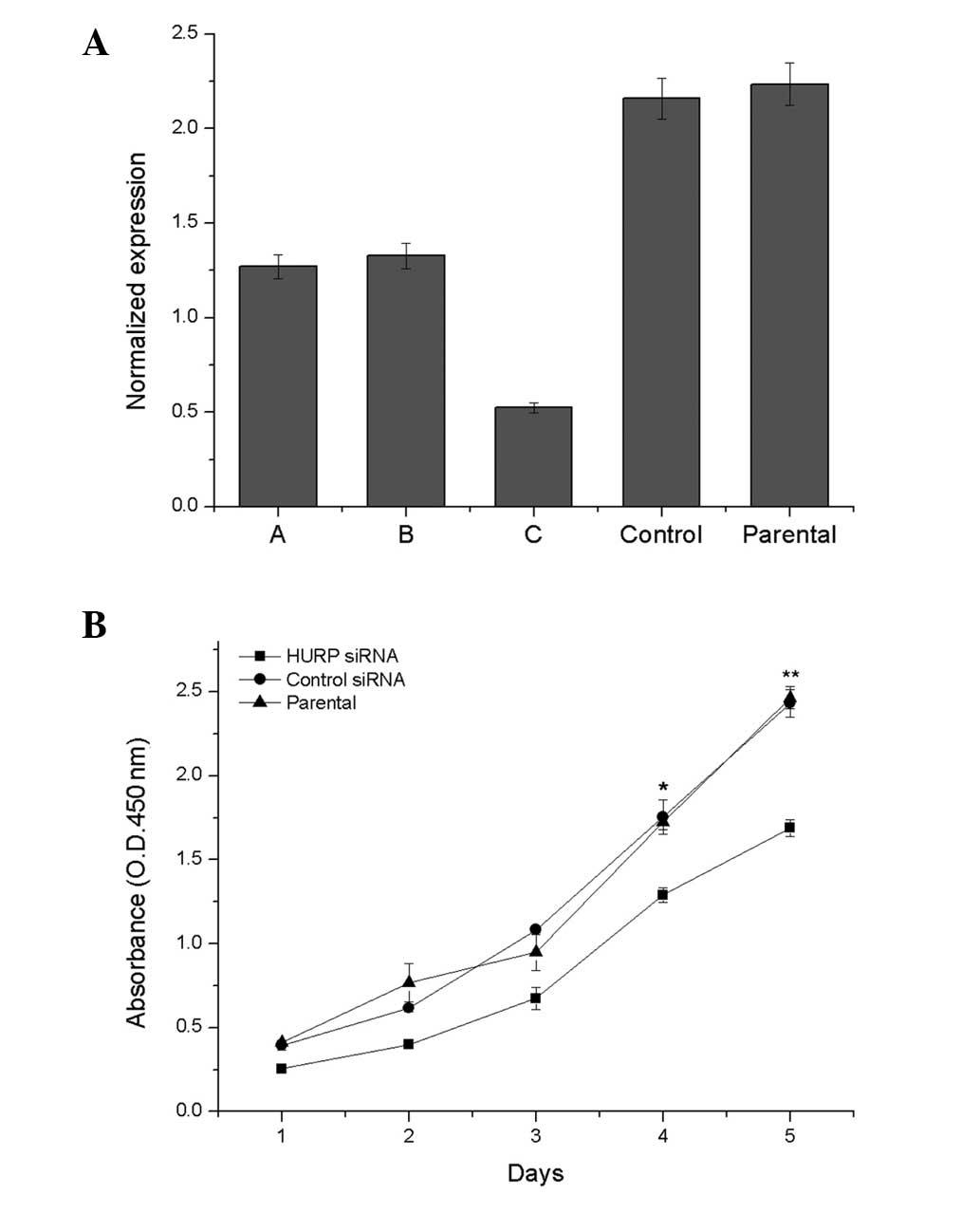
qPCR analysis confirmed that the HURP mRNA
expression level was lower in the HURP DsiRNA3-transfected
MDA-MB-231 cells compared with the MDA-MB-231 cells transfected
with DsiRNA1 or DsiRNA2, the negative control siRNA duplex and the
non-transfected cells (Fig. 2A).
Therefore, HURP DsiRNA3 was chosen as the inhibitor. The cell
proliferation analysis demonstrated that suppression of HURP by
HURP DsiRNA3 significantly inhibited cell growth. HURP DsiRNA3
cells grew slower than the parent or control cells in the CCK-8
assay (Fig. 2B).
Ab-mediated disruption of HURP function
in breast cancer cells inhibits proliferation
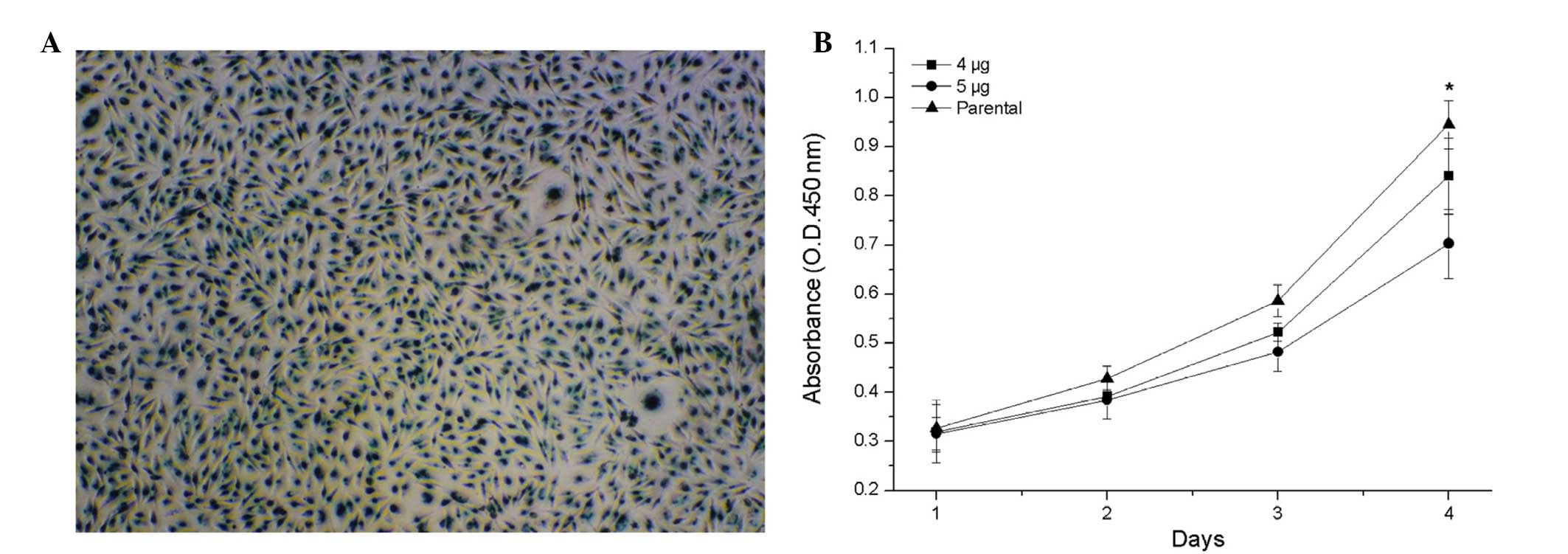
To investigate the transient effects of HURP and the
possibility of McAb or polypeptide drug treatment, active anti-HURP
Ab was transfected directly into the MDA-MB-231 cells. As the
control transfection, the MDA-MB-231 cells were transfected with
2μg β-galactosidase (β-gal), which revealed a high efficiency and a
high amount of β-gal protein per MDA-MB-231 cell (Fig. 3A). Cell proliferation analysis
revealed that anti-HURP Ab-mediated disruption of HURP activity
significantly inhibited cell growth. The higher the amount of
anti-HURP Ab transfected into the MDA-MB-231 cells, the slower the
cells grew (Fig. 3B).
Discussion
In the present study, the involvement of HURP in
human breast cancer carcinogenesis was investigated. The results
demonstrated that HURP mRNA and protein expression were
significantly higher in the breast cancer tumors than in the paired
normal tissues. The overexpression of HURP was also more prevalent
in the breast cancer cells that exhibited increased proliferation
and invasion (MDA-MB-231, MDA-MD-435S and ZR-75-30). The
statistical analysis indicated that the HURP expression level was
higher in the tumors with advanced-grade metastasis and was
strongly associated with the tumor stage. This suggests that HURP
is overexpressed in human breast cancer and that such
overexpression is correlated with tumors in advanced-grade
metastasis, which may be prognostic of a poor survival rate. This
is the first study demonstrating the role of HURP in breast cancer
progression and its association with the clinicopathological
factors of the disease. In addition, these results are consistent
with the previous findings that HURP is overexpressed in
hepatocellular carcinoma (5),
adrenocortical tumors (19) and
urothelial carcinoma (20). All
these studies suggest that HURP plays an important role in human
cancer, particularly in tumor progression.
Through cell proliferation assays, the potential
role of HURP in tumor formation and progression was determined. The
present results revealed that the suppression of HURP expression by
siRNA or anti-HURP Abs in breast cancer cells inhibited cell
proliferation in vitro. These in vitro findings are
compatible with the high expression levels of HURP observed in
breast cancer tissues from patients with aggressive disease.
Previous studies demonstrated that the overexpression of HURP in
non-tumorigenic HEK293 cells increases their proliferative ability
and transformation activity (21),
in addition to enhancing the invasiveness of hepatocellular
carcinoma cells (22). More
recently, HURP has been demonstrated to be the direct target gene
of NOTCH3, as growth inhibition in ovarian cancer cells induced by
pharmacological or RNA interference-mediated NOTCH inhibition is
notably prevented by the enforced expression of HURP (23). The present results are consistent
with these findings, indicating that the deregulation of HURP
expression, such as overexpression, in tumor cells, inhibits cell
growth.
HURP is an essential component of the mitotic
apparatus, which can form a complex with RanGTP and localize
predominantly to the K-MTs in vivo. By stabilizing the MTs,
HURP promotes MT polymerization and bipolar spindle formation when
cells enter mitosis (7). Recent
studies have demonstrated that the modulation of kinesin Kif18A
function by HURP results in the regulation of chromosome
congression. A higher level of HURP expression leads to increased
Kif18A sequestration at the K-MTs and a chromosome congression
defect is more likely to occur (24). In other studies, HURP reduced the
levels of p53 in normal and cancerous cells, and is therefore
indicated to act as an oncogene. Thus, suppression of HURP may
interfere with the interphase dynamics of MTs, affect the growth or
stability of spindle MTs and inhibit tumor growth. MT-targeting
agents have made a noteworthy contribution to cancer therapy over
the past 50 years and include the vinca alkaloids and taxanes,
which have been used to treat a broad range of malignancies
(25,26). Therefore, HURP-targeted therapy may
be of potential benefit in treating breast cancer in the future.
The present study attempted, for the first time, to transfect
anti-HURP Abs in order for them to directly act on HURP in cancer
cells. The results of the anti-HURP Ab transfection demonstrate
that HURP-targeted therapy may be effective in blocking the
progression of breast cancer. McAbs or polypeptide drugs, which
have a more effective target area and less toxicity, will be the
focus of future studies.
In conclusion, the present study found that HURP
expression was significantly elevated in breast cancer tumors and
that elevated HURP expression was associated with the proliferation
of breast cancer and the degree of malignancy. In addition to being
a tumor biomarker for prognosis, HURP may serve as a potential
therapeutic drug target for human breast cancer.
References
|
1
|
Polyak K: Breast cancer: origins and
evolution. J Clin Invest. 117:3155–3163. 2007.
|
|
2
|
Bray F, McCarron P and Parkin DM: The
changing global patterns of female breast cancer incidence and
mortality. Breast Cancer Res. 6:229–239. 2004.
|
|
3
|
Downs-Holmes C and Silverman P: Breast
cancer: overview & updates. Nurse Pract. 36:20–26. 2011.
|
|
4
|
Sauer G, Körner R, Hanisch A, Ries A, Nigg
EA and Silljé HH: Proteome analysis of the human mitotic spindle.
Mol Cell Proteomics. 4:35–43. 2005.
|
|
5
|
Tsou AP, Yang CW, Huang CY, et al:
Identification of a novel cell cycle regulated gene, HURP,
overexpressed in human hepatocellular carcinoma. Oncogene.
22:298–307. 2003.
|
|
6
|
Silljé HH, Nagel S, Körner R and Nigg EA:
HURP is a Ran-importin beta-regulated protein that stabilizes
kinetochore microtubules in the vicinity of chromosomes. Curr Biol.
16:731–742. 2006.
|
|
7
|
Koffa MD, Casanova CM, Santarella R, et
al: HURP is part of a Ran-dependent complex involved in spindle
formation. Curr Biol. 16:743–754. 2006.
|
|
8
|
Yu CT, Hsu JM, Lee YC, et al:
Phosphorylation and stabilization of HURP by Aurora-A: implication
of HURP as a transforming target of Aurora-A. Mol Cell Biol.
25:5789–5800. 2005.
|
|
9
|
Sasai K, Parant JM, Brandt ME, et al:
Targeted disruption of Aurora A causes abnormal mitotic spindle
assembly, chromosome misalignment and embryonic lethality.
Oncogene. 27:4122–4127. 2008.
|
|
10
|
Wong J, Lerrigo R, Jang CY and Fang G:
Aurora A regulates the activity of HURP by controlling the
accessibility of its microtubule-binding domain. Mol Biol Cell.
19:2083–2091. 2008.
|
|
11
|
Wong J and Fang G: HURP controls spindle
dynamics to promote proper interkinetochore tension and efficient
kinetochore capture. J Cell Biol. 173:879–891. 2006.
|
|
12
|
Negrini S, Gorgoulis VG and Halazonetis
TD: Genomic instability - an evolving hallmark of cancer. Nat Rev
Mol Cell Biol. 11:220–228. 2010.
|
|
13
|
Bakhoum SF and Compton DA: Kinetochores
and disease: keeping microtubule dynamics in check! Curr Opin Cell
Biol. 24:64–70. 2012.
|
|
14
|
Fragoso MC, Almeida MQ, Mazzuco TL, et al:
Combined expression of BUB1B, DLGAP5, and PINK1 as predictors of
poor outcome in adrenocortical tumors: validation in a Brazilian
cohort of adult and pediatric patients. Eur J Endocrinol.
166:61–67. 2012.
|
|
15
|
Chiu AW, Huang YL, Huan SK, et al:
Potential molecular marker for detecting transitional cell
carcinoma. Urology. 60:181–185. 2002.
|
|
16
|
Lakhani and Sunil R: WHO Classification of
Tumours of the Breast. 4th edition. International Agency for
Research on Cancer; Lyon, France: 2012
|
|
17
|
Singletary SE and Greene FL: Revision of
breast cancer staging: The 6th edition of the TNM Classification.
Seminars in Surgical Oncology. 21. Wiley Subscription Services,
Inc., A Wiley Company; 2003
|
|
18
|
Kuo TC, Chang PY, Huang SF, Chou CK and
Chao CC: Knockdown of HURP inhibits the proliferation of
hepacellular carcinoma cells via downregulation of gankyrin and
accumulation of p53. Biochem Pharmacol. 83:758–768. 2012.
|
|
19
|
Betz MJ and Beuschlein F: Diagnosis: Novel
molecular signatures for adrenocortical carcinoma. Nat Rev
Endocrinol. 5:297–299. 2009.
|
|
20
|
Huang YL, Chiu AW, Huan SK, et al:
Prognostic significance of hepatoma-up-regulated protein expression
in patients with urinary bladder transitional cell carcinoma.
Anticancer Res. 23:2729–2733. 2003.
|
|
21
|
Wang YC, Lee YH, Huang GC, et al: Enhanced
transformation and chemosensitivity of NIH3T3 cells transduced with
hepatoma upregulated protein. Biochem Biophys Res Commun.
340:244–249. 2006.
|
|
22
|
Zhao L, Qin LX, Ye QH, et al: KIAA0008
gene is associated with invasive phenotype of human hepatocellular
carcinoma - a functional analysis. J Cancer Res Clin Oncol.
130:719–727. 2004.
|
|
23
|
Chen X, Thiaville MM, Chen L, et al:
Defining NOTCH3 target genes in ovarian cancer. Cancer Res.
72:2294–2303. 2012.
|
|
24
|
Ye F, Tan L, Yang Q, et al: HURP regulates
chromosome congression by modulating kinesin Kif18A function. Curr
Biol. 21:1584–1591. 2011.
|
|
25
|
Zelnak AB: Clinical pharmacology and use
of microtubule-targeting agents in cancer therapy. Methods Mol Med.
137:209–234. 2007.
|
|
26
|
Jordan MA and Wilson L: Microtubules as a
target for anticancer drugs. Nat Rev Cancer. 4:253–265. 2004.
|