Introduction
MutL homolog 1 (MLH1), a constituent gene in
the mismatch repair pathway, carries germline mutations in
individuals with Lynch syndrome, also termed hereditary
non-polyposis colorectal cancer (HNPCC). The gene is reported to
acquire 300 different germline mutations, which in addition to
mutations in other genes involved in mismatch repair pathway,
mainly mutS homolog 2 (MSH2), predispose individuals to the
disease (1). MLH1, a key protein of
the mismatch repair process, contains interaction domains for MutS
homologs, including MSH2, MSH3 and MSH6, postmeiotic segregation
increased 2 (PMS2), MLH3 and PMS1(2). The heterodimers formed by MLH1 recruit
proteins for the excision and repair synthesis.
The germline mutations in MLH1 in HNPCC
include nucleotide substitutions, which result in missense,
nonsense or splicing errors and also comprise insertions/deletions.
A number of founder mutations, which account for a high proportion
of mutations in families with HNPCC, which have been reported in
patients with Lynch syndrome (3).
In addition, certain non-pathogenic mutations in exons and introns
of the gene have also been reported (4,5). The
MLH1 gene is highly polymorphic, with >1,600 variants
reported to date (http://genecards.org/cgi-bin/carddisp.pl?gene=MLH1&search=mlh1%23snp).
In this study, a novel germline mutation in
MLH1 in a patient with sporadic colorectal cancer (CRC) is
reported, which was detected during the whole genome mutational
screening. In addition, a total of 1,095 sporadic CRC patients and
1,469 controls were tested for the detected mutation.
Materials and methods
Study population
This study included a group of 104 newly diagnosed
CRC patients with DNA extracted from their tumor tissues, adjacent
healthy mucosa and peripheral blood tissues. A replication group
included 1,095 CRC patients and 1,469 controls from whom DNA was
extracted from peripheral blood lymphocytes. The information
regarding the CRC cases and controls included in the replication
group is shown in Table I and has
been described previously (6,7).
Patients included in this study attended the General University
Hospital (Prague, Czech Republic), Thomayer Hospital (Prague, Czech
Republic), Central Military Hospital (Prague, Czech Republic),
Faculty Hospital (Brno, Czech Republic), Regional Hospital Benesov
(Benesov, Czech Republic), Regional Hospital Liberec (Liberec,
Czech Republic), Hospital Na Plesi (Nova Ves pod Plesi, Czech
Republic), Regional Hospital Pribram (Pribram, Czech Republic),
Masaryk Regional Hospital (Ústí nad Labem, Czech Republic), Tomas
Bata Regional Hospital (Zlín, Czech Republic) or Jihlava Regional
Hospital (Jihlava, Czech Republic). This study was approved by the
ethics commitee of the General University Hospital in Prague
(Prague, Czech Republic) and written informed consent was obtained
from all patients.
 | Table ICharacteristics of the study
population. |
Table I
Characteristics of the study
population.
| Characteristic | CRC cases
(n=1095) | Control group I, CFCC
(n=688) | Control group II,
HBDV (n=781) | All controls
(n=1469) | OR | 95% CI | P-valuea |
|---|
| Tumor
localization |
| Colon | 725 | - | - | - | | | |
| Rectum | 370 | - | - | - | | | |
| Age (years) |
| 47≤ | 94 | 164 | 427 | 591 | Ref. | | |
| 48–55 | 208 | 145 | 277 | 422 | 3.10 | 2.36–4.09 | ≤0.01 |
| 56–65 | 370 | 209 | 77 | 286 | 8.13 | 6.25–10.66 | ≤0.01 |
| >65 | 423 | 170 | 0 | 170 | 15.37 | 11.66–20.44 | ≤0.01 |
| Gender |
| Female | 435 | 317 | 343 | 660 | Ref. | | |
| Male | 660 | 371 | 438 | 809 | 1.23 | 1.05–1.45 | 0.01 |
| BMI |
| 23.7≤ | 184 | 154 | 215 | 369 | Ref. | | |
| 23.7–26.2 | 192 | 147 | 213 | 360 | 1.07 | 0.83–1.37 | 0.61 |
| 26.3–28.9 | 226 | 139 | 184 | 323 | 1.40 | 1.10–1.79 | 0.01 |
| >28.9 | 222 | 172 | 157 | 329 | 1.35 | 1.06–1.73 | 0.02 |
| Smoking history |
| No | 536 | 364 | 451 | 815 | Ref. | | |
| Yesb | 501 | 254 | 327 | 581 | 1.31 | 1.12–1.54 | ≤0.01 |
| Family history of
CRC |
| No | 726 | 486 | 718 | 1204 | Ref. | | |
| Yes | 144 | 90 | 52 | 142 | 1.68 | 1.31–2.16 | ≤0.01 |
| Address |
| City | 511 | 338 | 614 | 952 | Ref. | | |
| Suburbs | 128 | 118 | 53 | 171 | 1.39 | 1.08–1.79 | 0.01 |
| Countryside | 242 | 157 | 112 | 270 | 1.67 | 1.36–2.05 | ≤0.01 |
| Education |
| Basic | 266 | 171 | 53 | 224 | Ref. | | |
| Medium | 469 | 327 | 492 | 820 | 0.48 | 0.39–0.59 | ≤0.01 |
| High | 138 | 114 | 231 | 345 | 0.34 | 0.26–0.44 | ≤0.01 |
Mutation screening in CRC patients
DNA from tumor tissues, healthy mucosa and blood
tissues was extracted using QIAamp DNA Mini Kit and QIAcube
(Qiagen, Hilden, Germany). The extracted DNA of the 104 newly
diagnosed CRC patients were subjected to mutation detection by
high-resolution melting (HRM) using LightCycler® and a 480 High
Resolution Melting Master® kit (Roche Diagnostics GmbH, Mannheim,
Germany). Polymerase chain reaction amplicons were designed to scan
the MLH1 gene using HRM analysis. All 19 exons in the
MLH1 gene were screened. The region containing exon 2 was
amplified using primers with the following sequence: Forward,
5′-AGTTTGTTATCATTGCTTGGCTCAT-3′ and reverse,
5′-TCCAGAACAGAGAAAGGTCCTGACT-3′ (8). The 10-μl reaction mixture contained 20
ng genomic DNA, 0.4 mM of each primer and 3 mM MgCl2.
The reaction conditions were as follows: Activation step at 95°C
for 10 min followed by 45 cycles of 95°C for 15 sec, 60°C for 15
sec, 72°C for 25 sec and 72°C for 7 min. Samples with positive HRM
signals were further analyzed by sequencing. Sequencing reactions
were carried out in a total volume of 10 μl containing 2 μl Big
Dye® Terminator v1.1 Cycle Sequencing kit (Applied
Biosystems, Life Technologies, Foster City, CA, USA), 2.4 μl
H2O, 0.3 μl DNA template and 0.3 μl of one primer. The
reaction products were purified by ethanol precipitation and
sequenced by automated sequencing (Applied Biosystems®
3130 Genetic Analyzer; Applied Biosystems, Life Technologies). The
reference sequence of the MLH1 gene (NG_007109.1) was
obtained from the National Center for Biotechnology Information
database (http://www.ncbi.nlm.nih.gov/nuccore/NG_007109.1?from=4863&to=62359&report=genbank).
Genotyping of the replication group
DNA samples from the replication group were
genotyped for the novel germ-line variant. The DNA samples from CRC
patients and controls were analyzed at the K Bio Science facility
(K Bio Science UK Ltd., Hoddesdon, UK) (protocol available at
http://www.kbioscience.co.uk/reagents/KASP_manual.pdf)
under conditions described previously (6).
Results
In the initial group, 1/104 sporadic CRC patients
exhibited a single nucleotide variant at codon 204 within exon 2 of
MLH1 in tumor tissue and mucosa, as well as in blood
lymphocytes DNA. A change of the base C to G resulted in Ile68Met
change in the amino acid residue (Fig.
1). The carrier of the newly identified variant was a 59 year
old female with rectal cancer (T3N0M0 stage). Although the family
history of the patient was unavailable, the patient had been
treated for schizophrenia for 15 years. The tumor was
microsatellite-stable, the CpG sites on the MLH1 promoter
were not methylated and the expression of MLH1 protein was not
altered in the tumor, when compared with the adjacent mucosa (data
not shown). Genotyping of the replication revealed that there were
no carriers of the newly identified variant Ile68Met in the CRC or
control groups.
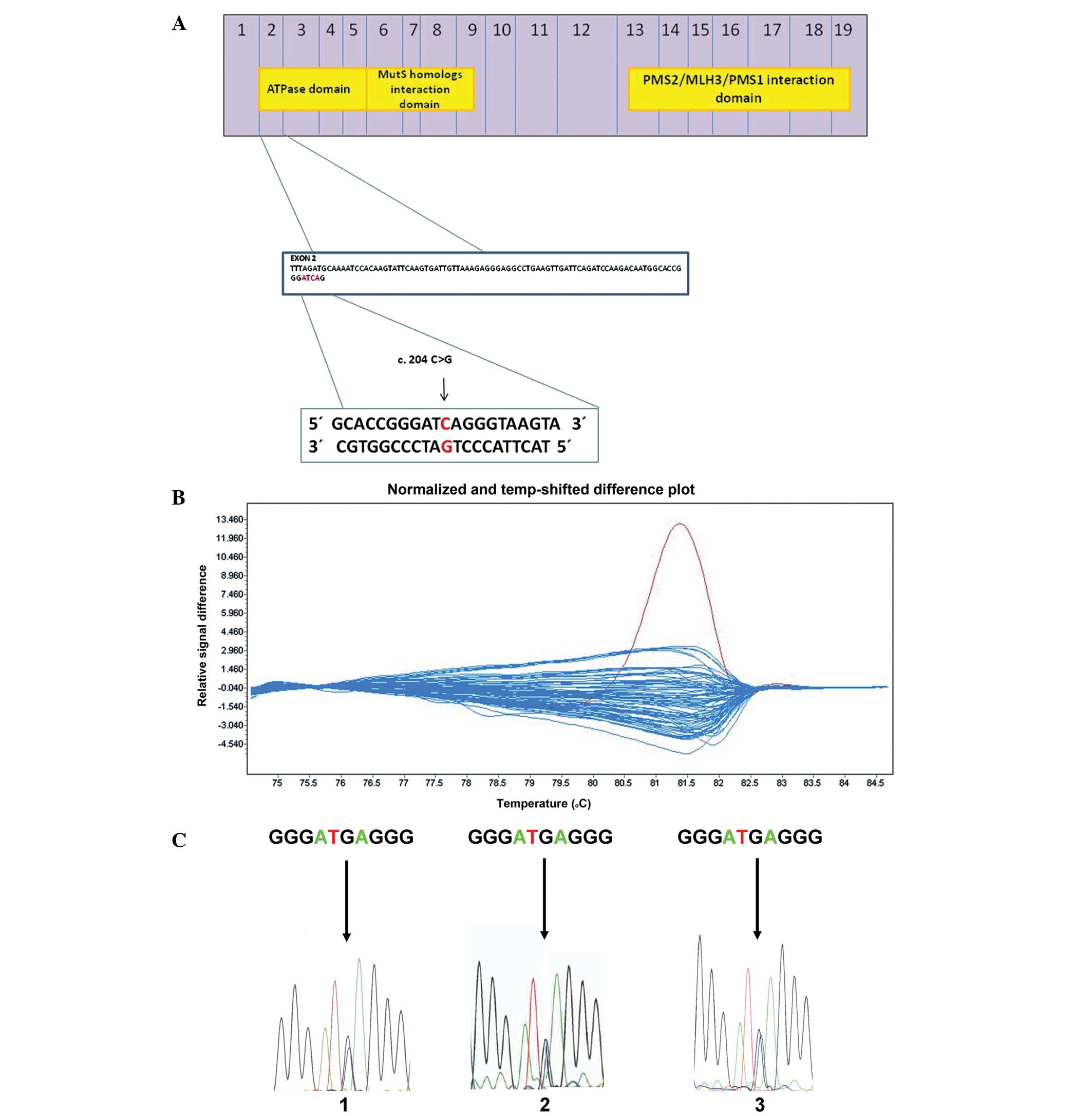 | Figure 1HRM analysis and DNA sequencing of
exon 2 of the MLH1 gene. (A) Diagram of the MLH1 protein in
scale. Numbers inside the blue boxes indicate the numbers of exons
from which each part of the protein is translated. The three yellow
boxes inside represent the ATPase domain, the MutS homolog
interaction domain and the PMS2/MLH3/PMS1 interaction domain. A new
germ-line variant in exon 2 is located at position c. 204 C>G,
p. Ile68Met, corresponding to the gene region coding a part of
ATPase domain of MLH1 protein. The position c. 204 is shown by the
black arrow. (B) HRM analysis of exon 2 in tumor DNA samples. The
difference plot chart shows relativity between DNA melting
temperature and relative intensity of emitted fluorescence. Each
curve represents the melting of one DNA sample. The DNA sample
bearing the novel gene variant exhibits a different melting
profile, which is highlighted by the red color in comparison with
the blue-colored melting profiles of wild-type DNA samples. This
discrepancy of different melting curves is caused by nucleotide
change in the analyzed DNA sequence. The red curve represents the
melting of the DNA sample which bears the newly identified variant,
c. 204 C>G, p. Ile68Met, in tumor tissue. To verify the possible
germ-line origin of the variant, HRM was performed in healthy
tissue and peripheral blood DNA samples. Both samples were positive
for the same variant (data not shown). (C) Comparison of DNA
sequencing plots of three different DNA samples obtained from the
same patient bearing new heterozygous germ-line gene variant c. 204
C>G, p. Ile68Met. 1, tumor DNA sample; 2, healthy mucosa tissue
DNA sample; and 3, peripheral blood DNA sample. MLH1, mutL homolog
1; PMS2, postmeiotic segregation increased 2;HRM, high-resolution
melting. |
Discussion
In this study, a unique variant c. 204 C>G, p.
Ile68Met in exon 2 of the MLH1 gene was identified in a
patient with sporadic CRC. The mutation was germline and was also
detectable in the DNA of tumor tissue, colon mucosal tissue and DNA
of the peripheral lymphocytes of the patient. Predictive algorithms
SIFT (9) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/)
indicated that the amino acid change from isoleucin to methionin
may influence the functionality of the MLH1 protein. A 3D model of
the MLH1 protein demonstrated that amino acid residue 68 isoleucin
is located within the enzymatic core that interacts with ATP
molecules (http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=90223).
Therefore, a substitution by methionine may decrease MLH1 activity.
Similarly, a variant in an HNPCC patient at the position c.203
T>A, p. Ile68Asn was previously shown to be deleterious
(10). Furthermore, a homologous
substitution in yeast was found to cause loss of function in a
mismatch repair assay (11).
The identification of a novel germline variant in
MLH1 with putative impact on the protein function is of
significant importance in CRC. MLH1 is the most frequently
mutated gene in HNPCC and is also often altered in sporadic forms
(12). The novel variant in
MLH1 gene presents a rare event in sporadic CRC, which was
identified in 1/1,199 patients. The effect of the mutation, whether
a causal germline variant or a rare polymorphism, remains to be
determined. Due to the patient history, an association between the
novel mutation and mental illness must not be excluded.
Neurodegenerative diseases, such as Huntington’s disease, are also
known to exhibit alterations in mismatch repair genes (13).
In conclusion, in the present study a novel
MLH1 mutation was detected in a patient with sporadic CRC.
The functionality of the novel c. 204 C>G, p.Ile68Met variant in
exon 2 of MLH1 gene remains to be determined experimentally,
along with its occurrence and relevance in other cancer types.
Acknowledgements
The study was supported by grants from the Grant
Agency of the Czech Republic (grant no. P304/11/P715) and the First
Medical Faculty, Charles University, Prague, Czech Republic (grant
nos. P304/12/1585 and Prvouk-P27/LF1/1).
References
|
1
|
Pritchard CC and Grady WM: Colorectal
cancer molecular biology moves into clinical practice. Gut.
60:116–129. 2011. View Article : Google Scholar
|
|
2
|
Cannavo E, Gerrits B, Marra G, Schlapbach
R and Jiricny J: Characterization of the interactome of the human
MutL homologues MLH1, PMS1, and PMS2. J Biol Chem. 282:2976–2986.
2007. View Article : Google Scholar
|
|
3
|
Nystrom-Lahti M, Wu Y, Moisio AL, et al:
DNA mismatch repair gene mutations in 55 kindreds with verified or
putative hereditary non-polyposis colorectal cancer. Hum Mol Genet.
5:763–769. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bianchi F, Raponi M, Piva F, et al: An
intronic mutation in MLH1 associated with familial colon and breast
cancer. Fam Cancer. 10:27–35. 2011. View Article : Google Scholar :
|
|
5
|
Kolodner RD, Tytell JD, Schmeits JL, et
al: Germ-line msh6 mutations in colorectal cancer families. Cancer
Res. 59:5068–5074. 1999.PubMed/NCBI
|
|
6
|
Naccarati A, Pardini B, Stefano L, et al:
Polymorphisms in miRNA-binding sites of nucleotide excision repair
genes and colorectal cancer risk. Carcinogenesis. 33:1346–1351.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pardini B, Rosa F, Barone E, et al:
Variation within 3′-UTRs of base excision repair genes and response
to therapy in colorectal cancer patients: A potential modulation of
microRNAs binding. Clin Cancer Res. 19:6044–6056. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rouleau E, Lefol C, Bourdon V, et al:
Quantitative PCR high-resolution melting (qPCR-HRM) curve analysis,
a new approach to simultaneously screen point mutations and large
rearrangements: application to MLH1 germline mutations in Lynch
syndrome. Hum Mutat. 30:867–875. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ng PC and Henikoff S: SIFT: Predicting
amino acid changes that affect protein function. Nucleic Acids Res.
31:3812–3814. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
annergard P, Lipford JR, Kolodner R,
Frodin JE, Nordenskjold M and Lindblom A: Mutation screening in the
hMLH1 gene in Swedish hereditary nonpolyposis colon cancer
families. Cancer Res. 55:6092–6096. 1995.
|
|
11
|
Ellison AR, Lofing J and Bitter GA: Human
MutL homolog (MLH1) function in DNA mismatch repair: a prospective
screen for missense mutations in the ATPase domain. Nucleic Acid
Res. 32:5321–5338. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Boland CR and Goel A: Microsatellite
instability in colorectal cancer. Gastroenterology. 138:2073–2087
e2073. PubMed/NCBI
|
|
13
|
Kantartzis A, Williams GM, Balakrishnan L,
Roberts RL, Surtees JA and Bambara RA: Msh2–Msh3 interferes with
Okazaki fragment processing to promote trinucleotide repeat
expansions. Cell Rep. 2:216–222. 2012. View Article : Google Scholar : PubMed/NCBI
|















