Introduction
Mitochondrial succinate dehydrogenase (SDH) is a
tetrameric iron-sulfur flavoprotein of the tricarboxylic acid (TCA)
cycle and respiratory chain. The SDH complex, subunit C (SDHC) is a
membrane-anchoring subunit of SDH that is tethered to the
mitochondrial matrix (1). SDH
oxidizes succinate to fumarate in the eighth step of the TCA
respiratory cycle (2), generating
two hydrogen ions and two electrons, which are transferred to
ubiquinone through the iron-sulfur cluster and flavin adenine
dinucleotide. SDHC is involved in the TCA cycle and electron
transport chain (3–5).
Hereditary paragangliomas (PGLs) are neuroendocrine
tissue tumors that arise symmetrically along the spinal axis
between the skull and the pelvis (6). The pathogenic mechanism of PGLs has
not yet been elucidated; however, mRNA splicing deficiencies of
proteins lacking normal function have been implicated in various
diseases, including PGLs (7–9).
Alternative splicing of primary transcripts is a fundamental
biological process involved in gene expression. In general,
alternative splicing produces variant proteins and expression
patterns as products of different genes. Up to one-third of human
genes are alternatively spliced (10), and several alternative splicing
variants (ASVs) have been associated with human diseases (11,12).
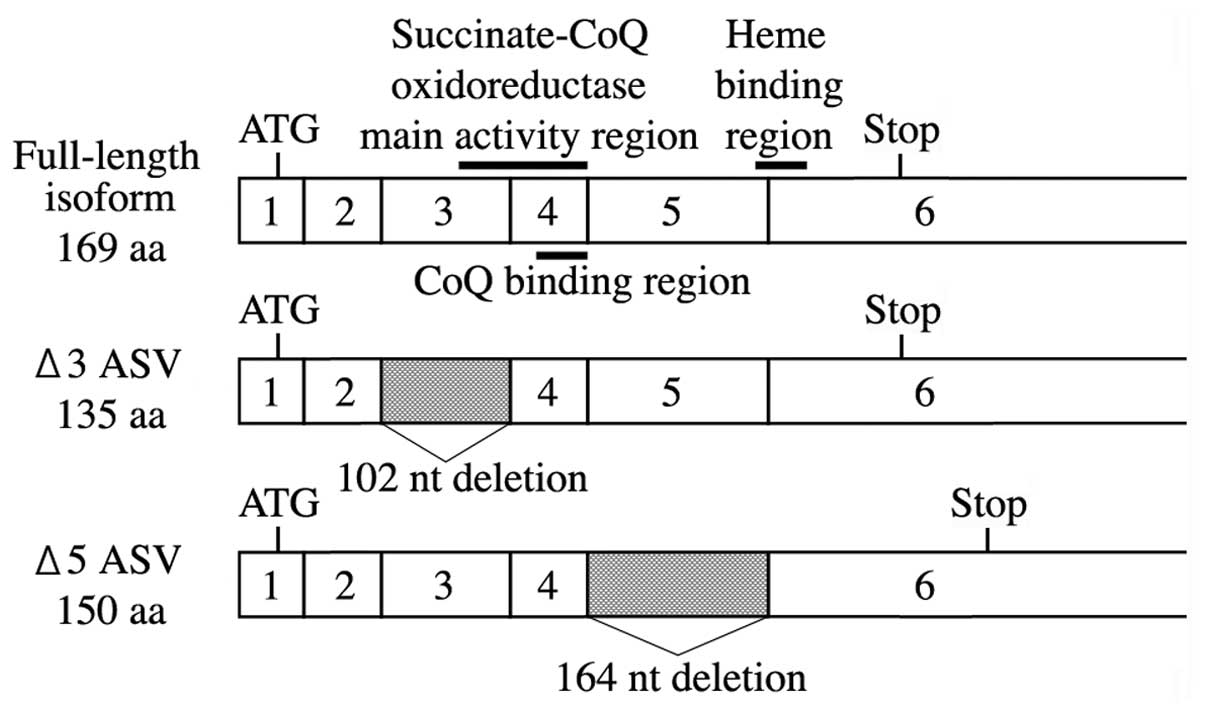
The human SDHC gene maps to q23.3 on the long arm of
chromosome 1, spanning >50 kb, and is composed of seven exons
separated by intronic sequences (13). The complete cDNA sequence
encompasses full-length SDHC, encoding a 169-amino acid (aa)
polypeptide. Deleted SDHC transcripts have been
characterized, comprising two isoforms generated by in-frame
deletions of 102 nucleotides corresponding to the complete loss of
exon 3, and a frameshift deletion of 164 nucleotides corresponding
to complete loss of exon 5. The exon 3-deleted variant (Δ3 isoform)
lacks exon 3, resulting in partial loss of the succinate-Coenzyme Q
(CoQ) oxidoreductase main activity region (14). In the exon 5-deleted variant (Δ5
isoform), the frameshift moves the stop codon to the 3′ non-coding
region, adding an extra 70 aa to the final SDHC protein. However,
frameshift mutations within this region abolish enzyme activity due
to the loss of the heme binding region (13,14).
Certain SDH mutations cause the enzyme to become a
significant source of mitochondrial superoxide production, which
may contribute directly to disease progression (15). Of particular interest are diseases
associated with reactive oxygen species (ROS) generated in the
electron transport system (15,16).
In the present study, to explore the mechanism of the PGL tumor
development, the occurrence of SDHC gene ASVs was
investigated, in particular, deleted exon 5 ASVs, which may induce
frameshift mutations. The correlation between ROS and SDHC
ASVs was also investigated.
Materials and methods
Cell culture
The HCT-15 (colorectal adenocarcinoma) cell line
(Japanese Collection of Research Bioresources, Osaka, Japan) was
maintained in Dulbecco’s modified Eagle’s medium (Gibco-BRL,
Carlsbad, CA, USA) supplemented with 10% heat-inactivated fetal
bovine serum under standard culture conditions (in a humidified
atmosphere of 5% CO2 at 37°C), until cells reached
80–90% confluence. Cells were treated with Accumax (Innovative Cell
Technologies, Inc., Logan, UT, USA) and counted using the Cell Lab
Quanta SC flow cytometer (Beckman Coulter, Brea, CA, USA),
according to the manufacturer’s instructions.
Cells were incubated for 2 h with or without 10 mM
2,2′-azobis(2-amidinopropane) dihydrochloride (AAPH), 1 mM
H2O2 or 200 μM thallium trifluoroacetate
(TTFA) (all Sigma-Aldrich, St. Louis, MO, USA). Apoptosis was
determined by Annexin V (Beckman Coulter, Brea, CA, USA) and
propidium iodide (Sigma-Aldrich) staining.
Reverse transcription-polymerase chain
reaction (RT-PCR) and expression vector generation
To identify SDHC gene ASVs, total RNA was
extracted from normal lung tissue purchased from Clontech
Laboratories (Mountain View, CA, USA). Total RNA from HTC-15 cells
was extracted using the ReliaPrep™ RNA cell miniprep system
(Promega Corporation, Madison, WI, USA), following the
manufacturer’s instructions. To prepare cDNA, DNase-treated total
RNA (0.1 μg) was incubated with M-MLV reverse transcriptase
(Invitrogen Life Technologies, Carlsbad, CA, USA) and random
primers (Invitrogen Life Technologies). The primer set for the
amplification of SDHC cDNA was designed according to GenBank
sequences: NM_003001.3 (full-length isoform), AB211234.1 (Δ3 ASV),
and AB211235.1 (Δ5 ASV) (Table I;
Fig. 1). PCR parameters were 95°C
for 20 sec, 60°C for 30 sec and 45 cycles of 72°C for 20 sec,
followed by a 10-min extension at 72°C, using AmpliTaq Gold DNA
polymerase (Applied Biosystems, Foster City, CA, USA). PCR products
were separated by electrophoresis on 2.0% agarose gels in
Tris-borate-EDTA buffer, stained with ethidium bromide, and then
detected under ultraviolet light. The PCR products were purified
using the High Pure PCR Product purification kit (Roche Applied
Science, Upper Bavaria, Germany), cloned into the pTriEX-3 neo
expression vector (Merck Millipore, Darmstadt, Germany), and
sequenced using the BigDye Terminator v3.1 Cycle sequencing kit
(Applied Biosystems) with the ABI PRISM® 3130 genetic
analyzer (Applied Biosystems). Finally, sequences were compared
with the full-length SDHC mRNA sequence. HCT-15 cells at
densities of ~1.0–3.0×105 were precultured with 2 mg of
plasmid DNA and 3 ml of FuGENE HD transfection reagent (Promega
Corporation). Following 15 min of incubation at room temperature,
cells were incubated at 37°C and 5% CO2 for 24 h.
SDHC-pTriEX-3 neo vector-transfected cells were selected
using geneticin (Roche Applied Science).
 | Table IDetails of the primers used. |
Table I
Details of the primers used.
| Gene | Accession no. | Deleted exon
(nt) | Primer | Sequence | Product size, bp |
|---|
| Full-length
isoform | NM_003001.3 | - | Forward |
5′-TATAGGTTCAAACCGTCCTCTG-3′ | 217 |
| | | Reverse |
5′-GGATCAGTGCTGGACCTAAGC-3′ | |
| Δ3 ASV | AB211234.1 | Exon 3 (102) | Forward |
5′-CTCAGCTCTGTATCAGAAATTGGT-3′ | 175 |
| | | Reverse |
5′-TGCAAACTTAGCTGTGTGGATCAGTGC-3′ | |
| Δ5 ASV | AB211235.1 | Exon 5 (164) | Forward |
5′-TATAGGTTCAAACCGTCCTCTG-3′ | 201 |
| | | Reverse |
5′-TTCCTAGGTCCCACATCTGCA-3′ | |
| GAPDH | X01677 | - | Forward |
5′-CGAGCCACATCGCTCAGACACC-3′ | 118 |
| | | Reverse |
5′-GGCAACAATATCCACTTTACCAGAG-3′ | |
Real-time RT-PCR
Real-time PCR reaction mixtures were prepared using
Kapa Sybr® Fast qPCR Master Mix (Kapa Biosystems,
Woburn, MA, USA). The primers used for the amplification of the
full-length and ASV isoforms of SDHC, as well as the
GAPDH mRNAs are shown in Table
I. Real-time PCR reactions were performed for 45 cycles (95°C
for 20 sec, 60°C for 25 sec and 72°C for 20 sec) using a real-time
PCR system (iCycler iQ™ real-time detection system; Bio-Rad,
Hercules, CA, USA). Following extension, an additional step was
added whereby the sample was maintained at 75°C for 5 sec to allow
for the fluorescence signal to be read. GAPDH housekeeping
mRNA was amplified for normalization purposes, and mRNA expression
levels are presented as the mRNA copy number per microgram of total
RNA. Data are presented as the mean of three measurements.
SDH activity
The transformed cells were pre-cultured at densities
of ~1.0–3.0×105 at 37°C and 5% CO2. The
culture medium was removed and the cells were incubated for 1 h
with 2.5 mM 3-(4,5-di-methylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide in 50 mM KPi (pH 8.0), 2 mM KCN and 350 mM
1-methoxy-5-methylphenazinium methylsulfate with 10 mM succinic
acid. Following incubation, the reagent absorbance at 530 nm was
measured using a FoodMark microplate reader (Bio-Rad). Data are
presented as the mean of three measurements.
Statistical analysis
All samples in the experiments were tested in
triplicate or quadruplicate. All data are presented as the mean ±
standard deviation. Differences between the mean values were
evaluated using Student’s t-test. P<0.05 was considered to
indicate a statistically significant difference.
Results
RT-PCR and real-time RT-PCR
Using RT-PCR with the primer sets for exons 1 and 6
of human SDHC mRNA, amplification products of three sizes
were obtained; specifically, a major amplification product of 566
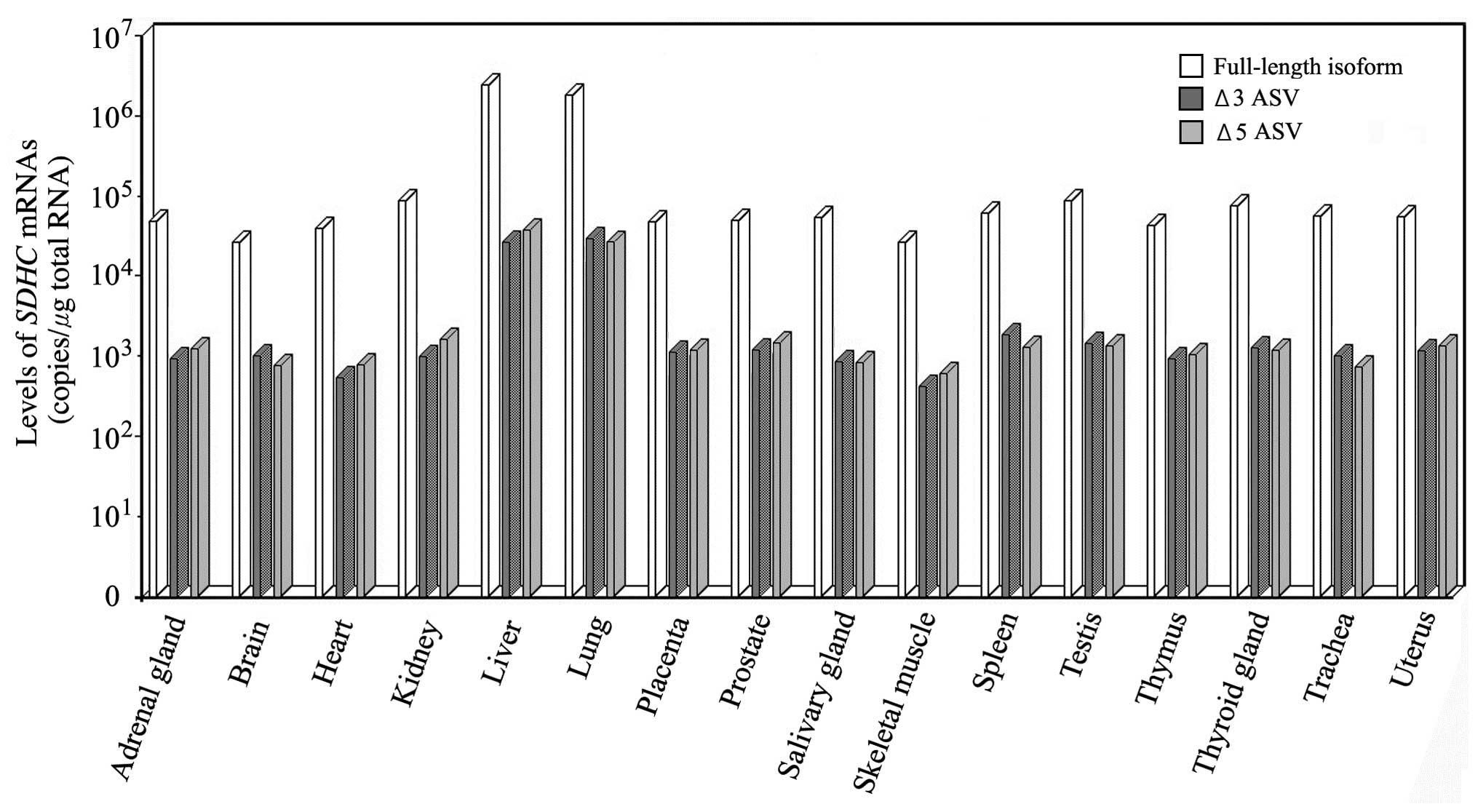
bp, and minor products of 464 and 402 bp (Fig. 1). To determine isoform expression
in vivo, real-time RT-PCR was used to examine normal cells
derived from 16 tissues, which included the adrenal gland, brain,
heart, kidney, liver, lung, placenta, prostate, salivary gland,
skeletal muscle, spleen, testis, thymus, thyroid gland, trachea and
uterus. Full-length SDHC mRNA was expressed in every tissue
examined. Δ3 and 5 ASV mRNAs were also ubiquitously expressed, but
by two orders of magnitude lower than that of the full-length mRNA
(Fig. 2).
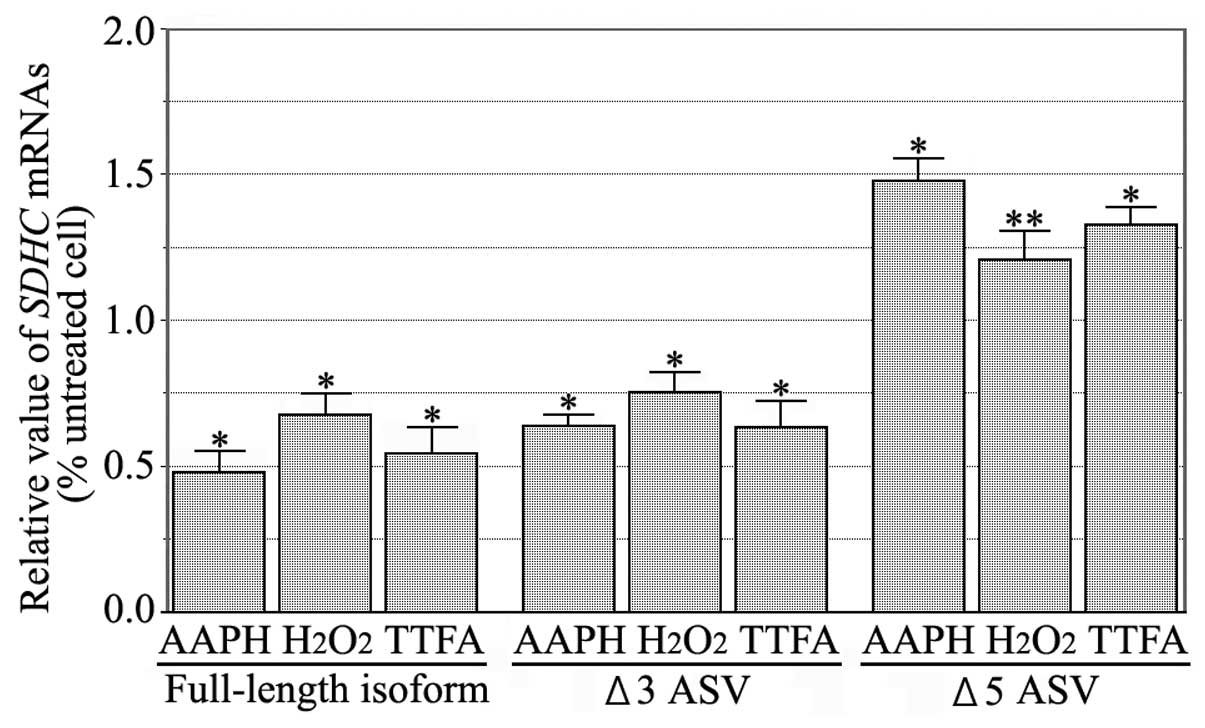
The treatment of HCT-15 cells with ROS reagents
produced a detectable apoptosis signal, as determined by Annexin V.
The incubation of HCT-15 cells with AAPH,
H2O2 or TTFA exhibited no synchronization
with the G0/G1 or G2/M phases of
the cell cycle, suggesting that none of the reagents cause changes
in state with regard to the cell cycle. The expression levels of
SDHC ASV mRNAs following treatment with ROS reagents, were
measured by real-time RT-PCR. The SDHC Δ5 ASV mRNA levels of
the treated cells were marginally elevated compared with the
untreated cells (P<0.05) (Fig.
3).
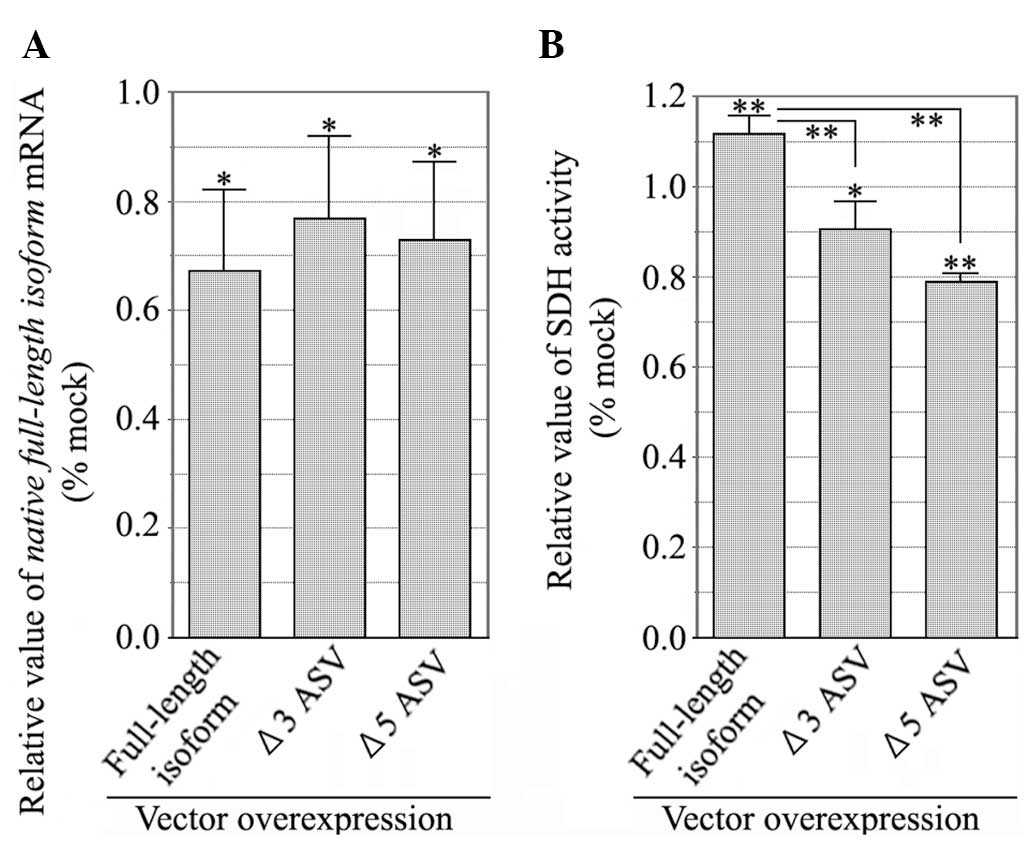
The expression levels of endogenous SDHC mRNA
in SDHC ASV overexpressing cells were measured by real-time
RT-PCR. Compared with the mock-transfected cells (transfected using
empty pTriEX-3 neo vector), the endogenous SDHC mRNA levels
in cells transfected with SDHC ASV overexpression vectors,
were significantly decreased to ~30%. These results suggested that
SDHC ASVs act as potent dominant-negative inhibitors of the
full-length isoform (Fig. 4A).
SDH activity
Full-length isoform-expressing cells significantly
increased SDH activity, compared with the control cells expressing
empty vector. However, cells overexpressing the Δ3 and Δ5 isoforms
exhibited reduced SDH activity. Compared with SDH activity from
full-length isoform-expressing cells, SDHC Δ5 ASV expression
significantly decreased the SDH activity to ~40% (P<0.05)
(Fig. 4B). These results
corroborate the finding that SDHC ASVs act as potent
dominant-negative inhibitors of the full-length isoform.
Discussion
Genes involved in cellular respiration, such as
SDH, are known to act as tumor suppressors, encoding
proteins that inhibit tumor formation and cell proliferation
(17,18). Mutations in tumor-suppressing genes
promote carcinogenesis through DNA replication, causing events such
as dysregulation in cell proliferation as well as the induction of
apoptosis. Furthermore, cancer is a disease leading to tumor
formation, and alternative splicing is triggered by disease onset.
A number of mutations in human SDH genes are responsible for
the development of PGLs, gastrointestinal stromal tumors and other
types of cancer (5,18). In the current study, SDHC ASV
was found to have a dominant-negative effect on SDHC activity,
which provides a new possible target for cancer therapy. With
improved understanding of the factors regulating SDHC
alternative splicing, it may be possible to limit the tumor cell
growth potential by promoting the production of non-functional or
dominant-negative types of SDHC ASVs. During development,
the dominant-negative function of the Δ5 variant may be significant
when certain cells undergo differentiation, yet exhibit incomplete
repression of SDHC transcription. During tumorigenesis,
cancer cells may evolve from differentiated cell types, in which
alternative splicing mechanisms may already be in place. Thus,
SDHC alternative splicing in tumor cells may reflect
pre-existing alternative splicing mechanisms present in the parent
cells. In this study, the Δ5 isoform was not detected in high
concentrations in the HCT-15 cell line protein extracts by western
blot analysis (data not shown). As the stability of these variants
is unknown, the Δ3 and 5 isoforms may not have been translated to
sufficient levels to alter the SDHC activity. Therefore, the
possibility that the Δ5 isoform causes dominant negative inhibition
of the full-length isoform cannot be excluded. It appears likely
that there is a more significant function for SDHC ASVs.
In cultured cells from mev-1 mice expressing
a mutated SDHC gene, mitochondrial ROS levels were increased
(19,20). Essentially, the amount of active
oxygen is tightly controlled by a balance between production and
elimination in vivo. However, control failure of the
SDHC gene induces the formation of tumors and cell death,
due to a significant increase in the production rate of active
oxygen (20). Similarly, in the
current study, a partial correlation was observed between
SDHC ASVs and ROS. This is the first example of active
oxygen generated from an SDHC ASV, which directly affects
cancer cell formation. ROS are widely known as ‘toxic factors’,
which induce cellular toxicity and dysfunction through the
oxidative damage of biomolecules. However, as a signal transduction
factor, ROS are responsible for controlling cell differentiation,
proliferation, cell death and cytoprotection (21,22).
Therefore, it is imperative to balance the supporting ROS
activities (a second messenger in intracellular signaling, as well
as escape from excessive active oxygen species production) against
their destructive behavior.
In the current study, to investigate the production
of the superoxide anion (O2−),
SDHC-overexpressing cells were stained with MitoSOX.
Compared with the control cells (transfected with empty vector), Δ5
isoform-expressing cells produced excessive
O2− (data not shown). Therefore, although SDH
activity is reduced in Δ5 isoform-overexpressing cells, the
resulting metabolic byproduct generates excessive superoxide. The
Δ5 isoform-overexpressing cells lack the heme binding region in the
SDHC protein, and are unlikely to transfer electrons properly.
Consequently, the accumulation and leakage of electrons may lead to
coupling with nearby oxygen, subsequently generating excessive
superoxide (23–25). In the present study, the Δ3 isoform
did not generate excessive superoxide, which may be due to the
incomplete inhibition of SDHC function, as the deletion of exon 3
only affects part of the succinate-CoQ oxidoreductase main activity
region. Thus, expression of the Δ5 isoform is likely to
preferentially cause superoxide production, leading to a variety of
ROS. In turn, these ROS may react to oxidative stress genes,
potentially altering the expression levels of cancer-related genes.
Studies are currently underway to clarify the manner in which
SDHC ASVs affect genes, leading to tumor formation. The
results of this study provide a basis for more detailed future
studies regarding the regulation of SDHC, and may lead to the
development of clinical trials investigating SDHC and ROS-related
diseases.
References
|
1
|
López-Jiménez E, de Campos JM, Kusak EM,
et al: SDHC mutation in an elderly patient without familial
antecedents. Clin Endocrinol (Oxf). 69:906–910. 2008. View Article : Google Scholar
|
|
2
|
Zaunmüller T, Kelly DJ, Glöckner FO and
Unden G: Succinate dehydrogenase functioning by a reverse redox
loop mechanism and fumarate reductase in sulphate-reducing
bacteria. Microbiology. 152:2443–2453. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Selak MA, Armour SM, MacKenzie ED, et al:
Succinate links TCA cycle dysfunction to oncogenesis by inhibiting
HIF-alpha prolyl hydroxylase. Cancer Cell. 7:77–85. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dong LF, Low P, Dyason JC, et al:
Alpha-tocopheryl succinate induces apoptosis by targeting
ubiquinone-binding sites in mitochondrial respiratory complex II.
Oncogene. 27:4324–4335. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
van Nederveen FH, Gaal J, Favier J, et al:
An immunohistochemical procedure to detect patients with
paraganglioma and phaeochromocytoma with germline SDHB, SDHC, or
SDHD gene mutations: a retrospective and prospective analysis.
Lancet Oncol. 10:764–771. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peczkowska M, Cascon A, Prejbisz A, et al:
Extra-adrenal and adrenal pheochromocytomas associated with a
germline SDHC mutation. Nat Clin Pract Endocrinol Metab. 4:111–115.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aratake K, Kamachi M, Iwanaga N, et al: A
cross-talk between RNA splicing and signaling pathway alters Fas
gene expression at post-transcriptional level: alternative splicing
of Fas mRNA in the leukemic U937 cells. J Lab Clin Med.
146:184–191. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Niemann S and Müller U: Mutations in SDHC
cause autosomal dominant paraganglioma, type 3. Nat Genet.
26:268–270. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schiavi F, Boedeker CC, Bausch B, et al;
European-American Paraganglioma Study Group. Predictors and
prevalence of paraganglioma syndrome associated with mutations of
the SDHC gene. JAMA. 294:2057–2063. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hanke J, Brett D, Zastrow I, et al:
Alternative splicing of human genes more the rule than the
exception? Trends Genet. 15:389–390. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu W, Qian C and Francke U: Silent
mutation induces exon skipping of fibrillin-1 gene in Marfan
syndrome. Nat Genet. 16:328–329. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Stallings-Mann ML, Ludwiczak RL, Klinger
KW and Rottman F: Alternative splicing of exon 3 of the human
growth hormone receptor is the result of an unusual genetic
polymorphism. Pro Natl Acad Sci USA. 93:12394–12399. 1996.
View Article : Google Scholar
|
|
13
|
Hirawake H, Taniwaki M, Tamura A, et al:
Cytochrome b in human complex II (succinate-ubiquinone
oxidoreductase): cDNA cloning of the components in liver
mitochondria and chromosome assignment of the genes for the large
(SDHC) and small (SDHD) subunits to 1q21 and 11q23. Cytogenet Cell
Genet. 79:132–138. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Maklashina E, Rajagukguk S, McIntire WS
and Cecchini G: Mutation of the heme axial ligand of Escherichia
coli succinate-quinone reductase: implications for heme ligation in
mitochondrial complex II from yeast. Biochim Biophys Acta.
1797:747–754. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ishii N: Role of oxidative stress from
mitochondria on aging and cancer. Cornea. 26(9 Suppl 1): S3–S9.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Y, McMillan-Ward E, Kong J, Israels
SJ and Gibson SB: Mitochondrial electron-transport-chain inhibitors
of complexes I and II induce autophagic cell death mediated by
reactive oxygen species. J Cell Sci. 120:4155–4166. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pasini B and Stratakis CA: SDH mutations
in tumorigenesis and inherited endocrine tumours: lesson from the
phaeochromocytoma-paraganglioma syndromes. J Intern Med. 266:19–42.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pantaleo MA, Astolfi A, Urbini M, et al:
GIST Study Group: Analysis of all subunits, SDHA, SDHB, SDHC, SDHD,
of the succinate dehydrogenase complex in KIT/PDGFRA wild-type
GIST. Eur J Hum Genet. 22:32–39. 2014. View Article : Google Scholar
|
|
19
|
Miyazawa M, Ishii T, Kirinashizawa M, et
al: Cell growth of the mouse SDHC mutant cells was suppressed by
apoptosis throughout mitochondrial pathway. Biosci Trends. 2:22–30.
2008.PubMed/NCBI
|
|
20
|
Ishii T, Miyazawa M, Onodera A, et al:
Mitochondrial reactive oxygen species generation by the SDHC V69E
mutation causes low birth weight and neonatal growth retardation.
Mitochondrion. 11:155–165. 2011. View Article : Google Scholar
|
|
21
|
Lluis JM, Buricchi F, Chiarugi P, Morales
A and Fernandez-Checa JC: Dual role of mitochondrial reactive
oxygen species in hypoxia signaling: activation of nuclear
factor-{kappa}B via c-SRC and oxidant-dependent cell death. Cancer
Res. 67:7368–7377. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sumimoto H: Structure, regulation and
evolution of Nox-family NADPH oxidases that produce reactive oxygen
species. FEBS J. 275:3249–3277. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tietze F: Enzymic method for quantitative
determination of nanogram amounts of total and oxidized
glutathione: applications to mammalian blood and other tissues.
Anal Biochem. 27:502–522. 1969. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yankovskaya V, Sablin SO, Ramsay RR, et
al: Inhibitor probes of the quinone binding sites of mammalian
complex II and Escherichia coli fumarate reductase. J Biol Chem.
271:21020–21024. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dröse S and Brandt U: The mechanism of
mitochondrial superoxide production by the cytochrome bc1 complex.
J Biol Chem. 283:21649–21654. 2008. View Article : Google Scholar : PubMed/NCBI
|