Introduction
Hepatocyte growth factor (HGF) is a widely-expressed
multifunctional growth and angiogenic factor (1). The activity of HGF is mediated by
binding to its receptor, a tyrosine kinase MET, and HGF transduces
multiple biological effects in target cells, including adhesion,
motility, growth, survival and morphogenesis (2).
The HGF level is frequently increased in advanced
cancer patients. HGF is a poor prognostic factor of patients with
various types of cancer, including hepatocellular carcinoma and
breast cancer (3–5). In addition, HGF is reported to be
associated with resistance to molecular target drugs, including
EGFR-specific tyrosine kinase inhibitors, in lung cancer (6,7). HGF
is secreted by tumor cells, vascular smooth muscle cells, pericytes
and fibroblasts. The HGF gene promoter in humans and mice
has been structurally and functionally analyzed (8–10).
Inflammatory cytokines, including interleukin-1- and
interleukin-6-responsive elements, are present in the human
HGF gene (8), which is
activated transcriptionally by these cytokines. However, little is
known about genomic alteration associated with the expression of
HGF. Ma et al reported that high expression of HGF is
regulated by a short deletion in the poly(dA) repeat sequence in
the HGF promoter region in breast cancer cells (11). The present study investigated the
association between the expression levels of HGF in those cells and
additional regulation mechanisms of HGF expression.
Materials and methods
Cells and cell culture
Human non-small cell lung cancer (NSCLC) PC-6, PC-7,
PC-9 and PC-14 cell lines were provided by Tokyo Medical University
(Tokyo, Japan) (12,13). The NSCLC SBC-3 cell line was
provided by Okayama University School of Medicine (Okayama, Japan)
(13). The NSCLC N231, LK-2, Ma-1
and 11_18 cell lines were provided by the National Cancer Research
Institute (Tokyo, Japan) (14–17).
The NSCLC A549, H1299, H69, Calu-1, Calu-6, H292, H358, H441, H460,
H2087, H1650, H1838, H1975 and HCC827 cell lines were obtained from
the American Type Culture Collection (Mannassas, VA, USA). The
NSCLC EBC-1 cell line was obtained from the Japanese Collection of
Research Bioresources Cell Bank (Osaka, Japan). Human gastric
cancer HSC38, HSC43 and HSC58 cell lines were provided by the
National Cancer Center Research Institute (18,19).
Human gastric cancer cell line, OKAJIMA, was provided by Osaka City
University (Osaka, Japan). Human gastric cancer IM95, MKN1, MKN7
and MKN74 cell lines were obtained from the Japanese Collection of
Research Bioresources Cell Bank. Human gastric cancer N87 and SNU16
cell lines and pancreatic cancer MIAPaCa cell lines were obtained
from the American Type Culture Collection. Human pancreatic cancer
Sui87, Sui68, Sui70 and Sui73 cell lines were provided by the
National Cancer Center Research Institute (20). Human colon cancer HCC56, SW837,
CCK-81, Colo201, Colo320 and WiDr cell lines and human
hepatocellular cancer HLE, HLF and Huh7 cell lines were obtained
from the Japanese Collection of Research Bioresources Cell Bank.
Human hepatocellular cancer HepG2, human breast cancer MDAMB-468
and BT-549 cell lines, the human glioma U251 cell line, the human
prostate PC-3 cancer cell line and human mesothelioma H28 and MSTO
cell lines were obtained from the American Type Culture Collection.
The cell lines were maintained in RPMI-1640 medium supplemented
with 10% heat-inactivated fetal bovine serum (FBS; Equitech-Bio,
Inc., Kerrville, TX, USA). All cell lines were maintained in a 5%
CO2, humidified atmosphere at 37°C.
Sample preparation
Total RNA was extracted from cells using ISOGEN
(Nippon Gene Co., Ltd., Tokyo, Japan), according to the
manufacturer’s instructions. The cDNA templates were synthesized
from 1 μg of total RNA using the GeneAmp® RNA polymerase
chain reaction (PCR) kit (Applied Biosystems, Foster City, CA,
USA).
DNA was extracted from cells using the QIAamp DNA
mini kit (Qiagen, Valencia, CA, USA), according to the
manufacturer’s instructions. For the 10 mM stock solution,
5-Aza-2′-deoxycytidine (5-Aza-dC; Sigma-Aldrich, St. Louis, MO,
USA) was dissolved in dimethylsulfoxide. Aliquots were prepared and
frozen at −80°C. The cells were treated with 5-Aza-dC for 48 h
prior to the cells being collected and total RNA extracted.
Reverse transcription-quantitative PCR
(RT-qPCR)
The methods used in the present section have been
previously described (21).
Briefly, RT-qPCR was performed by using a Premix Ex Taq and Smart
Cycler system (Takara Bio, Inc., Shiga, Japan), according to the
manufacturer’s instructions. The primers used for RT-qPCR were
purchased from Takara Bio, Inc. and were as follows: Forward,
5′-GTAAATGGGATTCCAACACGAACAA-3′ and reverse,
5′-TGTCGTGCAGTAAGAACCCAACTC-3′ for HGF; forward,
5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse, 5′-ATGGTGGTGAAGACGCCAGT-3′
for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The
cDNA was then used as the template for the qPCR reaction. The PCR
conditions were as follows: one cycle at 95°C for 5 min, followed
by 40 cycles at 95°C for 10 sec and 60°C for 30 sec. The threshold
cycle (Ct) values were determined using Thermal Cycler Dice Real
Time System (Takara Bio, Inc.). The experiment was independently
performed in triplicate using GAPDH as a reference to
normalize the data.
Enzyme-linked immunosorbent assay
(ELISA)
The cells were seeded at a density of
2×106 cells per 10-cm dish in medium supplemented with
10% FBS and were cultured for 24 h. The medium was then changed to
serum-free medium. Following 48 h of incubation, the conditioned
medium was collected to measure HGF production.
HGF concentrations in the cultured medium were
determined using a Human HGF Quantikine ELISA kit (R&D Systems,
Inc., Minneapolis, MN, USA) according to the manufacturer’s
instructions. The absorbance of the samples at 450 nm was measured
using VERSAmax (Molecular Devices Japan K.K, Tokyo, Japan).
Duplicate examinations of 50 μl of the cell-conditioned medium were
performed.
DNA amplification and fragment
sizing
DNA amplification was performed with Ex Taq
polymerase (Takara Bio, Inc.). The cycling program was one cycle of
98°C for 1 min and then 30 cycles of 98°C for 10 sec, 60°C for 30
sec and 72°C for 10 sec, followed by one cycle of 72°C for 2 min.
PCR fragments of 88 bp were analyzed using the Agilent 2100
bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). The primers
used for PCR amplification were as follows, forward,
5′-GGTAAATGTGTGGTATTTCGTGAG-3′ and reverse,
5′-GCTGCCTGCTCTGAGCCCAT-3′.
Sequencing analysis
DNA sequencing was performed directly on purified
PCR products using the BigDye terminator v 3.1 sequencing kit
(Applied Biosystems). DNA amplification was performed with Ex Taq
polymerase (Takara Bio, Inc.). The cycling program was one cycle at
98°C for 1 min and then 30 cycles at 98°C for 10 sec, 60°C for 30
sec and 72°C for 1 min, followed by one cycle at 72°C for 2 min.
Following PCR, the product was purified using the QIAquick PCR
purification kit (Qiagen), and then sequenced using the ABI BigDye
3.1 dye terminator V3.1 kit (Applied Biosystems) on an ABI
Prism® 3100 DNA Analyzer automated sequencer (Applied
Biosystems). The primer and probe sequences used were as follows:
Forward primer, 5′-TGTGATTCTTCTCCTCGTGGGGT-3′, reverse primer,
5′-AGCCTGACCGTGACCCTGAA-3′ and sequencing primer,
5′-AGCCTGACCGTGACCCTGAA-3′, for rs11763015 and rs78601897; forward
primer, 5′-TGTGATTCTTCTCCTCGTGGGGT-3′, reverse primer,
5′-CCAAGAAACAGTCATTGTCCATAGCCTGTCCC-3′ and sequencing primer,
5′-CCTGGGGACACCAGACAGAGGCTG-3′, for rs3735520 and rs3735521;
forward primer, 5′-GCATATTCAGTACTCACGAATTCAA-3′, reverse primer,
5′-TGGGACGGGGCTTGGGTTGGA-3′ and sequencing primer,
5′-CCAGGCATCTCCTCCAGAGGGATCCG-3′, for rs72525097.
Results
Sequencing of the poly(dA) repeat in the
HGF promoter region
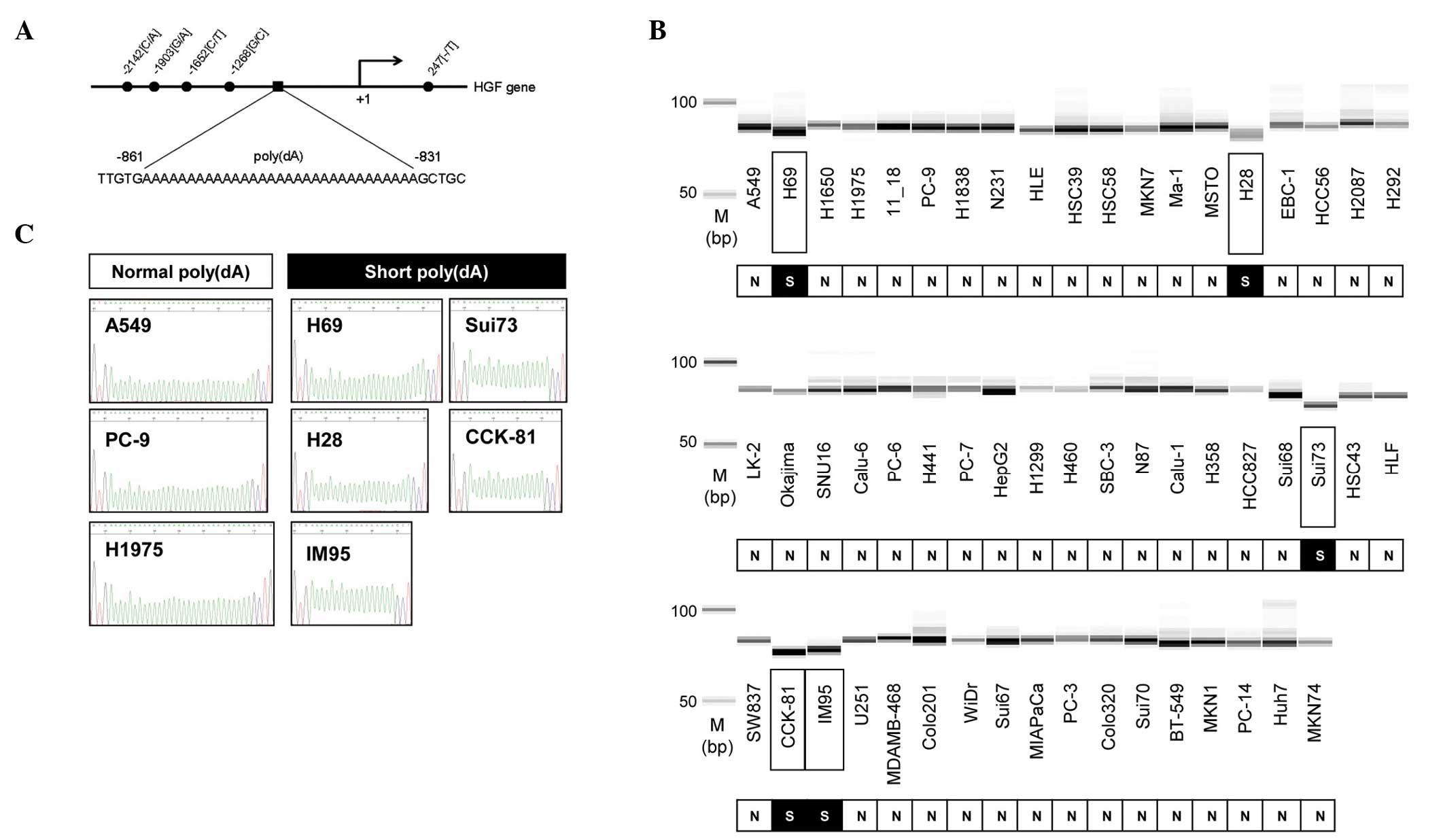
The HGF promoter region contains a poly(dA) repeat
at ~800 bp upstream from the translation initiation site (Fig. 1A). Based on a previous study
(11), the poly(dA) lengths were
analyzed by fragment sizing in 55 human cancer cell lines. Shorter
fragments were detected in the Sui73, CCK81, IM95, H69 and H28
cells, but not in the remaining cell lines (Fig. 1B). To confirm the fragment size, the
poly(dA) region of eight cell lines was sequenced (Fig. 1C). The number of mononucleotide
repeats in these cell lines matched with the result of the fragment
analysis.
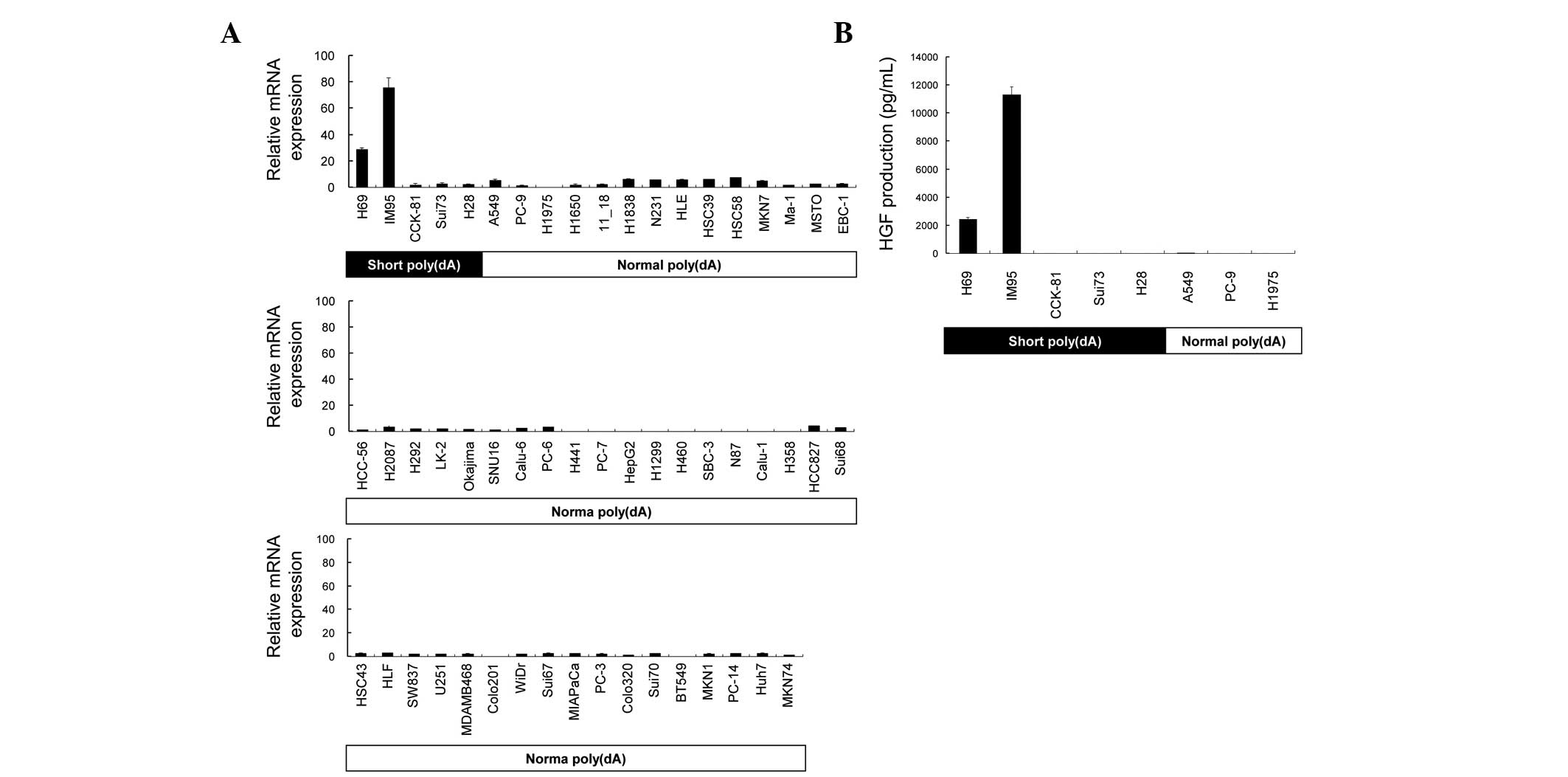
HGF expression in human cancer cells
The levels of HGF expression in 55 cell lines
were then examined by RT-qPCR (Fig.
2A). RT-qPCR analysis revealed a low expression in the majority
of the cell lines. On the other hand, the H69 and IM95 cells
expressed high levels of HGF mRNA. The expression of HGF
mRNA in the H69 and IM95 cells was >12.3- and >32.4-fold
higher compared with the average of any cell line other than H69 or
IM95, respectively. HGF protein secretion was examined in the eight
cell lines (Fig. 2B). The HGF
protein was highly secreted by H69 and IM95 cells in the
conditioned medium, which was consistent with mRNA expression.
The pattern of poly(dA) length (Fig 1B) and HGF expression (Fig. 2A) was compared in 55 cell lines. The
expression level of HGF was low in all the cell lines with a
normal poly(dA) length in the HGF promoter region. By
contrast, HGF expression in the five cell lines with short
poly(dA) length in the HGF promoter region differed in
pattern. High expression of HGF was observed in the H69 and
IM95, but not in the CCK-81, Sui73, and H28 cell lines. These
results suggest that the cell lines with a normal poly(dA) promoter
express low levels of HGF, and all cell lines with a short
poly(dA) do not express high levels of HGF. It was
hypothesized that the HGF expression was suppressed in the
CCK-81, Sui73 and H28 cells with short poly(dA) by other
mechanisms.
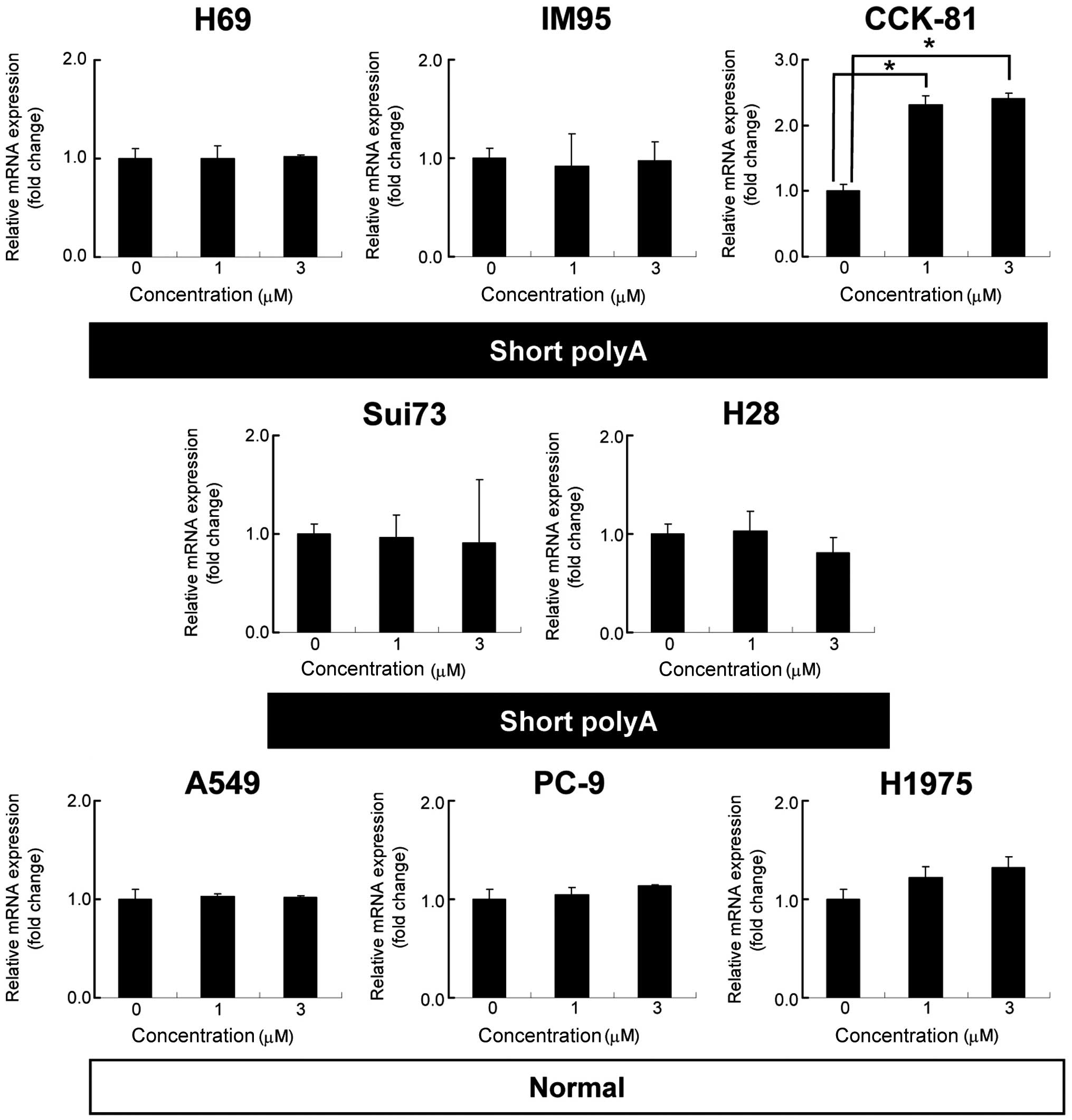
Effect of 5-Az-dC on HGF expression
The present study explored the cause of HGF
gene suppression in certain cell lines with shorter poly(dA)
sequences. It was hypothesized that DNA methylation may silence
HGF expression in these cell lines. The change in the
HGF mRNA expression in three cell lines was examined
following treatment with 1 or 3 μM 5-Aza-dC for 48 h (Fig. 3). A 2.3- and 2.4-fold increase in
HGF mRNA was observed in CCK-81 cells when treated with 1
and 3 μM 5-Aza-dC, respectively (P<0.01). No significant change
was observed in the Sui73 and H28 cells. These results suggest that
DNA methylation may contribute to the silencing of the HGF
gene in the CCK-81 cells.
Genotyping of SNPs in the HGF gene
All increased expression of HGF in the Sui73
and H28 cells was due to DNA methylation. Next, it was examined
whether a specific polymorphism is observed in these cells. The
HGF genotypes were analyzed upstream of the transcript start
(−2200 bp) to intron1 (+300 bp) of 12 cell lines with short
poly(dA) (H69, IM95, CCK-81, Sui73, and H28 cells) or normal
poly(dA) (A549, PC-9, H1975, HCC827, H1650, 11_18, and Ma-1 cells)
by direct sequencing (Table I). The
-1903G and -1268G SNPs were detected in all 12 cell lines. The
-2142C/A SNP was detected in only the IM95 cell line with short
poly(dA). The -1652C/T SNP was detected in four cell lines with
shorter and normal poly(dA). The +247T SNP in intron 1 was detected
in only the Sui73 and H28 cells with a short poly(dA). These
results suggest that insertion of a single T nucleotide at position
247 may be associated with the expression of HGF in the Sui73 and
H28 cells with a short poly(dA).
 | Table IA list of single nucleotide
polymorphisms detected in the 12 cell lines. The poly(dA) length
represents the number of deoxyadenosine repeat sequences in the
HGF gene from the results of direct sequencing. HGF
production represents the results of the enzyme-linked
immunosorbent assay. |
Table I
A list of single nucleotide
polymorphisms detected in the 12 cell lines. The poly(dA) length
represents the number of deoxyadenosine repeat sequences in the
HGF gene from the results of direct sequencing. HGF
production represents the results of the enzyme-linked
immunosorbent assay.
| Cell line | poly(dA) length | HGF production
(pg/ml) |
rs11763015-2142C/A |
rs78601897-1903G/A |
rs3735520-1652C/T |
rs3735521-1268G/C | rs72525097
247(−/T) |
|---|
| H69 | 24 | 2443.0 | C | G | C | G | - |
| IM95 | 17 | 11297.6 | C/A | G | T | G | - |
| Sui73 | 17 | 3.6 | C | G | C/T | G | T |
| CCK-81 | 16 | 10.0 | C | G | C | G | - |
| H28 | 20 | 6.2 | C | G | C/T | G | T |
| A549 | 27 | 26.2 | C | G | C | G | - |
| PC-9 | 29 | 3.1 | C | G | C | G | - |
| H1975 | 28 | 14.4 | C | G | C | G | - |
| HCC827 | 28 | NT | C | G | C/T | G | - |
| H1650 | 29 | NT | C | G | C | G | - |
| 11_18 | 29 | NT | C | G | C/T | G | - |
| Ma-1 | 28 | NT | C | G | C | G | - |
Discussion
HGF has been identified as a natural ligand of the
MET receptor and belongs to the plasminogen family (1). In cancer cells, activation of the MET
receptor increases invasion and metastasis, and allows the survival
of cancer cells in the bloodstream in the absence of anchorage
(2).
In the present study, the deletion mutation on the
HGF promoter region was detected in lung, colon, stomach,
pancreatic and mesothelial cancer cell lines. The cell lines with
short poly(dA) express high levels of HGF, whereas all the
cell lines with normal poly(dA) do not. This result is consistent
with a previous study (11) and
short poly(dA) in the HGF promoter region has been detected
in various types of cancer cell lines and breast cancer. H69
[poly(dA), 24 bp] and IM95 [poly(dA), 17 bp] highly express
HGF. When comparing these two cell lines, IM95 exhibits a
higher HGF expression compared with H69 (2.6- and 4.6-fold
higher at the mRNA and protein levels, respectively). This is also
consistent with the study by Ma et al, suggesting that the
cells with shorter poly(dA) exhibit higher HGF (11).
By contrast, certain cell lines exhibited a short
poly(dA) and low HGF expression. We considered that the low
HGF expression in these cell lines may be explained by the
methylation or polymorphisms of the HGF promoter. There has
been no previous paper reporting epigenetic regulation of HGF
expression. Recently, DNA methylation analysis of HGF
promoters was reported. Analysis of bisulfite DNA from BNL 1ME
A.7R.1 cells indicated that HGF has 35% of CpGs methylated
at two sites (22). This finding is
considered to be important for elucidating the detailed mechanisms
of epigenetic regulation of HGF expression, in order to develop
epigenetic therapy for HGF-related cancers.
An increase (by 2.4-fold) in HGF expression
was induced by 5Aza-dC in Sui73 and H28 cell lines, suggesting that
other mechanisms such as acetylation and transcription factors may
influence the full expression of HGF.
T insertion in intron 1 (247T) was detected in two
cell lines with short poly(dA). It is well-known that polymorphisms
in the promoter, but not in introns, influence the transcription of
a gene. However, Onouchi et al reported that an SNP in
intron 1 of 1,4,5-trisphosphate 3-kinase C influenced the
transcription of this gene (23).
Yamada et al reported that polymorphism in intron 1 of
transcription factor EGR3 regulates transcription of this
gene (24). Subsequently, it was
hypothesized that 247(−/T) may influence the transcriptional levels
of HGF in these cell lines, although functional analysis is
necessary in future studies. Another SNP was detected in 4/12 cell
lines (−1652C/T); however, the influence of this SNP on HGF
expression remains unknown.
HGF is an important mediator of
epithelial-mesenchymal transition (EMT) (25). Fibroblastic changes in the cells
were often observed during EMT induced by HGF and other ligands.
The association between SNPs in HGF and EMT may be investigated in
future studies.
In conclusion, the present study found that the
deletion polymorphism of poly(dA) in the HGF promoter was
present in various cancer cell lines, including lung, stomach,
colorectal and pancreatic cancer, and mesothelioma cell lines.
Future experiments with functional analysis of intron 1
polymorphisms may provide a novel negative regulatory mechanism of
HGF expression.
Acknowledgements
The present study was supported by the Third-Term
Comprehensive 10-Year Strategy for Cancer Control and funds for
Health and Labor Scientific Research Grants (20-9), and a
Grant-in-Aid for Scientific Research (B) (23701106).
References
|
1
|
Naldini L, Vigna E, Narsimhan RP, Gaudino
G, Zarnegar R, Michalopoulos GK and Comoglio PM: Hepatocyte growth
factor (HGF) stimulates the tyrosine kinase activity of the
receptor encoded by the proto-oncogene c-MET. Oncogene. 6:501–504.
1991.PubMed/NCBI
|
|
2
|
Benvenuti S and Comoglio PM: The MET
receptor tyrosine kinase in invasion and metastasis. J Cell
Physiol. 213:316–325. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schoenleber SJ, Kurtz DM, Talwalkar JA,
Roberts LR and Gores GJ: Prognostic role of vascular endothelial
growth factor in hepatocellular carcinoma: systematic review and
meta-analysis. Br J Cancer. 100:1385–1392. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Seidel C, Børset M, Turesson I, Abildgaard
N, Sundan A and Waage A: Elevated serum concentrations of
hepatocyte growth factor in patients with multiple myeloma. The
Nordic Myeloma Study Group. Blood. 91:806–812. 1998.PubMed/NCBI
|
|
5
|
Toi M, Taniguchi T, Ueno T, Asano M,
Funata N, Sekiguchi K, Iwanari H and Tominaga T: Significance of
circulating hepatocyte growth factor level as a prognostic
indicator in primary breast cancer. Clin Cancer Res. 4:659–664.
1998.PubMed/NCBI
|
|
6
|
Yano S, Wang W, Li Q, Matsumoto K,
Sakurama H, Nakamura T, Ogino H, et al: Hepatocyte growth factor
induces gefitinib resistance of lung adenocarcinoma with epidermal
growth factor receptor-activating mutations. Cancer Res.
68:9479–9487. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kasahara K, Arao T, Sakai K, Matsumoto K,
Sakai A, Kimura H, Sone T, et al: Impact of serum hepatocyte growth
factor on treatment response to epidermal growth factor receptor
tyrosine kinase inhibitors in patients with non-small cell lung
adenocarcinoma. Clin Cancer Res. 16:4616–4624. PubMed/NCBI
|
|
8
|
Miyazawa K, Kitamura A and Kitamura N:
Structural organization and the transcription initiation site of
the human hepatocyte growth factor gene. Biochemistry.
30:9170–9176. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Okajima A, Miyazawa K and Kitamura N:
Characterization of the promoter region of the rat
hepatocyte-growth-factor/scatter-factor gene. Eur J Biochem.
213:113–119. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Plaschke-Schlütter A, Behrens J, Gherardi
E and Birchmeier W: Characterization of the scatter
factor/hepatocyte growth factor gene promoter. Positive and
negative regulatory elements direct gene expression to mesenchymal
cells. J Biol Chem. 270:830–836. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma J, DeFrances MC, Zou C, Johnson C,
Ferrell R and Zarnegar R: Somatic mutation and functional
polymorphism of a novel regulatory element in the HGF gene promoter
causes its aberrant expression in human breast cancer. J Clin
Invest. 119:478–491. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nishio K, Arioka H, Ishida T, Fukumoto H,
Kurokawa H, Sata M, Ohata M and Saijo N: Enhanced interaction
between tubulin and microtubule-associated protein 2 via inhibition
of MAP kinase and CDC2 kinase by paclitaxel. Int J Cancer.
63:688–693. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kawamura-Akiyama Y, Kusaba H, Kanzawa F,
Tamura T, Saijo N and Nishio K: Non-cross resistance of ZD0473 in
acquired cisplatin-resistant lung cancer cell lines. Lung Cancer.
38:43–50. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Iwasa T, Okamoto I, Suzuki M, Nakahara T,
Yamanaka K, Hatashita E, Yamada Y, et al: Radiosensitizing effect
of YM155, a novel small-molecule survivin suppressant, in non-small
cell lung cancer cell lines. Clin Cancer Res. 14:6496–6504. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Okabe T, Okamoto I, Tamura K, Terashima M,
Yoshida T, Satoh T, Takada M, et al: Differential constitutive
activation of the epidermal growth factor receptor in non-small
cell lung cancer cells bearing EGFR gene mutation and
amplification. Cancer Res. 67:2046–2053. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Takezawa K, Okamoto I, Yonesaka K,
Hatashita E, Yamada Y, Fukuoka M and Nakagawa K: Sorafenib inhibits
non-small cell lung cancer cell growth by targeting B-RAF in KRAS
wild-type cells and C-RAF in KRAS mutant cells. Cancer Res.
69:6515–6521. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sato M, Takahashi K, Nagayama K, Arai Y,
Ito N, Okada M, Minna JD, et al: Identification of chromosome arm
9p as the most frequent target of homozygous deletions in lung
cancer. Genes Chromosomes Cancer. 44:405–414. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yanagihara K, Seyama T, Tsumuraya M,
Kamada N and Yokoro K: Establishment and characterization of human
signet ring cell gastric carcinoma cell lines with amplification of
the c-myc oncogene. Cancer Res. 51:381–386. 1991.PubMed/NCBI
|
|
19
|
Yanagihara K, Tanaka H, Takigahira M, Ino
Y, Yamaguchi Y, Toge T, Sugano K and Hirohashi S: Establishment of
two cell lines from human gastric scirrhous carcinoma that possess
the potential to metastasize spontaneously in nude mice. Cancer
Sci. 95:575–582. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yanagihara K, Takigahira M, Tanaka H, Arao
T, Aoyagi Y, Oda T, Ochiai A and Nishio K: Establishment and
molecular profiling of a novel human pancreatic cancer panel for
5-FU. Cancer Sci. 99:1859–1864. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kaneda H, Arao T, Tanaka K, Tamura D,
Aomatsu K, Kudo K, Sakai K, et al: FOXQ1 is overexpressed in
colorectal cancer and enhances tumorigenicity and tumor growth.
Cancer Res. 70:2053–2063. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ogunwobi OO, Puszyk W, Dong HJ and Liu C:
Epigenetic upregulation of HGF and c-Met drives metastasis in
hepatocellular carcinoma. PLoS One. 8:e637652013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Onouchi Y, Gunji T, Burns JC, Shimizu C,
Newburger JW, Yashiro M, Nakamura Y, et al: ITPKC functional
polymorphism associated with Kawasaki disease susceptibility and
formation of coronary artery aneurysms. Nat Genet. 40:35–42. 2008.
View Article : Google Scholar
|
|
24
|
Yamada K, Gerber DJ, Iwayama Y, Ohnishi T,
Ohba H, Toyota T, Aruga J, et al: Genetic analysis of the
calcineurin pathway identifies members of the EGR gene family,
specifically EGR3, as potential susceptibility candidates in
schizophrenia. Proc Natl Acad Sci USA. 104:2815–2820. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Grotegut S, von Schweinitz D, Christofori
G and Lehembre F: Hepatocyte growth factor induces cell scattering
through MAPK/Egr-1-mediated upregulation of Snail. EMBO J.
25:3534–3545. 2006. View Article : Google Scholar : PubMed/NCBI
|