Introduction
Gastric cancer (GC) is a major cause of
cancer-related mortalities worldwide (1). There are multiple pathogenic factors
that may promote the development and progression of gastric cancer.
Gastrin is a type of polypeptide hormone, secreted by G cells in
the stomach and upper-section of the small intestines. Its role in
the physiological regulation of gastric acid secretion has been
well established (2). Additionally,
gastrin contributes to the maintenance of gastric epithelial
architecture by regulating the expression of genes such as
plasminogen activator inhibitor 1 (PAI-1) (3) and regenerating gene 1 (Reg1)
(4). Increased expression of
gastrin has been demonstrated in the progression of intestinal
gastric cancer (5).
Reg1 encodes a β-cell growth factor, Reg1
protein, primarily observed during pancreatitis and pancreatic
islet regeneration (6,7). Reg1 has also been referred to as
pancreatic stone protein (PSP) (8),
or lithostathine (9), according to
different studies. The Reg family comprises of four subclasses
(Reg1–4) (10) across species, with
the majority of the orthologs belonging to the Reg1 and Reg3
groups. The Reg1 gene is approximately 3 kb in size,
containing six exons and five introns, and is located at chromosome
2p12. An increasing number of studies have provided evidence of the
participation of Reg1 protein in the proliferation and
differentiation of diverse cell types (11,12).
It has been demonstrated that Reg family members are associated
with various pathologies, including diabetes, epithelial
inflammation and a number of forms of cancer (13,14).
Reg1 is predominantly expressed in enterochromaffin-like
(ECL) cells, as well as pepsinogen-secreting chief cells, which are
also a target of gastrin within the gastric epithelium (15). It has been proposed that Reg1 and
gastrin may synergistically regulate gastric mucosal proliferation
during certain pathological settings (16,17).
Significantly less is known with regard to the transcriptional
mechanisms by which gastrin may regulate genes involved in the
maintenance of gastric epithelial architecture. It was previously
reported that a C-rich region of the proximal promoter sequence of
Reg1 is required for gastrin-stimulated expression in
gastric cancer cell line, AGS (15), and that the expression of
Reg1 is controlled through separate promoter elements −2111
and −104 bp by gastrin (4).
It has been demonstrated that intronic sequences in
eukaryotes have the potential to improve gene expression through a
variety of mechanisms. Human β-globin (hBG) introns can act as
enhancer-like elements for the expression of the human factor
IX in cultured Chinese hamster ovary cells, resulting in a
higher activity with respect to the second hBG intron compared with
the first one. The larger number of transcription factor binding
motifs in the second hBG intron accounts for its stronger effect
(18). The DNase I-hypersensitive
(HSS) sequences in intron 51 of the von Willebrand factor (VWF)
gene contain cis-acting elements that are necessary for the
VWF gene transcription in a subset of lung endothelial cells in
vivo (19). It was demonstrated
that Reg1 and gastrin may synergistically regulate gastric mucosal
proliferation during certain pathological settings such as wound
healing (16). To identify
additional cis-acting elements within the Reg1 gene
that may participate in transcriptional regulation and the effects
of gastrin on expression of Reg1, we investigated whether
introns of Reg1 gene can increase its expression and bind to
transcription factors in gastric cancer cells. This also
facilitated the exploration of the cellular mechanisms regulating
Reg1 expression in gastric cancer cells.
Materials and methods
Cell culture
The human gastric cancer cell line, SGC7901, was
provided by the Cell Bank of the Chinese Academy of Sciences
(Shanghai, China) and routinely cultured in RPMI 1640 medium
(Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine
serum, 100 U/ml penicillin (Sigma-Aldrich, St. Louis, MO, USA) and
100 μg/ml streptomycin (Sigma-Aldrich), at 37°C in a humidified
atmosphere of 5% CO2.
Cloning of the human Reg1 gene
introns
In total, five intron fragments of Reg1 were
cloned from genomic DNA of human blood cells by polymerase chain
reaction (PCR). PCR was performed in a final volume of 25 μl. The
PCR reaction was carried out at 94°C for 3 min, followed by 35
cycles at 95°C for 30 sec, 50–55°C for 45 sec and 72°C for 1 min.
PCR primers for the five introns are listed in Table I. The PCR fragments of five introns
were inserted into pbluescript II SK+, digested by Xho I/Hind III
(Takara Biotechnology Co., Ltd., Dalian, China) and subcloned into
luciferase reporter vector pGL3-Basic, which was used as a control
and digested with the same two enzymes. The five luciferase
reporter vectors containing the five introns of Reg1 were
constructed, and designated as pGL3-intron 1, pGL3-intron 2,
pGL3-intron 3, pGL3-intron 4 and pGL3-intron 5. All constructs were
confirmed by Xho I/Hind III digestion and DNA sequencing.
 | Table IPolymerase chain reaction primer
sequences for the five introns of the Reg1 gene. |
Table I
Polymerase chain reaction primer
sequences for the five introns of the Reg1 gene.
| Introns | Forward primer
5′-3′ | Reverse primer
5′-3′ | PCR products
length, bp | Tm, °C |
|---|
| Intron1 |
CCAACTCAGACTCAGCCAAC |
CATGCTGAGCTGCAATGAAT | 376 | 50 |
| Intron2 |
GCTGATCTCCTGCCTGATGT |
AACTCTGTCTGGGCCTCTTG | 700 | 55 |
| Intron3 |
TGCCTATCGCTCCTACTGCT |
AGGTTGCCCGAATTCATGT | 400 | 55 |
| Intron4 |
CCTTTGTGGCCTCACTGATT |
CAATGCCCCAGGACTTGTAG | 850 | 52 |
| Intron5 |
CTGTGTGAGCCTGACCTCAA |
CAAGGCACATCCTTCCATTT | 245 | 55 |
Luciferase assay
The SGC7901 gastric cancer cell line was plated on
96-well plates at a density of 2×103 cells/well. The
cells were transiently transfected with the above five luciferase
reporter plasmids using Lipofectamine Plus system according to the
manufacturer’s instructions (Invitrogen Life Technologies,
Carlsbad, CA, USA). To evaluate the effect of gastrin
(Sigma-Aldrich), the transfected cells were incubated with gastrin
(1×10−7 mol/l) for 24 h after 48 h transfection. The
luciferase activity of the transfected cells without or with
gastrin incubation was measured by Luciferase Assay System (Promega
Corporation, Madison, WI, USA) in a Glomax fluorescence detector
(Promega Corporation) according to the manufacturer’s instructions.
All assays were conducted in triplicate.
Southwestern blotting
Using genomic DNA from human blood cells as a
template, the five intron fragments of Reg1 gene were cloned
by PCR using the same primers (Table
I) and identified by 1% agarose gel electrophoresis.
Subsequently, the PCR products of the introns were randomly labeled
with digoxin (Roche Diagnostics Corporation, Indianapolis, IN, USA)
as a probe according to the manufacturer’s instructions. The
sensitivity of the probes was detected with DNA dot blotting by
using the DIG High Prime DNA Labeling and Detection Starter Kit I
(Roche Diagnostics Corporation). To observe the effect of gastrin,
the SGC7901 cells were cultured and incubated without or with
gastrin (1×10−7 or 1×10−8 mol/l) for 24 h.
Their nuclear proteins were subsequently extracted according to the
manufacturer’s instructions (Pierce Biotechnology, Inc., Rockford,
IL, USA). Protein concentration was determined by bicinchoninic
acid (BCA) protein assay (BCA assay kit; Beyotime Institute of
Biotechnology, Haimen, China) according to the manufacturer’s
instructions. Nuclear proteins of the cells were subjected to
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE) in a 10% polyacrylamide gel. The proteins were blotted
onto the PVDF membrane in a transfer buffer (Sigma-Aldrich). With
the five intron fragments of Reg1 as probes, the binding
activity of each intron to nuclear proteins was detected as
previously described (20). The
density of each band was analyzed using Image-Pro Plus Version
6.0.
Statistical analysis
All data are presented as the mean ± standard
deviation (SD). Statistical analysis was performed using an
unpaired two-tailed t test. P<0.05 was considered to indicate a
statistically significant difference. Data analysis was performed
using Statistical Product and Service Solutions software (version
15.0; SPSS, Inc., Chicago, IL, USA).
Results
Effects of introns of Reg1 on luciferase
activity
The five luciferase reporter vectors containing
introns of the Reg1 gene were identified by enzyme digestion
and DNA sequencing. The results demonstrated that the sequences of
inserted fragments were consistent with those of GenBank data with
the correct direction of transcription. The relative luciferase
activities in SGC7901 cells transfected with various recombinant
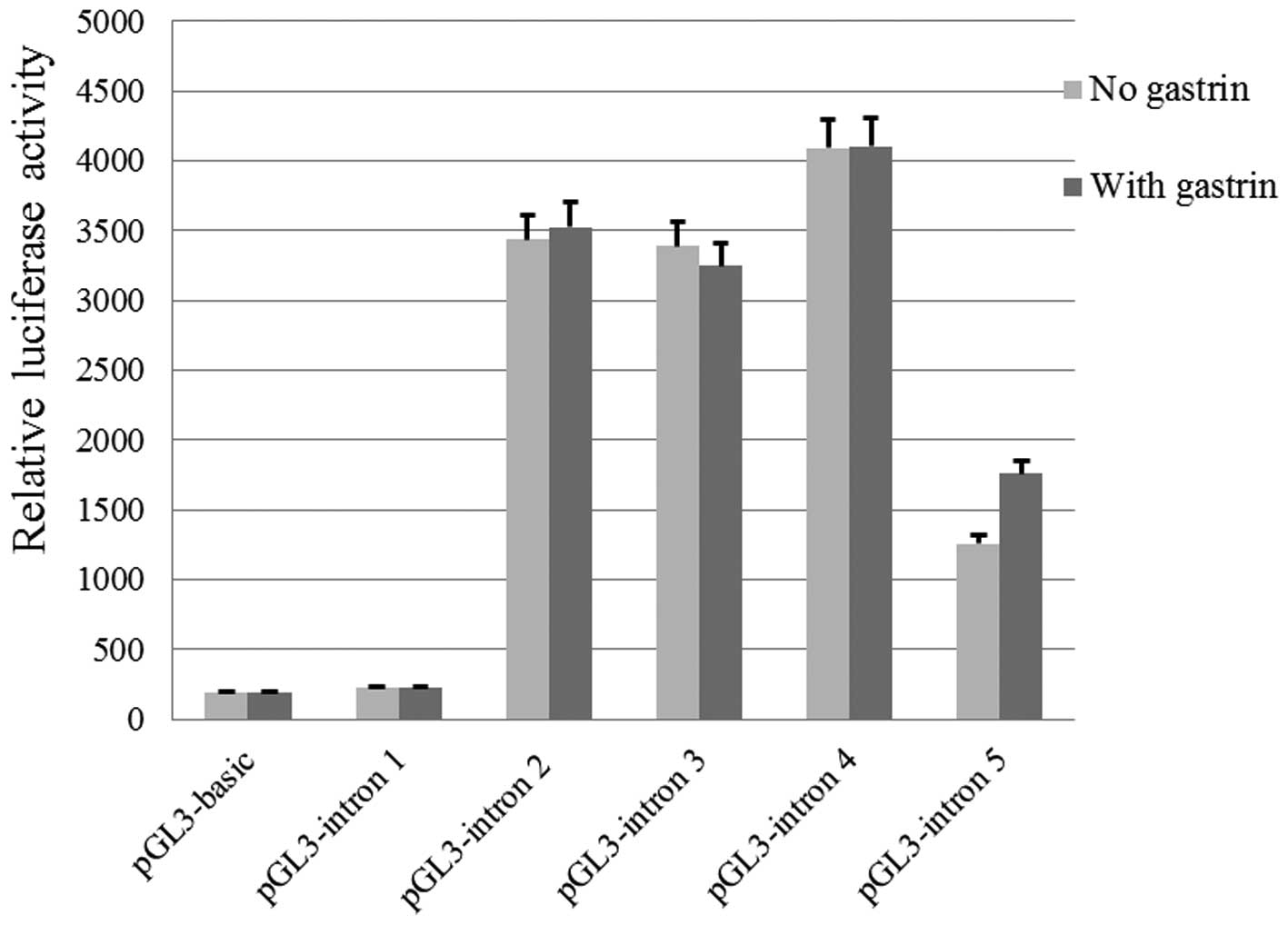
plasmids are shown in Fig. 1.
The relative luciferase activities of cells
transfected with pGL3-intron 2, pGL3-intron 3, pGL3-intron 4, and
pGL3-intron 5 were significantly higher when compared with that of
pGL3-Basic (P<0.05). No statistically significant difference in
luciferase activity was identified between cells transfected with
pGL3-intron 1 and those transfected with pGL3-Basic, which was used
as the control (P>0.05). After gastrin incubation, no
significant difference in luciferase activity was identified
between cells transfected with any of the above recombinant
plasmids (P>0.05).
Transcription factors (Tfs) binding
activity to five introns
The five intron fragments were cloned by PCR and
labeled with digoxin. The sensitivity of the five intron probes was
1 pg/μl, which was effective for detection. Results of the analysis
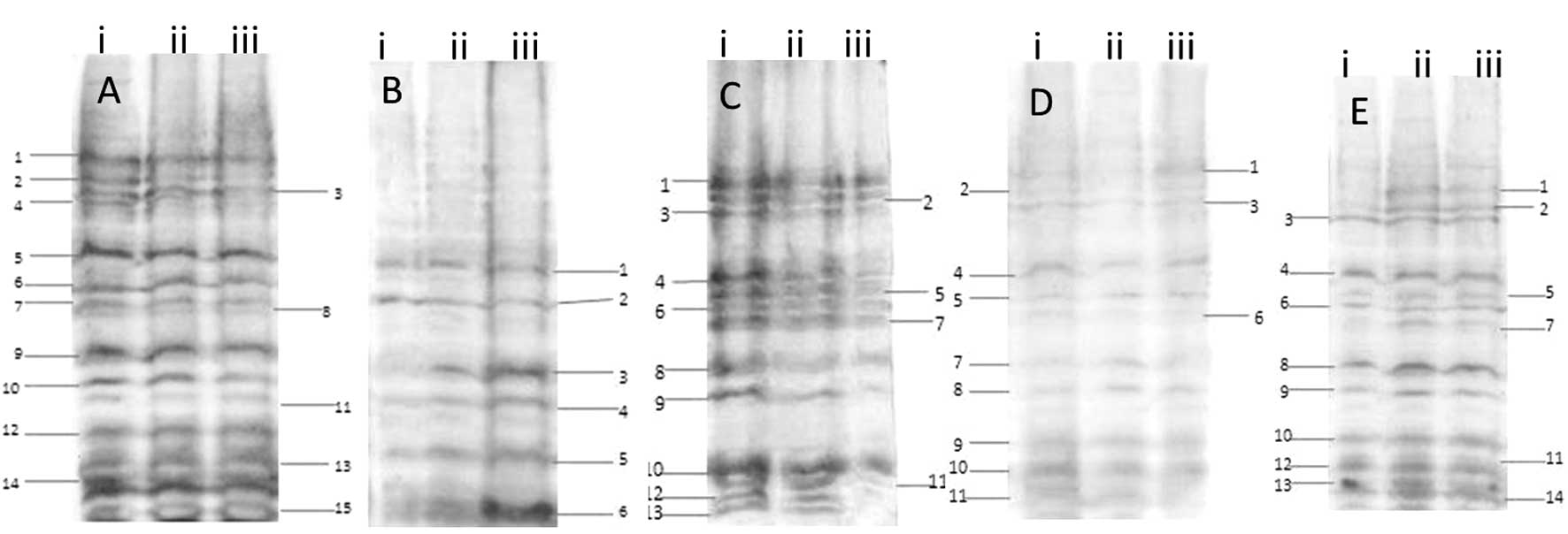
of transcription factors through Southwestern blotting are shown in
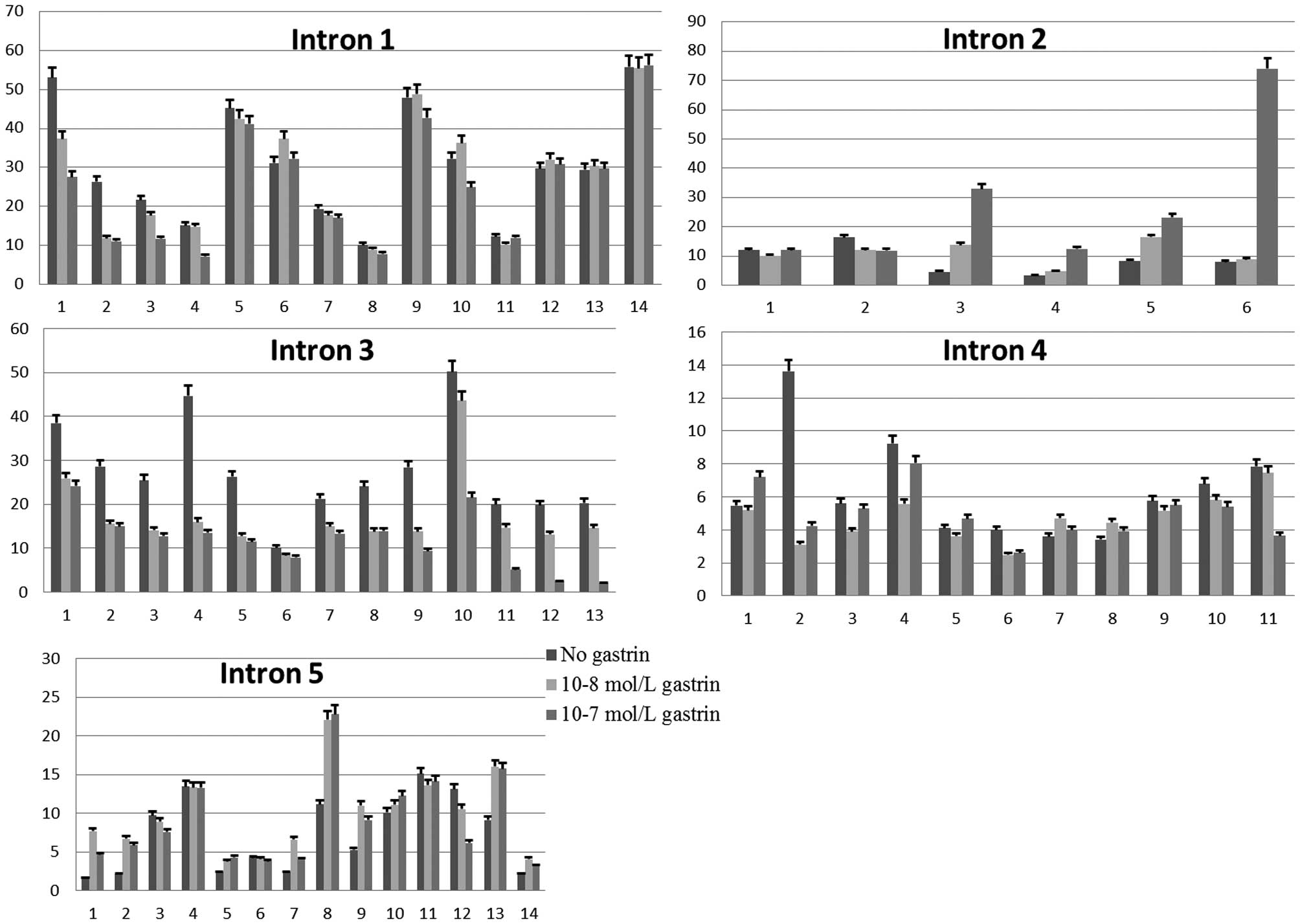
Figs. 2 and 3. For the probe of intron 1, 15 major
bands of Tfs with different molecular weights were detected.
Following gastrin treatment, the density of certain bands was
evidently changed. Compared with the control group (without gastrin
treatment), the density of bands 1, 2, 3, 4 and 7 was decreased
significantly, and that of band 6 was increased significantly
(P<0.05). For the cells incubated with 10−8 mol/l
gastrin, the density of bands 1, 3, 4, 6, 9 and 10 was increased
significantly when compared with that of the cells incubated with
10−7 mol/l gastrin (P<0.05).
With the intron 2 fragment as a probe, six different
major bands of Tfs were detected. Following gastrin treatment, the
density of band 2 was decreased significantly, and bands 3, 4, 5
and 6 were increased significantly (P<0.05). The density of
bands 3, 4, 5 and 6 was increased significantly in the cells
incubated with 10−7 mol/l gastrin compared with that of
10−8 mol/l gastrin (P<0.05).
The results of the intron 3 probe detection
identified 13 different major bands of Tfs. Compared with the
control group, the density of all 13 bands in the experimental
groups was decreased significantly (P<0.05). The density of
bands 4, 9, 10, 11, 12 and 13 were increased significantly in the
cells incubated with 10−8 mol/l gastrin compared with
that of the cells incubated with 10−7 mol/l gastrin
(P<0.05).
The result of the intron 4 probe detection showed 10
different major bands of Tfs. Compared with the control group, the
density of bands 2, 4, 6 and 11 were decreased significantly, and
the density of bands 7 and 8 were increased significantly
(P<0.05). When compared with those incubated with
10−8 mol/l gastrin, the density of bands 1, 3 and 4 was
increased significantly for cells incubated with 10−7
mol/l gastrin, and that of band 11 was decreased significantly
(P<0.05).
The result of the intron 5 probe detection showed 14
different major bands of Tfs. Compared with the control group, the
density of band 12 was decreased significantly, and that of bands
1, 2, 5, 7, 8, 9 and 13 was increased significantly (P<0.05).
The density of band 1, 7, 9 and 12 was increased significantly in
the cells incubated with 10−8 mol/l gastrin compared to
those incubated with 10−7 mol/l gastrin (P<0.05).
Discussion
Human Reg1 gene is a single copy gene that is
located on chromosome 2 and is composed of 6 exons and 5 introns
(21). One of the most
well-documented effects of Reg1 is on the proliferation of acinar
and islet cells of the pancreas. The expression of Reg1 is
increased in regenerating or hyperplastic islets. In addition to
its islet proliferation- and regeneration-promoting effects,
tumor-promoting activity of Reg1 protein has also been reported
(22). Aberrant Reg expression has
been detected in tissues from colorectal carcinoma and gastric
cancer (23,24). In gastric cancer tissues, expression
of Reg1 gene is associated with patient survival and numbers
of metastatic lymph nodes (25).
Reg1-deficient mice have normal gastric development, however Reg1
promotes gastric mucosal growth and restoration with gastrin. Reg1
and gastrin may synergistically regulate gastric mucosal
proliferation during certain pathological settings, such as wound
healing (16). The proliferative
efficiency of gastric cancer cell line SGC7901 decreases
significantly following Reg1 knock-down and incubation with
gastrin (26). It has been
suggested that Reg1 may be a critical downstream gene in the
process of gastrin stimulated gastric cancer development. To
further understand the molecular mechanism by which gastrin
stimulates the expression of Reg1 in gastric cancer cells,
the cis-regulatory function of the introns of Reg1
and the relationship with gastrin were explored.
In recent years, studies have shown that introns of
eukaryotic genes can promote transcriptional efficiency (27). The ability of introns to stimulate
gene expression is an extensively investigated subject area for a
wide range of organisms, including mammals and nematodes. Introns
may act as transcriptional enhancers or alternative promoters,
depending on cis-elements located within the intron spanning
sequence (28). For example,
uncoupling protein (Ucp) 2 and 3 expression is activated by
the peroxisome proliferator-activated receptors (PPARs). The most
prominent PPARγ binding site in the Ucp2 and Ucp3
loci was identified in intron 1 of the Ucp3 gene and was the
only site that facilitates PPARγ transactivation of a heterologous
promoter. The transactivation of Ucp2 and 3 is
mediated through this novel enhancer in Ucp3 intron 1
(29). A conserved Smad-binding
element (SBE1) in intron 1 of the follistatin gene can also
regulate the expression of this gene (30).
The present study demonstrated that the introns of
the Reg1 gene exhibited cis-regulating function, with
the exception of intron 1, indicating that introns 2, 3, 4 and 5 of
Reg1 may contain cis-regulatory elements. It has been
reported that a C-rich region of the rat Reg1 promoter is
critical for gastrin-stimulated Reg expression (15). It is hypothesized that
cis-regulatory elements in introns and promoters of
Reg1 may synergistically regulate the gene expression. The
present study also showed that, following gastrin incubation, the
luciferase activity was not significantly different, which appeared
to indicate that gastrin had no effect on regulating Reg1
gene expression. However, in physiological conditions, gene
expression is regulated by interaction of promoter and intron. The
luciferase activity was detected in the cells transfected with only
a single intron, which may contribute to the negative results
obtained in the current study.
In eukaryotic cells, transcriptional regulation is
executed by the interaction between trans-acting factors and
cis-acting regulatory elements. Trans-acting factors
are also known as transcription factors, and can recognize and bind
to specific cis-acting elements. For example, the
cis-elements of human ubiquitin C, able to bind in
vitro the ubiquitous Sp1 and YY1 transcription factors, are
involved in the stimulation of reporter gene transcription
(31). In the present study, the
effect of gastrin on Tfs that bind to the cis-acting
regulatory elements in introns of the Reg1 gene in gastric
cancer cells was also explored by Southwestern blotting. The
results revealed multiple Tfs binding to the five introns of
Reg1, which suggested that the introns may function via
binding to their Tfs.
The direct effect of intron-mediated transcriptional
regulation is often referred to as ‘intron-mediated enhancement’
(IME) (28). IME requires the
presence of an intron close to the 5′ end of the gene. It has been
hypothesized that a promoter proximal to the 5′ splice site
facilitates the recruitment of transcriptional machinery to the
promoter, which includes transcription factors such as c-Jun and
activating transcription factor 2 binding to the cyclic adenosine
monophosphate response element site and, therefore, aids in the
initiation of transcription (32–34).
The precise mechanism of intron-dependent enhancement of
transcription, however, remains unclear. It was recently reported
that the inclusion of an intron in INO1, which is a
nonintronic gene, resulted in the constitutive activation of the
gene. In the presence of the intron, the promoter of INO1
interacted with its terminator region to form a gene loop in yeast
(35). The intronic
cis-acting elements of the cystic fibrosis transmembrane
conductance regulator gene (CFTR) interact with the
CFTR promoter and contribute to the regulation of
CFTR gene expression (36).
In Reg1, the C-rich region in the gene promoter was found to
be critical for the response to gastrin (15,37),
and the expression of Reg1 was controlled through separate
promoter elements by gastrin (4).
The present study found that gastrin may alter the
density of certain Tf bands, and that the density of some Tf bands
was altered at different gastrin concentrations. This indicates
that gastrin can alter the ability of Tfs to bind to the
recognition sequences in introns to affect the formation of the
transcriptional complex, and may be significant in the interaction
between the promoter and introns to regulate the expression of
Reg1. Although the first intron of Reg1 has no
cis-regulatory function, it can bind to at least 14 types of
Tf. It was reported that the control of human Ucp3
transcription in skeletal muscle is not solely conferred by the
promoter, but depends on several cis-acting elements in
intron 1, suggesting a complex association between the promoter and
intronic sequences (38). Intron
removal, or replacement with a heterologous chimeric intron, caused
a significant reduction in promoter activity (27). The results of the current study
suggest that intron 1 of the Reg1 gene may function only as
a mediator in the formation of multiple molecule complexes for
regulating gene expression.
In conclusion, our data demonstrate that introns of
Reg1 can bind to many transcription factors and enhance gene
expression. However, we still did not identify the precise
transcription factors. The hormone gastrin can influence the
ability of Tf binding to introns. Gastrin may regulate Reg1
gene expression by binding to the transcription factors to form a
multiple molecule complex. Future research on the interaction of
the promoter and introns of the Reg1 gene and identifying
the transcription factors that bind to the introns of Reg1
gene will be useful in elucidating the mechanism of Reg1
gene expression.
References
|
1
|
Ferlay J, Shin HR, Bray F, et al:
Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int
J Cancer. 127:2893–2917. 2010. View Article : Google Scholar
|
|
2
|
Dockray G, Dimaline R and Varro A:
Gastrin: old hormone, new functions. Pflugers Arch. 449:344–355.
2005. View Article : Google Scholar
|
|
3
|
Nørsett KG, Steele I, Duval C, et al:
Gastrin stimulates expression of plasminogen activator inhibitor-1
in gastric epithelial cells. Am J Physiol Gastrointest Liver
Physiol. 301:G446–G453. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Steele IA, Dimaline R, Pritchard DM, et
al: Helicobacter and gastrin stimulate Reg1 expression in gastric
epithelial cells through distinct promoter elements. Am J Physiol
Gastrointest Liver Physiol. 293:G347–G354. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Henwood M, Clarke PA, Smith AM and Watson
SA: Expression of gastrin in developing gastric adenocarcinoma. Br
J Surg. 88:564–568. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Okamoto H: The Reg gene family and Reg
proteins: with special attention to the regeneration of pancreatic
beta-cells. J Hepatobiliary Pancreat Surg. 6:254–262. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Terazono K, Yamamoto H, Takasawa S, et al:
A novel gene activated in regenerating islets. J Biol Chem.
263:2111–2114. 1988.PubMed/NCBI
|
|
8
|
De Caro A, Lohse J and Sarles H:
Characterization of a protein isolated from pancreatic calculi of
men suffering from chronic calcifying pancreatitis. Biochem Biophys
Res Commun. 87:1176–1182. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sarles H, Dagorn JC, Giorgi D and Bernard
JP: Renaming pancreatic stone protein as ‘lithostathine’.
Gastroenterology. 99:900–901. 1990.PubMed/NCBI
|
|
10
|
Graf R, Schiesser M, Reding T, et al:
Exocrine meets endocrine: pancreatic stone protein and regenerating
protein - two sides of the same coin. J Surg Res. 133:113–120.
2006. View Article : Google Scholar
|
|
11
|
Miyaoka Y, Kadowaki Y, Ishihara S, et al:
Transgenic over-expression of Reg protein caused gastric cell
proliferation and differentiation along parietal cell and chief
cell lineages. Oncogene. 23:3572–3579. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sanchez D, Mueller CM and Zenilman ME:
Pancreatic regenerating gene I and acinar cell differentiation:
influence on cellular lineage. Pancreas. 38:572–577. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Parikh A, Stephan AF and Tzanakakis ES:
Regenerating proteins and their expression, regulation and
signaling. Biomol Concepts. 3:57–70. 2012.PubMed/NCBI
|
|
14
|
van Beelen Granlund A, Østvik AE, Brenna
Ø, et al: REG gene expression in inflamed and healthy colon mucosa
explored by in situ hybridisation. Cell Tissue Res. 352:639–646.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ashcroft FJ, Varro A, Dimaline R and
Dockray GJ: Control of expression of the lectin-like protein Reg-1
by gastrin: role of the Rho family GTPase RhoA and a C-rich
promoter element. Biochem J. 381:397–403. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peterson AJ, Nguyen N, Okamoto H, et al:
Loss of RegI in conjunction with gastrin deficiency in mice
facilitates efficient gastric ulcer healing but is dispensable for
hyperplasia and tumourigenesis. Regul Pept. 160:9–18. 2010.
View Article : Google Scholar
|
|
17
|
Fukuhara H, Kadowaki Y, Ose T, et al: In
vivo evidence for the role of RegI in gastric regeneration:
transgenic overexpression of RegI accelerates the healing of
experimental gastric ulcers. Lab Invest. 90:556–565. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Haddad-Mashadrizeh A, Zomorodipour A,
Izadpanah M, et al: A systematic study of the function of the human
beta-globin introns on the expression of the human coagulation
factor IX in cultured Chinese hamster ovary cells. J Gene Med.
11:941–950. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kleinschmidt AM, Nassiri M, Stitt MS, et
al: Sequences in intron 51 of the von Willebrand factor gene target
promoter activation to a subset of lung endothelial cells in
transgenic mice. J Biol Chem. 283:2741–2750. 2008. View Article : Google Scholar
|
|
20
|
Siu FK, Lee LT and Chow BK: Southwestern
blotting in investigating transcriptional regulation. Nat Protoc.
3:51–58. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Watanabe T, Yonekura H, Terazono K,
Yamamoto H and Okamoto H: Complete nucleotide sequence of human reg
gene and its expression in normal and tumoral tissues. The reg
protein, pancreatic stone protein, and pancreatic thread protein
are one and the same product of the gene. J Biol Chem.
265:7432–7439. 1990.PubMed/NCBI
|
|
22
|
Yamaoka T, Yoshino K, Yamada T, et al:
Diabetes and tumor formation in transgenic mice expressing Reg I.
Biochem Biophys Res Commun. 278:368–376. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zheng HC, Sugawara A, Okamoto H, et al:
Expression profile of the REG gene family in colorectal carcinoma.
J Histochem Cytochem. 59:106–115. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fukui H, Fujii S, Takeda J, et al:
Expression of reg I alpha protein in human gastric cancers.
Digestion. 69:177–184. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dhar DK, Udagawa J, Ishihara S, et al:
Expression of regenerating gene I in gastric adenocarcinomas:
correlation with tumor differentiation status and patient survival.
Cancer. 100:1130–1136. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cheng Sh, Ding Y and Zhang QX: Role of
RegI in the pathway of gastrin stimulating proliferation of gastrin
cancer cells in vitro. Acta Anatomica Sinica. 43:63–67. 2012.(In
Chinese).
|
|
27
|
Bianchi M, Crinelli R, Giacomini E,
Carloni E and Magnani M: A potent enhancer element in the 5′-UTR
intron is crucial for transcriptional regulation of the human
ubiquitin C gene. Gene. 448:88–101. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rose AB: Intron-mediated regulation of
gene expression. Curr Top Microbiol Immunol. 326:277–290.
2008.PubMed/NCBI
|
|
29
|
Bugge A, Siersbaek M, Madsen MS, et al: A
novel intronic peroxisome proliferator-activated receptor gamma
enhancer in the uncoupling protein (UCP) 3 gene as a regulator of
both UCP2 and -3 expression in adipocytes. J Biol Chem.
285:17310–17317. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Blount AL, Vaughan JM, Vale WW and
Bilezikjian LM: A Smad-binding element in intron 1 participates in
activin-dependent regulation of the follistatin gene. J Biol Chem.
283:7016–7026. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bianchi M, Crinelli R, Giacomini E, et al:
Yin Yang 1 intronic binding sequences and splicing elicit
intron-mediated enhancement of ubiquitin C gene expression. PloS
One. 8:e659322013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kamo K, Kim AY, Park SH and Joung YH: The
5′UTR-intron of the Gladiolus polyubiquitin promoter GUBQ1 enhances
translation efficiency in Gladiolus and Arabidopsis. BMC Plant
Biol. 12:792012. View Article : Google Scholar
|
|
33
|
Damgaard CK, Kahns S, Lykke-Andersen S, et
al: A 5′ splice site enhances the recruitment of basal
transcription initiation factors in vivo. Mol Cell. 29:271–278.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tanabe A, Konno J, Tanikawa K and Sahara
H: Transcriptional machinery of TNF-α-inducible YTH domain
containing 2 (YTHDC2) gene. Gene. 535:24–32. 2014. View Article : Google Scholar
|
|
35
|
Moabbi AM, Agarwal N, El Kaderi B and
Ansari A: Role for gene looping in intron-mediated enhancement of
transcription. Proc Natl Acad Sci USA. 109:8505–8510. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ott CJ, Suszko M, Blackledge NP, et al: A
complex intronic enhancer regulates expression of the CFTR gene by
direct interaction with the promoter. J Cell Mol Med. 13:680–692.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
O’Hara A, Howarth A, Varro A and Dimaline
R: The role of proteasome beta subunits in gastrin-mediated
transcription of plasminogen activator inhibitor-2 and regenerating
protein1. PloS One. 8:e599132013. View Article : Google Scholar
|
|
38
|
Girousse A, Tavernier G, Tiraby C, et al:
Transcription of the human uncoupling protein 3 gene is governed by
a complex interplay between the promoter and intronic sequences.
Diabetologia. 52:1638–1646. 2009. View Article : Google Scholar : PubMed/NCBI
|